Abstract
Ribosomal 30S protein S1 causes disruption of the secondary structure of certain pyrimidine-containing polynucleotides. Helical poly(U), poly(C, U), and neutral and acidic poly(C) are stoichiometrically converted by S1 to structures indistinguishable from their partially or completely thermally denatured forms, as revealed by circular dichroism. Of the several double- and triple-stranded helical polynucleotides tested that contain one polypurine strand and at least one polypyrimidine strand, only the conformation of the DNA.RNA hybrid, poly(A)-poly(dT), is perturbed. In the presence of S1, this hybrid undergoes a transition to a new structure that has a circular dichroism spectrum unlike either the native or thermally denatured forms. Intercalated ethidium bromide is released from poly(A)-poly(dT) by S1, confirming the occurrence of a conformational rearrangement. The translation inhibitor, autintricarboxylic acid, completely inhibits the action of S1 on polypyrimidines, but has no effect on the conformational perturbation of poly(A(-poly(dT). The possible relation between these observations and the biological function of protein S1 is discussed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AKINRIMISI E. O., SANDER C., TS'O P. O. Properties of helical polycytidylic acid. Biochemistry. 1963 Mar-Apr;2:340–344. doi: 10.1021/bi00902a028. [DOI] [PubMed] [Google Scholar]
- Adler A. J., Grossman L., Fasman G. D. Polyriboadenylic and polydeoxyriboadenylic acids. Optical rotatory studies of pH-dependent conformations and their relative stability. Biochemistry. 1969 Sep;8(9):3846–3859. doi: 10.1021/bi00837a051. [DOI] [PubMed] [Google Scholar]
- Brahms J., Maurizot J. C., Michelson A. M. Conformation and thermodynamic properties of oligocytidylic acids. J Mol Biol. 1967 May 14;25(3):465–480. doi: 10.1016/0022-2836(67)90199-4. [DOI] [PubMed] [Google Scholar]
- Brahms J., Michelson A. M., Van Holde K. E. Adenylate oligomers in single- and double-strand conformation. J Mol Biol. 1966 Feb;15(2):467–488. doi: 10.1016/s0022-2836(66)80122-5. [DOI] [PubMed] [Google Scholar]
- Carmichael G. G. Isolation of bacterial and phage proteins by homopolymer RNA-cellulose chromatography. J Biol Chem. 1975 Aug 10;250(15):6160–6167. [PubMed] [Google Scholar]
