Abstract
Glioblastoma multiforme is the most aggressive primary tumor of the central nervous system. Glioma stem cells (GSCs), a small population of tumor cells with stem-like properties, are supposedly responsible for glioblastoma multiforme relapse after current therapies. In approximately thirty percent of glioblastoma multiforme tumors, telomeres are not maintained by telomerase but through an alternative mechanism, termed alternative lengthening of telomere (ALT), suggesting potential interest in developing specific therapeutic strategies. However, no preclinical model of ALT glioma was available until the isolation of TG20 cells from a human ALT glioma. Herein, we show that TG20 cells exhibit a high level of telomeric recombination but a stable karyotype, indicating that their telomeres retain their protective function against chromosomal instability. TG20 cells possess all of the characteristic features of GSCs: the expression of neural stem cell markers, the generation of intracerebral tumors in NOD-SCID-IL2Rγ (NSG) mice as well as in nude mice, and the ability to sustain serial intracerebral transplantations without expressing telomerase, demonstrating the stability of the ALT phenotype in vivo. Furthermore, we also demonstrate that 360B, a G-quadruplex ligand of the pyridine derivative series that impairs telomere replication and mitotic progression in cancer cells, prevents the development of TG20 tumors. Together, our results show that intracerebral grafts of TG20 cells in immunodeficient mice constitute an efficient preclinical model of ALT glioblastoma multiforme and that G-quadruplex ligands are a potential therapy for this specific type of tumor.
What's new?
All cancerous cells share the need to maintain and elongate their telomeres. In approximately 30% of glioblastoma multiforme tumors, telomeres are not maintained by telomerase but through an alternative mechanism, termed ALT, making specific therapeutic strategies potentially interesting. Here, the authors showed that intracerebral grafts of an ALT glioma cell line called TG20 in immunodeficient mice constitute an efficient preclinical model of ALT glioblastoma multiforme, which could aid the understanding of the underlying pathogenesis. This model could also support the development of specific therapies, with the study revealing G-quadruplex ligands as a potential therapy for this specific type of tumor.
Keywords: glioma stem cells, telomerase, alternative lengthening of telomeres, in vivo model, G-quadruplex ligands
Telomerase is activated in most tumor cells to maintain telomere length, which is required for long-term proliferative capability.1 However, tumors lacking telomerase can rely on a different mechanism for telomere elongation, referred to as alternative lengthening of telomeres (ALT).2 While several studies have shown that ALT depends on homologous recombination between telomeres,3 it is not yet clear how the ALT machinery is activated and what mechanisms are involved. The ALT pathway is predominant in osteosarcoma4 and is detected in approximately 30% of glioblastoma multiforme,5 the most common and malignant primary tumor of the adult central nervous system. While several ALT cell lines have been derived from osteosarcoma patients, we were the first to describe an ALT glioma cell line, TG20, which was obtained from an ALT glioma patient.6 We have shown that TG20 cells display markers and characteristics of ALT cells, such as the lack of telomerase expression, the presence of ALT-associated PML bodies, and heterogeneous telomere length.6 A second ALT glioma cell line has recently been reported.7
Gliomas have been shown to contain a small population of cancer cells, termed glioma stem cells (GSCs), which share some properties with neural stem cells. These cells are more resistant to current treatments than the other “differentiated” cancer cells and are able to regenerate the tumor because of their stem properties.8 Understanding the biology of GSCs is thus crucial for developing specific therapies to prevent tumor relapse. We have shown that TG20 cells exhibit the phenotype and properties of GSCs, such as the expression of neural stem cell markers, the capacity for long-term proliferation in vitro, and the ability to form intracranial tumors in immunodeficient mice.6
All cancerous cells share the need to maintain and elongate their telomeres. Therefore, telomeres and their maintenance machinery constitute an appealing target for anticancer therapies. Various strategies for telomerase inhibition have been emerging based on chemical inhibitors, antisense oligonucleotides, and immunotherapies.9 However, telomerase inhibition is inadequate for tumors lacking telomerase and is potentially capable of activating ALT as a mechanism of drug resistance.10 A more common putative target for telomere-targeting therapies could be G-quadruplexes, which are secondary structures commonly formed by telomeres.11 Several G-quadruplex ligands (e.g., BRCAO19, RHPS4, telomestatin and 360A) have been designed and have shown efficiency in reducing cancer cell proliferation and/or tumor growth,12–17 but limited data are available regarding the effects of G-quadruplex ligands on ALT cells.
In the present work, we further characterized the ALT phenotype and the GSC properties of TG20 cells. We showed that intracerebral grafts of TG20 cells represent an efficient preclinical model of ALT glioma that helps reveal the sensitivity of these cells to the G-quadruplex ligand 360B, a more soluble derivative of 360A, known to impair telomere replication and mitotic progression in cancer cells.17–19
Materials and Methods
Cell culture
Telomerase-positive GSCs, including the TG16, TG1N, TG10 and OB1 cell lines, and the ALT GSC TG20 line were derived from tumor samples obtained from patients at Sainte Anne Hospital (Paris, France). This procedure was approved by the Institutional Review Board and patients provided an informed consent. GSCs were grown in neurospheres in serum-free Dulbecco's modified eagle medium (DMEM)/F12 supplemented with B27 (Gibco, Life Technologies), heparin (Stem Cell Technologies), human recombinant epidermal growth factor (EGF), and basic fibroblast growth factor (FGF2; Sigma), as previously described.6,20 eGFP-positive TG20 cells were obtained through lentiviral transfection, as previously described.6 The osteosarcoma ALT cell line SAOS-2 was grown in DMEM (Gibco, Life Technologies) supplemented with 10% fetal bovine serum (PAA Laboratories).
5-Azacytidine treatment
GSCs were grown as adherent cultures on laminin (Millipore)-coated flasks. Medium was supplemented with DMSO or with 2.5 or 5 μM 5-azacytidine (A2385; Sigma). Cells were treated three times every 24 hr. At 48 hr after the last treatment, the cells were collected as dry pellets.
WST-1 cell proliferation assay
A total of 5,000 TG20 cells were plated in 96-well plates that were previously coated with laminin (Millipore) to allow cell adherence. The WST-1 assay (11644807001; Roche) was then performed at different time points according to the manufacturer's instructions.
Telomeric sister chromatid exchange
Chromosome orientation FISH (CO-FISH) was performed as previously described.21 Briefly, cells were grown as adherent cultures in culture medium supplemented with 10 μM BrdU for one cell cycle. Metaphase spreads were stained with Hoechst 33258, exposed to ultraviolet (UV) light, and digested with exonuclease III (Promega). Successive hybridizations with a FITC-labeled (TTAGGG)3 PNA probe (Applied Biosystems) and a Cy-3-labeled (CCCTAA)3 PNA probe allowed the detection of the parental telomere C and G strands. Metaphase images were captured and analyzed using an Axio Imager Z.2 (Zeiss, Germany) coupled to a Metafer Image Analysis System (MetaSystems, Germany).
Sister chromatid exchange
Cells were cultured in medium supplemented with 10 µM BrdU for two cell cycles. After the addition of 10 µg/mL colchicine for 2 h, cells were collected and incubated in KCl (0.075 M) and human serum (1:6 v/v) for 20 min at 37°C, fixed in ethanol/acetic acid (3:1 v/v), and spread onto cold, clean slides. The slides were incubated in 50 µg/mL Hoechst (33258) dye for 20 min and then denatured by exposure to UV light at 365 nm (Fisher Bioblock Scientific) for 30 min in the presence of 2× SSC. After washing in double-distilled water, the slides were stained with 1.5% Giemsa and 1.5% phosphate buffer in double-distilled water. Metaphases were observed under a microscope (Olympus AX70) and analyzed using the Cytovision system. The sister chromatid exchange (SCE) frequency was calculated as the total number of SCEs divided by the total number of chromosomes.
Multi-FISH analysis
Metaphase spreads were prepared as for CO-FISH experiments. Metaphases were hybridized with a 24 XCyte probe kit (MetaSystems, Altlussheim, Germany) according to the manufacturer's instructions. Images were captured and analyzed using an Axioplan fluorescence microscope (Zeiss, Germany) with the appropriate filters and M-FISH software (ISIS, MetaSystems).
2D gel electrophoresis and genomic blotting
DNA was phenol-chloroform extracted, ethanol precipitated, resuspended in nuclease-free water, and digested with MboI (New England Biolabs) for 4 hr at 37°C. For the first dimension, 25 µg of total DNA was subjected to electrophoresis in a 0.4% agarose gel at 1 V/cm for 18 hr at RT. The lanes were excised, and a 1% agarose gel in 1× TBE containing ethidium bromide was poured around the excised slab. The second dimension of electrophoresis was performed at 5 V/cm for 3 hr. Gels were treated with 5 M NaOH and 1 M NaCl for 20 min to denature the DNA, transferred overnight onto nitrocellulose membranes (Whatman), and UV cross-linked. Hybridization with a telomeric C-rich digoxigenin probe was performed in Church's buffer at 42°C for 12 hr. Telomeric DNA was visualized with Digoxigenin DNA Detection Kits (Roche) using an LAS-4000 camera (Fuji).
Quantitative real-time polymerase chain reaction
Total RNA was extracted using the RNeasy Plus Mini kit (Qiagen) according to the manufacturer's instructions. Isolated RNA was transcribed to cDNA using the High-Capacity RNA-to-cDNA Master Mix (Applied Biosystems). Quantitative real-time polymerase chain reactions (RT-PCRs) were performed in 96-well plates in duplicate using SYBR Green Master Mix (Applied Biosystems). The primers used are listed in Supporting Information Table S1.
Methylation-specific PCR
Cellular genomic DNA was isolated as previously described.22 Bisulfite modification and DNA purification were performed using the EZ DNA methylation kit (Zymo Research, Proteigene, Saint-Marcel, France) following the manufacturer's instructions. Primers specific for the unmethylated and methylated alleles of hTERT were used to amplify bisulfite-modified DNA as previously described.23 Methylation-specific PCR (MSP) primers were designed for a proximal region encompassing the transcription and translation start sites and the WT1 consensus binding site. The primer sequences and conditions are presented in Supporting Information Table S2. PCR was performed on bisulfite-treated DNA using ZymoTaq™ DNA Polymerase (Zymo Research, Proteigene, Saint-Marcel, France). PCR products were separated on 2% agarose gels and visualized after ethidium bromide staining.
Telomerase activity assay
Telomerase activity was measured after neurosphere growth, using the TRAPeze ELISA Telomerase Detection Kit (Chemicon) according to the manufacturer's instructions. Proteins were extracted using the lysis buffer (CHAPS) provided in the kit, and 0.5 or 1 μg of protein extract was used.
Western blot
Cells were collected as dry pellets and then lysed using RIPA buffer (R0278; Sigma) supplemented with protease (11873580001; Roche) and phosphatase inhibitors (78420; Thermo Scientific). Thirty micrograms of protein was diluted in RIPA buffer supplemented with LDS Sample Buffer (NP0007; Invitrogen by Life technologies) and Sample Reducing Agent (NP004; Novex by Life Technologies). Samples were heated for 5 min at 95°C and loaded into NuPAGE 4–12% Bis-Tris gels (NP0322BOX; Novex by Life Technologies). For electrophoresis, NuPAGE running buffer (NP001; Novex by Life Technologies) was used. Protein transfer was performed using nitrocellulose membranes (LC2000; Invitrogen by Life Technologies) and NuPAGE Transfer Buffer (NP0006-1; Invitrogen by Life Technologies). WT1 (Wilm's tumor protein 1) antibody (1:400, NB-110–60011; Novus Biologicals) was used to detect the WT1 protein.
Mice
Three-month-old male NOD-SCID-IL2Rγ (NSG) mice (25–30 g) and 3-month-old nude mice (Swissnu/nu; 25–30 g) from Charles River Laboratories (L'Arbresle, France) were used in this study. Animals were provided with food and water ad libitum and housed in a colony isolator maintained at a constant temperature of 19–22°C and humidity (40–50%) on a 12:12 hr light/dark cycle. The experiments were performed in compliance with the European Communities Council Directive of November 24, 1986 (86/609/EEC) and the principles of laboratory animal care (NIH publication No. 85-23, revised 1985) and were approved by our institutional committee on animal welfare (CETEA-CEA DSV IdF, saisine number #12–029). All surgical procedures were performed under anesthesia with ketamine (75 mg/kg, Imalgen; Merial, Lyon, France) and medetomidine (1 mg/kg, Domitor; Pfizer, Paris, France). After the surgery, paracetamol (1.64 mg/mL, Doliprane; Sanofi, France) was administered in the drinking water for 1 week.
Serial intracranial transplantations
70,000–100,000 TG20-eGFP GSCs were injected stereotaxically into the striatum of 3-month-old NSG mice, as previously described.6 After 2 or 3 months, mice were sacrificed by cervical dislocation, and brain tissues containing eGFP-positive cells were micro-dissected using a Carl-Zeiss Lumar fluorescence stereomicroscope. The dissected tissues were pelleted and dissociated in 0.5% trypsin (Gibco, Life Technologies) and 0.5 mg/mL DNase I (Roche) for 15 min at 37°C. eGFP-positive cells were sorted by FACS (INFLUX cell sorter, BD), and dead cells were excluded by propidium iodide incorporation (10 µg/mL). Sorted cells were resuspended in PBS (0.15% BSA) and reinjected (100,000 cells in 2 µL) into 3-month-old NSG mice.
G-quadruplex (G4) ligand (360B) treatment
360B is a close derivative of the previously described G-quadruplex ligand 360A. 360B was prepared in two steps from 2,6-pyridine-dicarboxylic acid and uinolone-3-amine, in an overall 72% yield after recrystallization from ethanol. 1H-NMR (300 MHz, DMSO d6, δ en ppm): 4.82 (s); 8.12 (t, J = 8 Hz); 8.27 (t, J = 8 Hz); from 8.45 to 8.65 (mt); 9.68 (s); 10.14 (s); 11.93 (bs; as shown in Scheme 1). In vitro, 360A and 360B exhibit exactly the same properties for either their ability to stabilize DNA G-quadruplex structures, such as telomeric 22AG, or to inhibit cell growth of tumor cells, such as lung adenocarcinoma cell lines H460 and A549. However, 360B is more soluble in aqueous media (27.6 mg/mL instead of 0.35 mg/mL for 360A in phosphate buffer at pH 7.4).