- Dahlberg A. E., Dahlberg J. E. Binding of ribosomal protein S1 of Escherichia coli to the 3' end of 16S rRNA. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2940–2944. doi: 10.1073/pnas.72.8.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C., Stiegler P., Ebel J. P. Sequence analysis of the 3'-T1 oligonucleotide of 16S ribosomal RNA from Escherichia coli. FEBS Lett. 1974 Dec 1;49(1):47–48. doi: 10.1016/0014-5793(74)80628-9. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Stiegler P., Mackie G. A., Zimmermann R. A., Ebel J. P., Fellner P. Primary sequence of the 16S ribosomal RNA of Escherichia coli. Nucleic Acids Res. 1975 Feb;2(2):265–278. doi: 10.1093/nar/2.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FASMAN G. D., LINDBLOW C., GROSSMAN L. THE HELICAL CONFORMATIONS OF POLYCYTIDYLIC ACID: STUDIES ON THE FORCES INVOLVED. Biochemistry. 1964 Aug;3:1015–1021. doi: 10.1021/bi00896a002. [DOI] [PubMed] [Google Scholar]
- FRESCO J. R., KLEMPERER E. Polyriboadenylic acid, a molecular analogue of ribonucleic acid and desoxyribonucleic acid. Ann N Y Acad Sci. 1959 Sep 4;81:730–741. doi: 10.1111/j.1749-6632.1959.tb49354.x. [DOI] [PubMed] [Google Scholar]
- Goodman H. M., Billeter M. A., Hindley J., Weissmann C. The nucleotide sequence at the 5'-terminus of the Q RNA minus trand. Proc Natl Acad Sci U S A. 1970 Oct;67(2):921–928. doi: 10.1073/pnas.67.2.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman A. P., Stewart M. L. Inhibition of the attachment of messenger ribonucleic acid to ribosomes. Proc Natl Acad Sci U S A. 1968 Oct;61(2):719–725. doi: 10.1073/pnas.61.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARTMAN K. A., Jr, RICH A. THE TAUTOMERIC FORM OF HELICAL POLYRIBOCYTIDYLIC ACID. J Am Chem Soc. 1965 May 5;87:2033–2039. doi: 10.1021/ja01087a031. [DOI] [PubMed] [Google Scholar]
- Inouye H., Pollack Y., Petre J. Physical and functional homology between ribosomal protein S1 and interference factor i. Eur J Biochem. 1974 Jun 1;45(1):109–117. doi: 10.1111/j.1432-1033.1974.tb03535.x. [DOI] [PubMed] [Google Scholar]
- KATZ S., COMB D. G. A NEW METHOD FOR THE DETERMINATION OF THE BASE COMPOSITION OF RIBONUCLEIC ACID. J Biol Chem. 1963 Sep;238:3065–3067. [PubMed] [Google Scholar]
- Kamen R., Kondo M., Römer W., Weissmann C. Reconstitution of Q replicase lacking subunit with protein-synthesis-interference factor i. Eur J Biochem. 1972 Nov 21;31(1):44–51. doi: 10.1111/j.1432-1033.1972.tb02498.x. [DOI] [PubMed] [Google Scholar]
- Kenner R. A. A protein-nucleic acid crosslink in 30S ribosomes. Biochem Biophys Res Commun. 1973 Apr 16;51(4):932–938. doi: 10.1016/0006-291x(73)90016-8. [DOI] [PubMed] [Google Scholar]
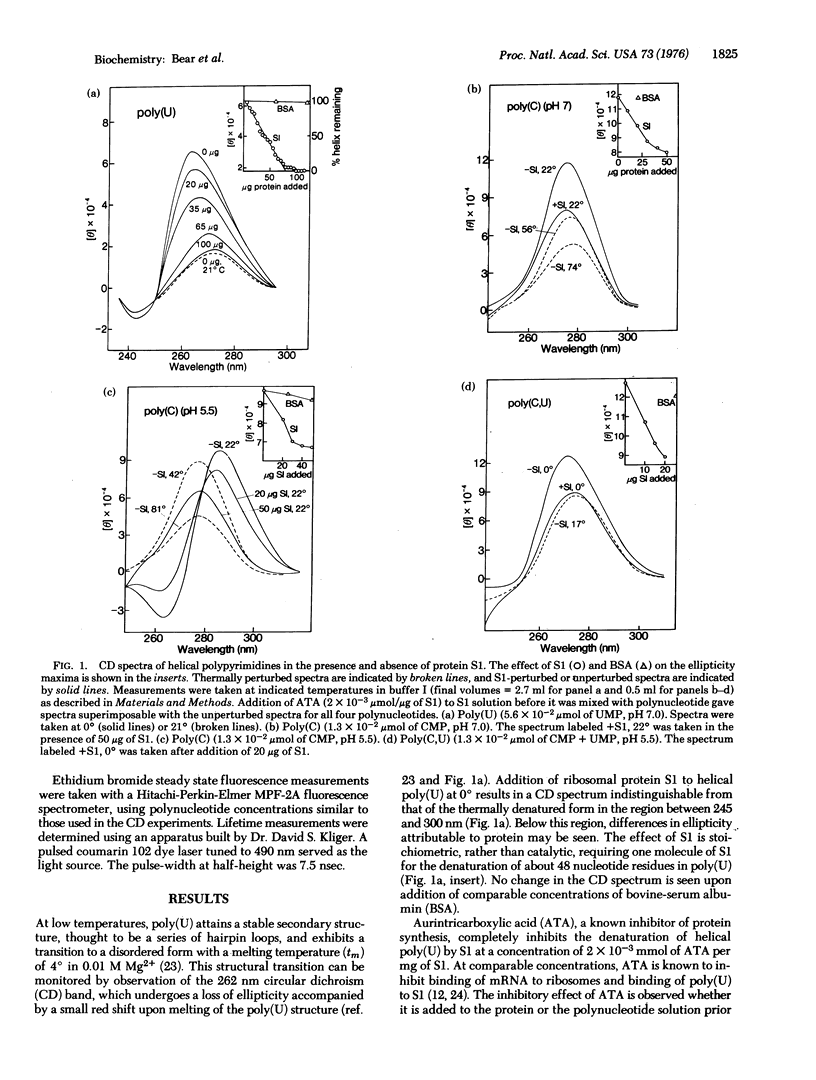