Sch 1.
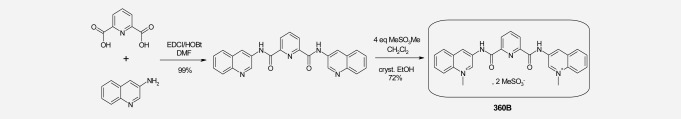
Synthesis of “360B.”
For grafting, cells in the active growth phase were washed and collected in serum-free media. Two microliters containing 100,000 cells was injected (0.5 µL/min) bilaterally at the following coordinates (from bregma): AP = +0.5, L = ±1.5, and DV = −3. One week after engraftment, 2 μL of 10 µM 360B or 2 µl of DMSO diluted in DMEM/F12 medium were microinjected close to the initial position at 3 mm under the dural surface with the incisor bar at 5 mm above the interaural line.
Immunostaining of brain sections
Animals were deeply anesthetized in a CO2 chamber and transcardially perfused with a 4% paraformaldehyde solution in 0.1 M sodium phosphate, pH 7.2. Brains were removed, postfixed for 2 hr in the same solution, cryoprotected with increasing 10–30% PBS-sucrose solutions, and frozen in dry ice-cooled isopentane. Serial coronal cryostat sections (30 µm) were processed for hematoxylin and eosin staining or immunofluorescence. For immunofluorescence, brain sections were permeabilized with PBS containing 0.3% Triton X-100 and 1% BSA at room temperature for 1 hr and incubated overnight at 4°C with primary antibodies raised against cleaved-caspase 3 (mAb9579, 1:200; Cell Signaling), human Nestin (MAB1259, 1:200; R&D systems), or laminin (L939, 1:200; Sigma). After three washes, sections were incubated for 2 hr at room temperature with secondary antibodies coupled to fluorochromes (FITC or cyanine-3; 1:200; Life Technologies), rinsed, and mounted with DAPI Fluoromount-G (Southern Biotech). Mosaic images were captured using a motorized microscope (Pathfinder, Imstar S.A., France) equipped with a Hamamatsu C8484-05G camera (Hamamatsu Photonics, France) with the NIS Elements software v3.1 (Nikon Instruments). Tumor volumes and cell counts were calculated by multiplying the tumor area or the number of cells by the distance between successive sections. Confocal images were captured using the Leica DM 2500 microscope.
Results
TG20 cells exhibit a high level of telomeric recombination
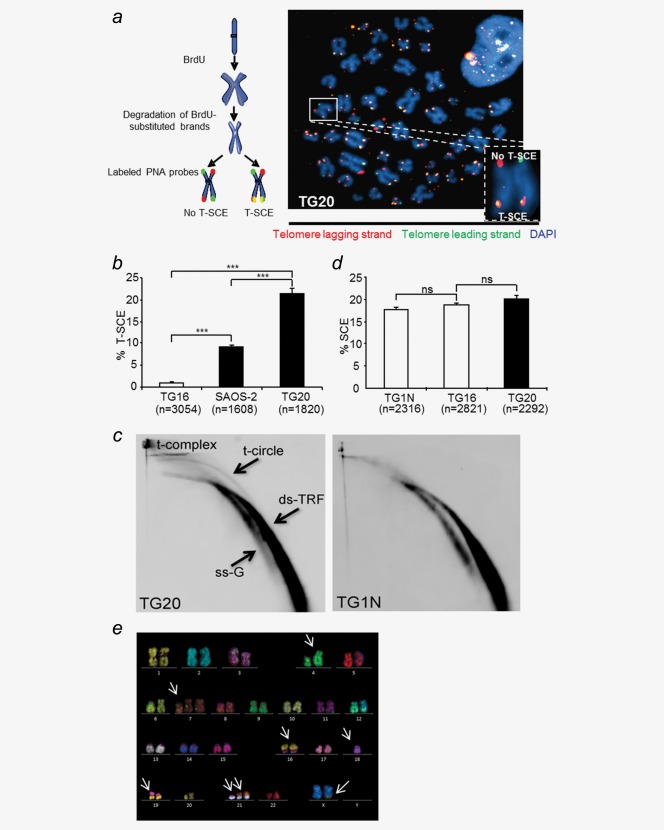
The ALT-mediated elongation of telomeres has been associated with abundant recombination events between telomeres of sister chromatids, commonly referred to as telomeric sister chromatid exchanges (T-SCEs), which are rare or absent in non-ALT cells.24 As shown by CO-FISH, TG20 cells exhibited a higher rate of T-SCE than the telomerase-positive GSC line TG16 and the ALT osteosarcoma cell line SAOS-2 (21.4% in TG20 cells vs. 1% in TG16 cells and 9.1% in SAOS-2 cells, p < 0.001, Figs. 1a and 1b).
Figure 1.
TG20 cells exhibit a high level of telomeric recombination. (a) The chromosome oriented (CO)-FISH procedure was used for T-SCE analysis. Successive hybridizations with FITC or Cy3-labeled PNA probes allowed detection of the parental telomere C and G strands, respectively. Telomere lagging strands are stained in red, telomeric leading strands are stained in green, and yellow-stained telomeres are scored as T-SCEs. The representative metaphase image on the right is an example from the TG20 ALT cell line. The inset shows a chromosome with extremities with or without T-SCE. (b) T-SCE results of the depicted cell lines. Values are presented as T-SCE events per 100 chromosome extremities, and “n” indicates the number of chromosome extremities counted (***p < 0.001, Student's t-test). (c) 2D gel electrophoresis and analysis of telomeric DNA after hybridization with a telomeric C-rich probe in TG1N and TG20 GSCs. ds-TRF, double-stranded telomeric restriction fragments; ss-G, linear single-G-strand telomeric DNA. (d) SCE analysis on TG1N and TG16 telomerase-positive cell lines and the TG20 ALT cell line. Values are presented as SCE events per 100 chromosomes, and “n” indicates the number of analyzed chromosomes (ns = non-significant, p > 0.05, Student's t-test). (e) Representative M-FISH results from late-passage TG20 cells. Arrows indicate the anomalies.
Telomeric circles, referred to as t-circles, have been reported to represent part of the extrachromosomal telomeric DNA found in ALT cells and could serve as a copy template for the rolling circle-dependent DNA amplification of telomere sequences.25,26 As shown by 2D gel electrophoresis, TG20 but not TG1N cells exhibited abundant t-circles (Fig. 1c). Furthermore, signals at the origin of the gel migration front in the first but not the second dimension were also detected in TG20 cells. As reported previously,27 these signals correspond to the t-complex, a mixture of highly branched telomeric DNAs, which are supposed Holliday junction intermediates of telomeric homologous recombination (HR) in ALT cells.
Contrary to these indicators of a high level of telomere recombination in TG20 cells, the frequency of exchanges between SCE was similar in these cells to the frequency observed in the telomerase-positive GSC lines TG1N and TG16 (Fig. 1d). Furthermore, karyotypic analysis showed a single pseudodiploid clone corresponding to 47(XX), del4qter, +7p, −18, +21, t(8;21), t(16;19), t(X;9) in metaphases from three different passages of TG20 cells (p11, p26, and p30; Fig. 1e). Other anomalies involving chromosomes #8; 9; 21 and X were detected in some metaphases but did not lead to the emergence of new clones. Therefore, these cytogenetic data strongly suggest that TG20 cells have a rather stable karyotype.
Together, our data show that TG20 cells are characterized by a high rate of telomere recombination that is not associated with a high level of chromosome instability.
Methylation is not directly involved in the absence of hTERT expression in TG20 cells
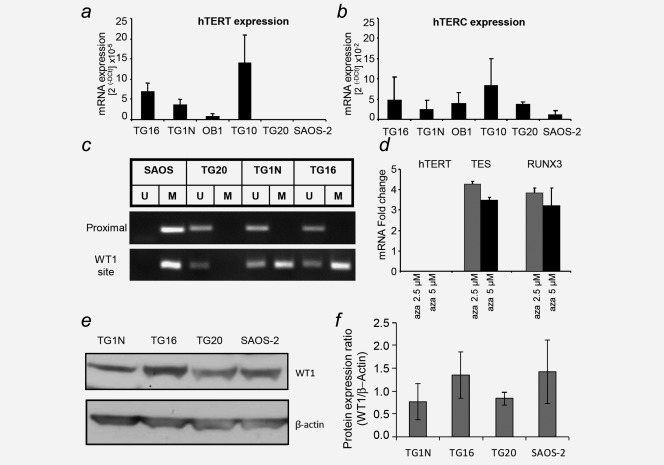
TG20 cells do not exhibit telomerase activity.6 Consistently, the transcripts of the catalytic subunit of the human telomerase hTERT were not detected by qRT-PCR in TG20 cells, which was similar to the findings in SAOS-2 cells (Fig. 2a). In contrast, transcripts of the RNA subunit hTERC were detected in both ALT cell lines (Fig. 2b), indicating that the absence of telomerase activity in TG20 cells was related to the absence of hTERT expression but not to the expression of hTERC. Importantly, karyotyping demonstrated that TG20 cells possessed two stable copies of the distal portion of chromosome arm 5p, the location of the htert gene (Fig. 1e).
Figure 2.
Absence of hTERT expression in TG20 ALT cells. (a) hTERT mRNA expression in telomerase-positive GSCs (TG16, TG1N, OB1 and TG10) and ALT cells (TG20 and SAOS-2). Values are presented as the mean ± SEM of two independent experiments performed in duplicate. (b) The same cell lines as in (a) were evaluated but for hTERC mRNA expression. (c) Representative methylation-specific PCR (MSP) results for the proximal region and WT1 binding site of the TERT promoter in SAOS-2 and TG20 ALT cells and in TG1N and TG16 telomerase-positive cells. Bands in the M and U lanes represent methylated and unmethylated PCR products, respectively. (d) mRNA expression of hTERT, TES, and RUNX3 in TG20 cells after treatment with 2.5 or 5 µM 5-azacytidine (5-aza), relative to their expression in control DMSO-treated cells. (e) WT1 protein levels determined by Western blot analysis for the indicated cell lines. β-Actin was used as a loading control. (f) Comparison of expression ratios of WT1 protein vs. β-actin in the different cell lines. Experiment was repeated twice using samples from independent cell cultures.
The htert promoter contains clusters of CpG islands where methylation can occur, suggesting that promoter methylation might be a key regulator of htert expression.28 Consistently, two studies have previously shown that htert core promoter methylation inhibits its expression.29,30 To assess whether methylation was involved in htert silencing in ALT cells, we determined the methylation status of a proximal region of the htert promoter using a MSP-based assay in TG20 cells, SAOS-2 cells, and the telomerase-positive TG16 and TG1N cells. In accordance with the role of the methylation of this region in htert expression regulation, this region was methylated in the telomerase-negative SAOS-2 cells and unmethylated in the telomerase-positive GSCs (Fig. 2c). However, this region was also unmethylated in TG20 cells, indicating that the absence of hTERT expression in an ALT GSC line was not directly related to the methylation status of the htert proximal core promoter.
To determine whether the methylation of other regions in this promoter is involved in the repression of htert, we tested the effects of 5-azacytidine (5-aza), a chemical analogue of the cytosine nucleoside that causes global DNA demethylation, on htert expression. To verify that 5-aza was capable of demethylating DNA in TG20 cells, we also measured the expression of two genes known to be downregulated by hypermethylation in glioblastoma, TES and RUNX3.31 As shown in Figure 2d, the expression levels of both TES and RUNX3 but not hTERT were significantly increased in TG20 cells following 5-aza treatment. These results confirm that the absence of telomerase activity in TG20 cells is not directly related to htert promoter methylation.
Methylation has been shown to prevent the binding of WT1, a known htert repressor, to a distal region of the htert promoter in promyelocytic leukemia cells, thereby favoring hTERT expression.32,33 Because all our telomerase-positive and ALT cell lines expressed WT1 (Fig. 2e), we investigated the methylation of one distal promoter region encompassing the WT1 binding site using MSP, as described above. The results showed that this region of the promoter was half methylated in telomerase-positive GSCs, whereas it was completely unmethylated in TG20 cells. These results support the previous observation that the methylation of the distal region of the htert promoter can prevent WT1 binding and thereby favor hTERT expression (Fig. 2c). These findings raise the possibility that the binding of WT1 to this region plays a role in the repression of htert in the TG20 ALT cell line. However, the WT1-binding domain of the htert promoter was fully methylated in SAOS-2 ALT and telomerase-positive cells, thereby preventing the potential binding of WT1 and excluding a putative WT1-mediated htert repression as a mechanism common to all ALT cells.