- LANGRIDGE R., RICH A. Molecular structure of helical polycytidylic acid. Nature. 1963 May 25;198:725–728. doi: 10.1038/198725a0. [DOI] [PubMed] [Google Scholar]
- Leng M., Felsenfeld G. A study of polyadenylic acid at neutral pH. J Mol Biol. 1966 Feb;15(2):455–466. doi: 10.1016/s0022-2836(66)80121-3. [DOI] [PubMed] [Google Scholar]
- Miller M. J., Niveleau A., Wahba A. J. Inhibition of synthetic and natural messenger translation. I. Purification and properties of a protein isolated from Escherichia coli MRE 600 ribosomes. J Biol Chem. 1974 Jun 25;249(12):3803–3807. [PubMed] [Google Scholar]
- Miller M. J., Wahba A. J. Inhibition of synthetic and natural messenger translation. II. Specificity and mechanism of action of a protein isolated from Escherichia coli MRE 600 ribosomes. J Biol Chem. 1974 Jun 25;249(12):3808–3813. [PubMed] [Google Scholar]
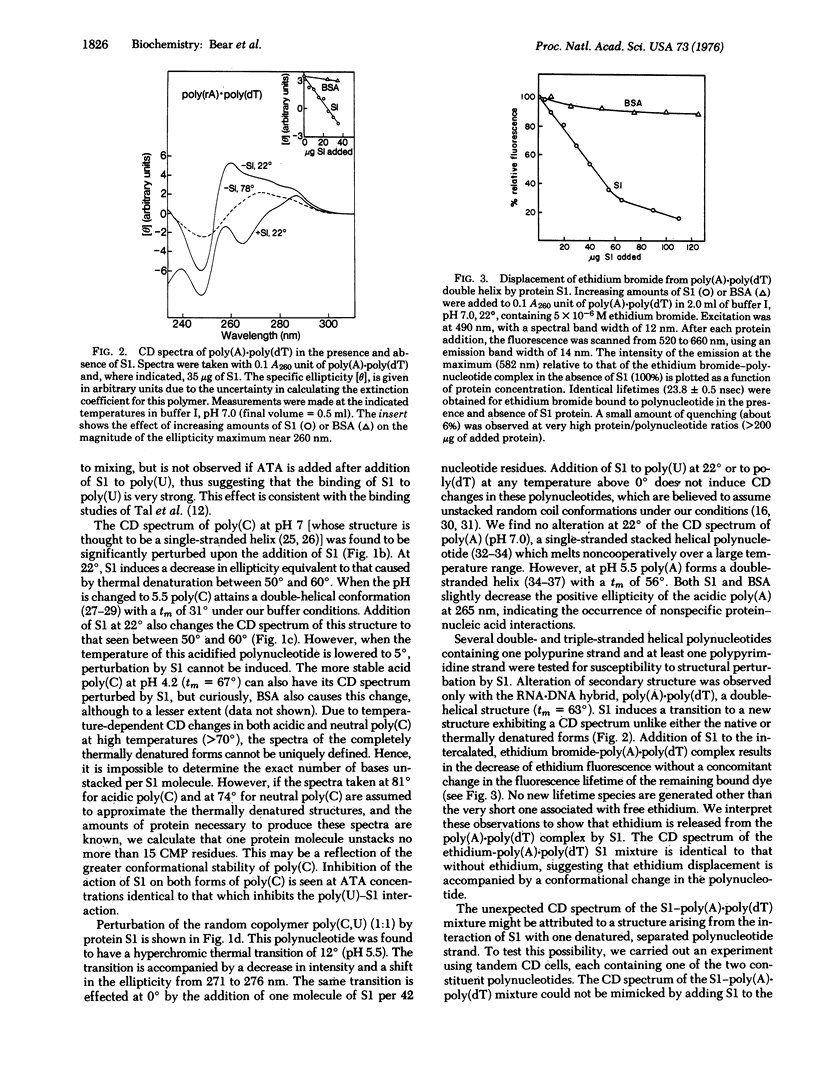
- Moore P. B., Traut R. R., Noller H., Pearson P., Delius H. Ribosomal proteins of Escherichia coli. II. Proteins from the 30 s subunit. J Mol Biol. 1968 Feb 14;31(3):441–461. doi: 10.1016/0022-2836(68)90420-8. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Herr W. Nucleotide sequence of the 3' terminus of E. coli 16S ribosomal RNA. Mol Biol Rep. 1974 Dec;1(8):437–439. doi: 10.1007/BF00360668. [DOI] [PubMed] [Google Scholar]
- RICH A., DAVIES D. R., CRICK F. H., WATSON J. D. The molecular structure of polyadenylic acid. J Mol Biol. 1961 Feb;3:71–86. doi: 10.1016/s0022-2836(61)80009-0. [DOI] [PubMed] [Google Scholar]
- Riley M., Maling B. Physical and chemical characterization of two- and three-stranded adenine-thymine and adenine-uracil homopolymer complexes. J Mol Biol. 1966 Sep;20(2):359–389. doi: 10.1016/0022-2836(66)90069-6. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K. U., Steitz J. A. The 3' terminal oligonucleotide of E. coli 16S ribosomal RNA: the sequence in both wild-type and RNase iii- cells is complementary to the polypurine tracts common to mRNA initiator regions. Nucleic Acids Res. 1975 Jun;2(6):787–798. doi: 10.1093/nar/2.6.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A., Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szer W., Leffler S. Interaction of Escherichia coli 30S ribosomal subunits with MS2 phage RNA in the absence of initiation factors. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3611–3615. doi: 10.1073/pnas.71.9.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal M., Aviram M., Kanarek A., Weiss A. Polyuridylic acid binding and translating by Escherichia coli ribosomes: stimulation by protein I, inhibition by aurintricarboxylic acid. Biochim Biophys Acta. 1972 Oct 27;281(3):381–392. doi: 10.1016/0005-2787(72)90452-2. [DOI] [PubMed] [Google Scholar]