TG20 cells display a stable ALT phenotype after serial intracranial transplantations into NOD-SCID-IL2Rγ (NSG) mice
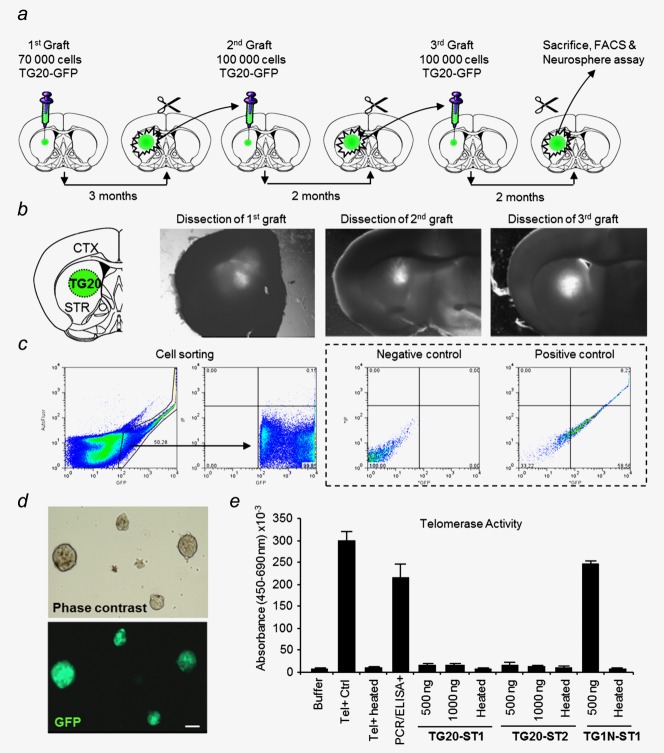
We have previously shown that TG20 cells can generate intracranial tumors in non-obese diabetic severe combined immunodeficient NOD-SCID-IL2Rγ (NSG) mice.6 To further characterize the GSC potential of TG20 cells and the stability of their ALT phenotype, we investigated whether TG20 cells could sustain tumor formation upon sequential transplantation into the brain of NSG mice. Thus, TG20 cells and the telomerase-positive GSC line TG1N were engineered by lentiviral infection to stably express eGFP (TG20-eGFP and TG1N-eGFP cells, respectively). Three months after the first engraftment, the tumor (eGFP-positive) cells were sorted by FACS (Figs. 3a–3c) and re-injected into the brains of tumor-naive mice (Fig. 3a). This experiment was successfully repeated for 2 consecutive transplantations, with a 2-month interval, further demonstrating that TG20 cells possess the main characteristics of GSCs (Fig. 3a). After each transplantation, we verified that sorted eGFP+ cells maintained the capacity to generate neurospheres in vitro, another characteristic of stemness (Fig. 3d). Importantly, using the TRAPeze ELISA assay, we also demonstrated that the sorted cells from TG1N tumors remained telomerase positive (Fig. 3e) and that cells from TG20 tumors retained the ALT phenotype and remained telomerase negative (Fig. 3e). These data indicate that TG20 cells have a stable ALT phenotype, which allows them to sustain a high number of divisions in vivo as well as the global self-renewal of GSCs in tumors.
Figure 3.
Stable ALT phenotype in TG20 cells upon serial transplantation. (a) Serial intracranial transplantations of TG20-eGFP cells were performed in immunodeficient mice. Two or three months after each graft, the brain was removed and sliced coronally. (b and c) Grafted eGFP+ tumors were microdissected using a Zeiss Lumar V12 fluorescence stereomicroscope. After disaggregation from tissue chunks, TG20-eGFP tumor cells were sorted by FACS, excluding autofluorescence of non-eGFP cells and dead cells by PI. Appropriate gates were determined using negative (TG20) and positive (TG20-eGFP) controls from in vitro cultures. Immediately after sorting, 100,000 cells were re-grafted. (d) After dissection and selection, noninjected eGFP+ cells were cultured to check the formation of eGFP+ neurospheres (scale bar = 100 µm). (e) After seven days in culture, telomerase activity was measured in the formed neurospheres, using the TRAP assay; 500 or 1000 ng of total protein extracts from the first and the second transplantations (TG20-TS1 and TG20-TS2, respectively) was used. Similar experiments were performed with the telomerase+ TG1N cells grafted in parallel mice (TG1N-TS1). For each condition, negative controls were performed by heating samples for 10 min at 95°C.
TG20 cells are sensitive to the G-quadruplex ligand 360B in vivo
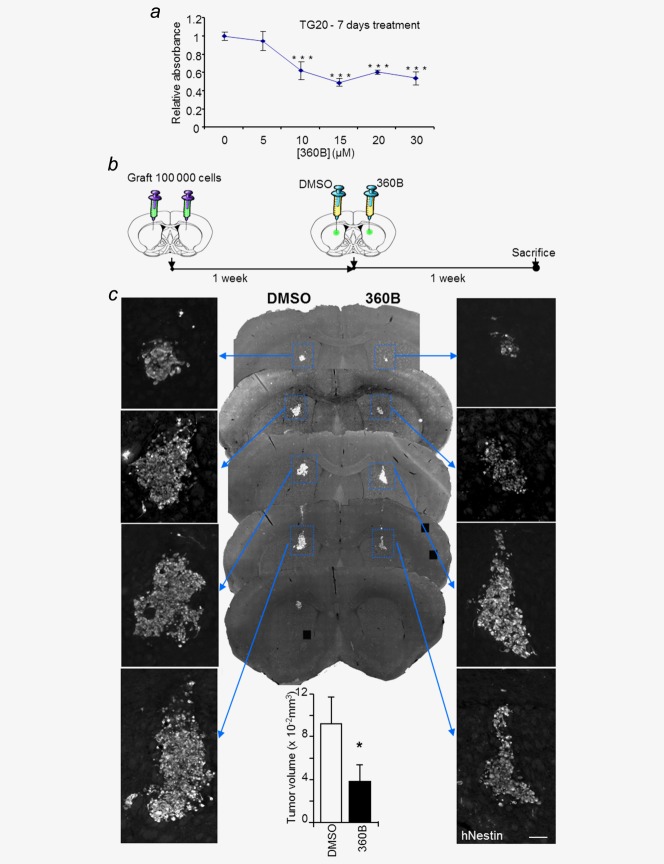
As the first preclinical model of ALT glioma using human cells, TG20 tumors generated in mice could be used as a preclinical model to test the efficiency of different therapies specific to this type of tumor, particularly that of therapies targeting telomere maintenance mechanisms. G-quadruplex (G4) ligands have been shown to be promising new compounds in cancer therapy.12–17 We tested the effects of the G-quadruplex ligand 360B, a close derivative of 360A, that has been shown to destabilize telomeres and to induce cell death in cancer cell lines in vitro, including the death of ALT cell lines.17,19 360B was used instead of 360A, because of its improved solubility in aqueous media. As shown in Figure 4a, 360B decreased TG20 cell viability in vitro after 7 days of treatment (Fig. 4a).
Figure 4.
TG20 cells are sensitive to the G-quadruplex ligand 360B in vitro and in vivo. (a) WST-1 proliferation assay for TG20 cells, 7 days after treatment with the G-quadruplex ligand 360B at different concentrations. Values were calculated relative to the WST-1 absorbance value of the control cells treated with DMSO. Error bars represent the SEM from four replicates (***p < 0.001, Student's t-test). (b) For 360B testing in vivo, 100,000 TG20 cells were bilaterally injected intracranially into NOD-SCID mouse brains. One week later, DMSO or 360B (2 µL at 10 µM) was injected in the tumor cell injection site. Mice were sacrificed one week after treatment. (c) Representative images of successive equidistant sections from a brain injected with TG20 cells and treated with either DMSO or 360B, as in b. Panels on the right and left sides show a magnified image of the TG20 tumor, stained with human Nestin (hNestin; scale bar = 100 µm). The lower graph represents the mean of DMSO- or 360B-treated tumor volumes from n = 3 animals (*p < 0.05, unpaired Student's t-test).
To test the effects of 360B on the ALT GSCs in vivo, 100,000 TG20 cells were bilaterally injected into the striatum of the brain in three NSG mice. One week later, 2 µL of 360B (10 µM) or 2 µL of DMSO (0.1%, vehicle) were injected at the site of tumor cell implantation. The animals were sacrificed after either 7 days (Fig. 4b) or one month (Fig. 5a). Variants of this drug delivery method have been previously used for the delivery of neurodegenerative chemical agents.34,35 Immunostaining for the human nestin protein was used to detect TG20 cells on serial brain sections and revealed a significant 42% reduction of the mean volume occupied by the tumor cells in the hemispheres treated with 360B compared with the hemispheres treated with vehicle in mice sacrificed 7 days after treatment (0.039 ± 0.009 and 0.092 ± 0.015 mm3 for 360B and vehicle-treated hemispheres, respectively; n=3, unpaired Student's t-test, p value = 0.036; Fig. 4c).
Figure 5.
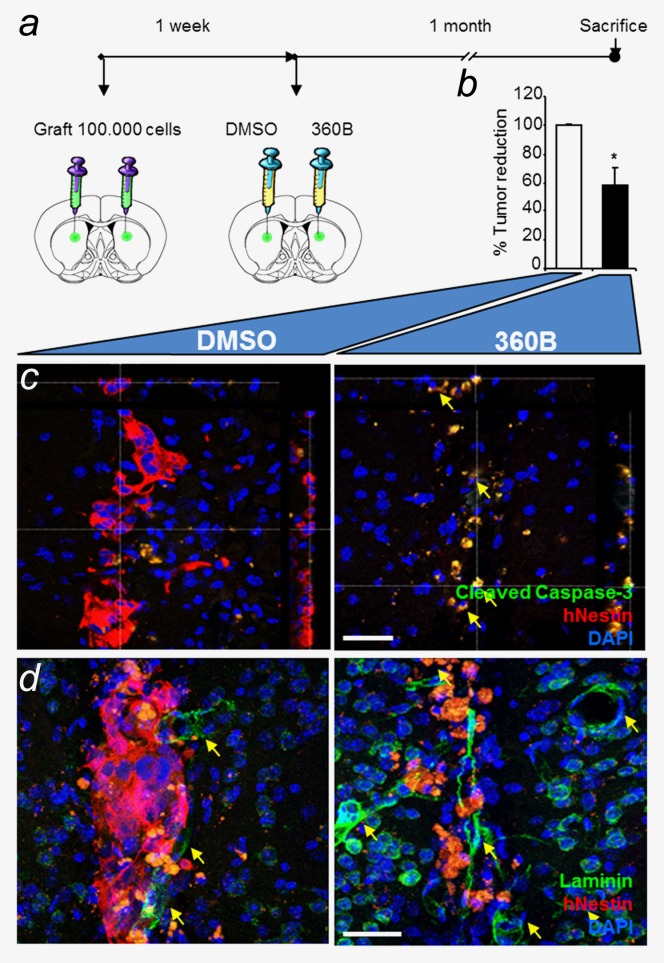
Long-term in vivo efficiency of the G-quadruplex ligand 360B on TG20 cells. (a) TG20 cells were injected intracranially into each hemisphere of NOD-SCID mouse brains, and drugs were added one week later, as in Figure 4b. Mice were sacrificed one month after treatment. (b) The percentage of tumor reduction in 360B-treated versus DMSO-treated hemispheres was evaluated by counting the remaining TG20 cells in each brain section (*p < 0.05, Student's t-test). (c and d) Immunofluorescence in representative brain sections shows human Nestin (hNestin) staining and cleaved caspase-3 or laminin staining. n = 2 animals (scale bar = 40 µm).
The effects of the compound were still detectable one month after treatment, as shown in two NSG mice by the 40% reduction in human nestin-positive cells in hemispheres treated with 360B compared with vehicle-treated hemispheres (p value = 0.0397, unpaired Student's t-test; Fig. 5b). Additionally, immunostaining for cleaved caspase-3 showed that the majority of TG20 cells were undergoing apoptosis in 360B-treated tumors, whereas only rare apoptotic cells were detected in controls (Fig. 5c). Notably, cleaved caspase-3 expression was detected only in human Nestin-positive cells, showing that 360B induced glioma cell apoptosis and spared normal cells. Together these data show the in vivo sensitivity of TG20 cells to the G-quadruplex ligand 360B, emphasizing the potential use of G-quadruplex ligands in ALT glioma therapy and TG20 cells in developing specific therapeutic approaches for ALT gliomas.
TG20 cells generate intracranial tumors in nude mice
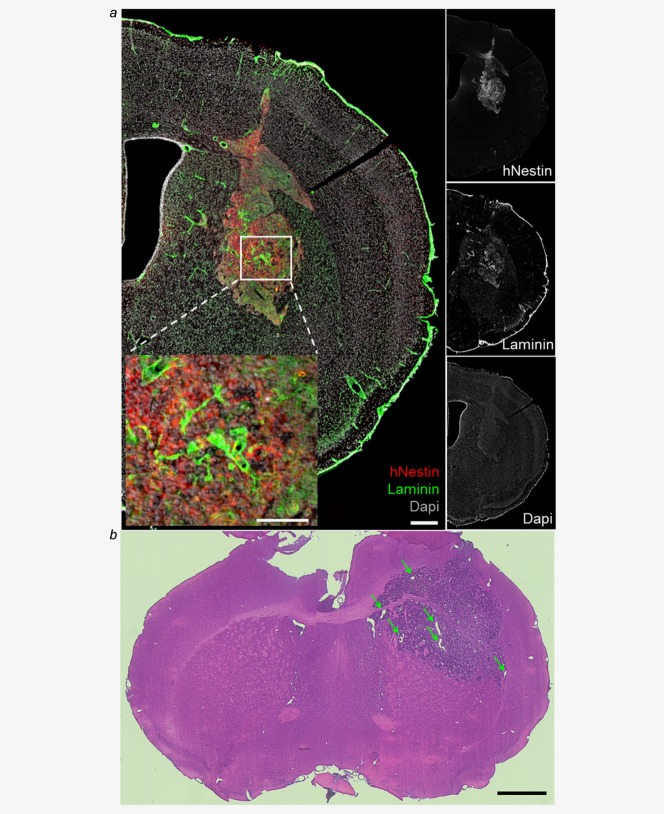
The use of isolator-reared NSG mice is not trivial, and the development of different types of experiments is therefore difficult. We thus assessed the tumorigenicity of TG20 cells in three nude mice (athymic, T- and B-cell deficient), which are less permissive to tumor engraftment because they retain innate immune cells, such as natural killer (NK) cells, but are easier to manipulate than NSG mice.36 As shown in Figure 6, TG20 cells generated tumors in nude mice. Immunostaining of brain sections for human nestin and for laminin, to reveal TG20 cells and blood vessels, respectively, revealed strong tumor vascularization 4 months after engraftment (Fig. 6a). Hematoxylin and eosin staining at 6 months after engraftment also showed microvascular hyperplasia (Fig. 6b).
Figure 6.
TG20 cells generate tumors in nude mice. (a) Immunofluorescence staining in a nude mouse brain slice containing a TG20 tumor 4 months after cell engraftment. The right panels show separate immunostaining of hNestin, laminin, and DAPI, and the left panel shows an enlarged merge image of these immunostained sections (scale bar = 0.3 mm). The inset shows high vascularization of the tumor (scale bar = 0.15 mm). (b) Representative hematoxylin & eosin staining of TG20 tumors 6 months after engraftment in nude mice shows large blood vessels (indicated by arrows), specifically in the tumor region. Representative image of n = 3 animals (scale bar = 1 mm).