- Thomas G., Sweeney R., Chang C., Noller H. F. Identification of proteins functionally altered by chemical modification of the transfer RNA and polyuridylic acid binding sites of 30 S ribosomal subunits. J Mol Biol. 1975 Jun 15;95(1):91–102. doi: 10.1016/0022-2836(75)90338-1. [DOI] [PubMed] [Google Scholar]
- Thrierr J. C., Dourlent M., Leng M. A study of polyuridylic acid. J Mol Biol. 1971 Jun 28;58(3):815–830. doi: 10.1016/0022-2836(71)90042-8. [DOI] [PubMed] [Google Scholar]
- Van Dieijen G., Van Der Laken C. J., Van Knippenberg P. H., Van Duin J. Function of Escherichia coli ribosomal protein S1 in translation of natural and synthetic messenger RNA. J Mol Biol. 1975 Apr 15;93(3):351–366. doi: 10.1016/0022-2836(75)90282-x. [DOI] [PubMed] [Google Scholar]
- Von Hippel P. H., McGhee J. D. DNA-protein interactions. Annu Rev Biochem. 1972;41(10):231–300. doi: 10.1146/annurev.bi.41.070172.001311. [DOI] [PubMed] [Google Scholar]
- Voynow P., Kurland C. G. Stoichiometry of the 30S ribosomal proteins of Escherichia coli. Biochemistry. 1971 Feb 2;10(3):517–524. doi: 10.1021/bi00779a026. [DOI] [PubMed] [Google Scholar]
- Wahba A. J., Miller M. J., Niveleau A., Landers T. A., Carmichael G. G., Weber K., Hawley D. A., Slobin L. I. Subunit I of G beta replicase and 30 S ribosomal protein S1 of Escherichia coli. Evidence for the identity of the two proteins. J Biol Chem. 1974 May 25;249(10):3314–3316. [PubMed] [Google Scholar]
- Weber H. J. Stoichiometric measurements of 30S and 50S ribosomal proteins from Escherichia coli. Mol Gen Genet. 1972;119(3):233–248. doi: 10.1007/BF00333861. [DOI] [PubMed] [Google Scholar]
- van Duin J., Kurland C. G., Dondon J., Grunberg-Manago M. Near neighbors of IF3 bound to 30S ribosomal subunits. FEBS Lett. 1975 Nov 15;59(2):287–290. doi: 10.1016/0014-5793(75)80394-2. [DOI] [PubMed] [Google Scholar]
- van Duin J., van Knippenberg P. H. Functional heterogeneity of the 30 S ribosomal subunit of Escherichia coli. 3. Requirement of protein S1 for translation. J Mol Biol. 1974 Mar 25;84(1):185–195. doi: 10.1016/0022-2836(74)90221-6. [DOI] [PubMed] [Google Scholar]
- van Knippenberg P. H., Hooykaas P. J., van Duin J. The stoichiometry of E. coli 30S ribosomal protein S1 on in vivo and in vitro polyribosomes. FEBS Lett. 1974 May 1;41(2):323–326. doi: 10.1016/0014-5793(74)81239-1. [DOI] [PubMed] [Google Scholar]