Discussion
Patients suffering from ALT glioblastomas survive slightly longer than patients suffering from telomerase-positive glioblastomas. The prognosis remains nevertheless dismal, and the development of specific new therapies is needed.37,38 Here, we further characterized the ALT phenotype and the GSC potential of TG20 cells and showed that intracranial grafts of TG20 cells in NSG and nude mice constituted efficient preclinical models of ALT gliomas, which could aid the understanding of ALT glioma pathogenesis and the development of specific therapies.
ALT cells replicate their telomeres in a recombination-dependent manner.3 Consistently, we found that TG20 cells exhibited a high rate of telomeric recombination, as revealed by high levels of T-SCEs, T-circles, and T complexes, all of which have been associated with the ALT mechanism.24–26 This high level of recombination concerns only telomeres because the frequencies of SCE elsewhere in the genome are similar in TG20 cells and telomerase-positive GSCs, suggesting that the dysregulation of recombination is not global in TG20 cells but is likely telomere specific, which could be a general feature of ALT cells as previously proposed by Bechter et al.39
TG20 cells, similarly to other ALT cells, form numerous telomere dysfunction-induced foci (TIF), a telomere-specific DNA damage response analogous to a double-strand break response, which indicates that telomeres are deprotected.6,40 The main function of telomeres is to protect chromosomes from end-to-end fusions. The relative stability of the TG20 cell karyotype suggests that despite abundant signals of dysfunction and a high rate of recombination, the telomeres of TG20 cells retain their protective function against chromosome instability. The shelterin complex plays an essential role in telomere capping, which protects the chromosome ends from being recognized as DNA damage sites and/or from fusions.41 Cesare et al.40 have proposed that the telomeres of ALT cells have an intermediate state of telomere capping, where telomeres are less saturated with shelterin proteins. Such capping is supposedly efficient enough to inhibit end-to-end telomere fusions, but it induces a DNA-damage response and subsequent telomeric recombination, as observed here in TG20 cells.
The potential interest of developing specific anticancer strategies against ALT mechanisms is largely dependent on the stability of the ALT phenotype. Furthermore, determining the mechanisms by which telomerase is repressed in ALT cells may provide a better understanding of the pathogenetic mechanisms that lead to the emergence of this type of cancer cell. We have shown that the ALT phenotype of TG20 cells is stable upon serial transplantation in mice and that TG20 cells do not reacquire telomerase activity after several transplantations in vivo. As for the majority of ALT cells, the absence of telomerase activity in TG20 cells is directly related to the lack of expression of the catalytic subunit hTERT. One proposed mechanism of tert gene repression in ALT cells is DNA methylation of the tert promoter, as global DNA-demethylating compounds have been shown to induce the expression of TERT mRNA in some ALT cells, such as SUSM-1, U2OS, and GM847 cells.42,43 Unlike those cells, tert gene repression in TG20 cells does not appear to be related to promoter methylation. Demethylation treatment did not stimulate TERT expression in TG20 cells, indicating that other stable mechanisms of repression are involved. One of these mechanisms could be mediated by WT1, a transcriptional repressor of hTERT. In TG20 cells, we showed that the WT1 binding site, which is present in the distal region of the tert promoter, was unmethylated, a condition required for WT1 repression of htert transcription.32,33
Serial transplantations of TG20 cells into NOD-SCID mice showed that GSCs did not absolutely require telomerase activation to maintain their telomeres in a selective in vivo environment and confirmed that the ALT pathway is capable of supporting the long-term proliferative potential and self-renewal capacity of these cells. GSCs are supposed to play an essential role in glioma relapse because of their putative resistance to current treatments and their capacity to regenerate tumors.8 As a GSC line, TG20 cells can be used to screen potential anticancer compounds during the initial phase of tumor generation in mice in the hopes of evaluating their effects more precisely on GSCs rather than on “differentiated” cancer cells. Because ALT-specific therapies have not yet been identified, we decided to test the G-quadruplex ligand 360B on TG20 cells. Given the importance of G-quadruplexes in telomere homeostasis, ligands stabilizing these structures have been developed for use as antitumor therapeutic strategies.12–17 360A, a pyridine derivative, has been shown to target G4 telomeric structures and to impair telomere replication and mitotic progression leading to cell death.17–19,21 While G-quadruplex ligands have been extensively tested in telomerase-positive cells, to our knowledge, no data exist on the effects of these ligands on ALT tumors in vivo. 360A has been previously shown to efficiently reduce the proliferation of telomerase-positive glioma cell lines and SAOS-2 cells in vitro.17 We showed here that TG20 cells were sensitive both in vitro and in vivo to 360B, demonstrating that this treatment impairs tumor development when performed at the initiation phase just after engraftment.
These encouraging results emphasize the potential interest of targeting G-quadruplex structures for ALT glioma treatments. However, the effects of 360B on established tumors and its potential use in combination with other treatments such as ionizing radiation and the alkylating agent temozolomide, which is currently used in glioblastoma patients after surgery, should be now investigated.44 Accordingly, intracerebral grafts of TG20 cells in nude mice could be of great interest because contrary to NSG mice, nude mice are not particularly radiosensitive.
To conclude, we have confirmed and further characterized the ALT phenotype and GSC potential of TG20 cells, and our findings indicate that intracerebral TG20 grafts in immunodeficient mice represent an attractive preclinical model of ALT glioma. TG20 cells have been isolated from a second-relapse glioblastoma in a 38-year-old patient that benefited from the STUPP protocols.6 This model could therefore be particularly adapted to evaluate the effects of treatment on resistant tumors.
Acknowledgments
We thank all members of LRP for helpful discussions and comments on this work. We are grateful for Dr. F. Pflumio for providing NOD-SCID mice and V. Neuville and C. Joubert for animal facility. We acknowledge Dr. P. Fouchet and O. Etienne for technical help, T. Kortulewski and L. Irbah for imaging facility and N. Dechamps and J. Baijer for Cell sorting facility.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information
Supplementary Information
References
- Kim NW, Piatyszek MA, Prowse KR. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–5. doi: 10.1126/science.7605428. et al. [DOI] [PubMed] [Google Scholar]
- Henson JD, Reddel RR. Assaying and investigating alternative lengthening of telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–11. doi: 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–30. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- Scheel C, Schaefer KL, Jauch A. Alternative lengthening of telomeres is associated with chromosomal instability in osteosarcomas. Oncogene. 2001;20:3835–44. doi: 10.1038/sj.onc.1204493. et al. [DOI] [PubMed] [Google Scholar]
- Henson JD, Hannay JA, McCarthy A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res. 2005;11:217–25. et al. [PubMed] [Google Scholar]
- Silvestre DC, Pineda JR, Hoffschir F. Alternative lengthening of telomeres in human glioma stem cells. Stem Cells. 2011;29:440–51. doi: 10.1002/stem.600. et al. [DOI] [PubMed] [Google Scholar]
- Heaphy CM, Schreck KC, Raabe E. A glioblastoma neurosphere line with alternative lengthening of telomeres. Acta Neuropathol. 2013;126:607–8. doi: 10.1007/s00401-013-1174-x. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. et al. [DOI] [PubMed] [Google Scholar]
- Ruden M, Puri N. Novel anticancer therapeutics targeting telomerase. Cancer Treat Rev. 2013;39:444–56. doi: 10.1016/j.ctrv.2012.06.007. [DOI] [PubMed] [Google Scholar]
- Hu J, Hwang SS, Liesa M. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell. 2012;148:651–63. doi: 10.1016/j.cell.2011.12.028. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffitzel C, Berger I, Postberg J. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci USA. 2001;98:8572–7. doi: 10.1073/pnas.141229498. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowan SM, Harrison JR, Patterson L. A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol Pharmacol. 2002;61:1154–62. doi: 10.1124/mol.61.5.1154. et al. [DOI] [PubMed] [Google Scholar]
- Burger AM, Dai F, Schultes CM. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–96. doi: 10.1158/0008-5472.CAN-04-2910. et al. [DOI] [PubMed] [Google Scholar]
- Tauchi T, Shin-Ya K, Sashida G. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways. Oncogene. 2003;22:5338–47. doi: 10.1038/sj.onc.1206833. et al. [DOI] [PubMed] [Google Scholar]
- Riou JF, Guittat L, Mailliet P. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc Natl Acad Sci USA. 2002;99:2672–7. doi: 10.1073/pnas.052698099. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatak P, Cookson JC, Dai F. Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Br J Cancer. 2007;96:1223–33. doi: 10.1038/sj.bjc.6603691. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennarun G, Granotier C, Gauthier LR. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005;24:2917–28. doi: 10.1038/sj.onc.1208468. et al. [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Granotier C, Hoffschir F. Rad51 and DNA-PKcs are involved in the generation of specific telomere aberrations induced by the quadruplex ligand 360A that impair mitotic cell progression and lead to cell death. Cell Mol Life Sci. 2012;69:629–40. doi: 10.1007/s00018-011-0767-6. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granotier C, Pennarun G, Riou L. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005;33:4182–90. doi: 10.1093/nar/gki722. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patru C, Romao L, Varlet P. CD133, CD15/SSEA-1, CD34 or side populations do not resume tumor-initiating properties of long-term cultured cancer stem cells from human malignant glio-neuronal tumors. BMC Cancer. 2010;10:66. doi: 10.1186/1471-2407-10-66. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennarun G, Granotier C, Hoffschir F. Role of ATM in the telomere response to the G-quadruplex ligand 360A. Nucleic Acids Res. 2008;36:1741–54. doi: 10.1093/nar/gkn026. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal-Bendirdjian E, Jacquemin-Sablon A. Cisplatin resistance in a murine leukemia cell line is associated with a defective apoptotic process. Exp Cell Res. 1995;218:201–12. doi: 10.1006/excr.1995.1148. [DOI] [PubMed] [Google Scholar]
- Zinn RL, Pruitt K, Eguchi S. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67:194–201. doi: 10.1158/0008-5472.CAN-06-3396. et al. [DOI] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Der-Sarkissian H, Cazes L. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–7. doi: 10.1158/0008-5472.can-03-4035. et al. [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–57. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosek J, Rycovska A, Makhov AM, Griffith JD. Amplification of telomeric arrays via rolling-circle mechanism. J Biol Chem. 2005;280:10840–5. doi: 10.1074/jbc.M409295200. et al. [DOI] [PubMed] [Google Scholar]
- Nabetani A, Ishikawa F. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol Cell Biol. 2009;29:703–13. doi: 10.1128/MCB.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyo S, Takakura M, Fujiwara T. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99:1528–38. doi: 10.1111/j.1349-7006.2008.00878.x. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KH, Kang MK, Dicterow E. Hypermethylation of the hTERT promoter inhibits the expression of telomerase activity in normal oral fibroblasts and senescent normal oral keratinocytes. Br J Cancer. 2003;89:1473–8. doi: 10.1038/sj.bjc.6601291. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatina NG, Poole JC, Saldanha SN. Control mechanisms in the regulation of telomerase reverse transcriptase expression in differentiating human teratocarcinoma cells. Biochem Biophys Res Commun. 2003;306:650–9. doi: 10.1016/s0006-291x(03)01033-7. et al. [DOI] [PubMed] [Google Scholar]
- Mueller W, Nutt CL, Ehrich M. Downregulation of RUNX3 and TES by hypermethylation in glioblastoma. Oncogene. 2007;26:583–93. doi: 10.1038/sj.onc.1209805. et al. [DOI] [PubMed] [Google Scholar]
- Azouz A, Wu YL, Hillion J. Epigenetic plasticity of hTERT gene promoter determines retinoid capacity to repress telomerase in maturation-resistant acute promyelocytic leukemia cells. Leukemia. 2010;24:613–22. doi: 10.1038/leu.2009.283. et al. [DOI] [PubMed] [Google Scholar]
- Oh S, Song Y, Yim J. The Wilms' tumor 1 tumor suppressor gene represses transcription of the human telomerase reverse transcriptase gene. J Biol Chem. 1999;274:37473–8. doi: 10.1074/jbc.274.52.37473. et al. [DOI] [PubMed] [Google Scholar]
- Perez-Navarro E, Arenas E, Reiriz J. Glial cell line-derived neurotrophic factor protects striatal calbindin-immunoreactive neurons from excitotoxic damage. Neuroscience. 1996;75:345–52. doi: 10.1016/0306-4522(96)00336-3. et al. [DOI] [PubMed] [Google Scholar]
- Pineda JR, Rubio N, Akerud P. Neuroprotection by GDNF-secreting stem cells in a Huntington's disease model: optical neuroimage tracking of brain-grafted cells. Gene Ther. 2007;14:118–28. doi: 10.1038/sj.gt.3302847. et al. [DOI] [PubMed] [Google Scholar]
- Habu S, Fukui H, Shimamura K. In vivo effects of anti-asialo GM1. I. Reduction of NK activity and enhancement of transplanted tumor growth in nude mice. J Immunol. 1981;127:34–8. et al. [PubMed] [Google Scholar]
- Hakin-Smith V, Jellinek DA, Levy D. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet. 2003;361:836–8. doi: 10.1016/s0140-6736(03)12681-5. et al. [DOI] [PubMed] [Google Scholar]
- McDonald KL, McDonnell J, Muntoni A. Presence of alternative lengthening of telomeres mechanism in patients with glioblastoma identifies a less aggressive tumor type with longer survival. J Neuropathol Exp Neurol. 2010;69:729–36. doi: 10.1097/NEN.0b013e3181e576cf. et al. [DOI] [PubMed] [Google Scholar]
- Bechter OE, Zou Y, Shay JW. Homologous recombination in human telomerase-positive and ALT cells occurs with the same frequency. EMBO Rep. 2003;4:1138–43. doi: 10.1038/sj.embor.7400027. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Kaul Z, Cohen SB. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol. 2009;16:1244–51. doi: 10.1038/nsmb.1725. et al. [DOI] [PubMed] [Google Scholar]
- Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–7. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessain SK, Yu H, Reddel RR. Methylation of the human telomerase gene CpG island. Cancer Res. 2000;60:537–41. et al. [PubMed] [Google Scholar]
- Devereux TR, Horikawa I, Anna CH. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59:6087–90. et al. [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. et al. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Information