Abstract
Human skin is constantly exposed to solar light containing visible and ultraviolet radiation (UVR), a powerful skin carcinogen. UVR elicits cellular responses in epidermal cells via several mechanisms: direct absorption of short-wavelength UVR photons by DNA, oxidative damage caused by long-wavelength UVR, and, as we recently demonstrated, via a retinal-dependent G protein-coupled signaling pathway. Because the human epidermis is exposed to a wide range of light wavelengths, we investigated whether opsins, light-activated receptors that mediate photoreception in the eye, are expressed in epidermal skin to potentially serve as photosensors. Here we show that four opsins—OPN1-SW, OPN2, OPN3 and OPN5—are expressed in the two major human epidermal cell types, melanocytes and keratinocytes, and the mRNA expression profile of these opsins does not change in response to physiological UVR doses. We detected two OPN3 splice variants present in similar amounts in both cell types and three OPN5 splice isoforms, two of which encode truncated proteins. Notably, OPN2 and OPN3 mRNA were significantly more abundant than other opsins and encoded full-length proteins. Our results demonstrate that opsins are expressed in epidermal skin cells and suggest that they might initiate light–induced signaling pathways, possibly contributing to UVR phototransduction.
Introduction
Phototransduction, the mechanism by which photons are absorbed and converted into a cellular response, occurs primarily through a class of proteins called opsins (1). Opsins are light-sensitive members of the superfamily of heptahelical G protein-coupled receptors (GPCRs), a large family of cell surface receptors that are activated by a wide variety of stimuli and mediate signaling across biological membranes (2). Functional opsin receptors require two components: an opsin apoprotein plus a covalently bound vitamin A-derived aldehyde chromophore (retinal), which binds to a highly conserved lysine residue (K296 in bovine rhodopsin) (3).
Visual pigments include rhodopsin (OPN2) and the cone opsins (OPN1), which are expressed in rod and cone cells of the retina and mediate dim-light and color vision, respectively (1). In addition to the classical visual opsins, a number of other opsins have been recently described, including panopsin or encephalopsin (OPN3), melanopsin (OPN4) and neuropsin (OPN5) (4). OPN4 is expressed in a subset of retinal cells and mediates circadian photoentrainment in mammals (5–8), whereas OPN3 and OPN5 have been identified in the brain and their function remains unknown (9–14).
We recently showed that human epidermal melanocytes (HEMs) use a G protein-coupled phototransduction mechanism to increase levels of the protective pigment melanin at early time points after exposure to long-wavelength UVR (15,16). In our previous study we used degenerate primers to investigate expression of opsin messenger RNA (mRNA) in melanocytes (15). These primers amplified a cDNA fragment containing OPN2 as well as a truncated OPN3 splice variant. This led us to further investigate the expression and functional role of OPN2/3 in melanocytes. OPN2, a blue–light-sensitive pigment in retinal rod cells, contributes to the UVR response in HEMs, as siRNA-mediated reduction of its expression level decreases retinal–dependent UVR responses (15). As rhodopsin is sensitive to blue-green light and couples to different G proteins in rod cells and melanocytes (17), it is likely that UVR sensitivity in HEMs involves other opsins or GPCRs. Evidence for receptor-mediated phototransduction in melanocytes raises the question of whether a similar mechanism exists in primary human epidermal keratinocytes (KERs).
Here, we used a more sensitive approach to detect expression of opsins in the two major human epidermal cell types, HEMs and KERs. We used multiple opsin-specific primers to test for expression of opsins using reverse transcription polymerase chain reaction (RT-PCR) and found that four different opsins express in both cell types. We showed that, in addition to being expressed in HEMs (15), OPN2 mRNA and protein are also present in primary KERs. We also found that the short-wavelength OPN1 (OPN1-SW), but not medium- or long-wavelength OPN1 (OPN1-M/LW) mRNA, is expressed in both HEMs and KERs. In addition, we showed that the nonvisual receptors encephalopsin (OPN3) and neuropsin (OPN5), as well as novel splice variants of both OPN3 and OPN5, are expressed in HEMs and KERs. Using quantitative PCR (qPCR) analysis, we showed that these two epidermal cell types differentially express the opsin receptors: while OPN1-SW, and OPN5 are expressed at similarly low levels, OPN2 and OPN3 mRNA are expressed at significantly higher levels in both HEMs and KERs. The expression of opsins mRNA in skin suggests that these proteins might mediate phototransduction pathways in human skin either singularly or in concert.
Material and methods
Cell culture, Primary human epidermal melanocytes (HEM) obtained from neonatal foreskin (Cascade Biologics/Invitrogen) were cultured in Medium 254 containing Human Melanocyte Growth Supplement (HMGS2, Cascade Biologics/Invitrogen) and propagated for a limited number of cell divisions (≤15). Primary human keratinocytes (KER; Lifeline Cell Technology) were cultured in keratinocyte media with keratinocyte growth factors (Lifeline Cell Technology) and propagated in an undifferentiated state for a limited number of cell divisions (usually ≤10).
HEK293 (ATCC) cells were cultured in DMEM containing 10% FBS and human lung fibroblasts CCD-19Lu (ATCC) cells were cultured in EMEM/FBS. Neutrophils and buffy coat cells extracted from blood were kindly provided by Dr. Reichner's lab (Brown University) and used without further culturing. Human Umbilical Vein Endothelial Cells (HUVEC) were cultured in EGM-2 (Lonza) and HL60 cells in IMDM with 20% FBS.
RNA extraction, reverse transcription, and PCRHEM and KER cDNA was obtained by reverse transcription (RT) using SuperScript III kit (Invitrogen) with total RNA extracted from cultured cells using the RNeasy Plus kit (Qiagen). PCR reactions were performed with the primers shown below:
| Gene | Primer (5′-3′) | Exon span |
|---|---|---|
| OPN1-SW | F: GTGGGGCCGTGGGATGGGCCTCAGTACCAC | e1–e5 |
| R: CCTCAGTTGGGGCCAACTTGGGTAGACGAGACAG | ||
| F: GGARGTGASSCGCATGGTGGTKGTGATG | e4–e5 | |
| R: CCTCAGTTGGGGCCAACTTGGGTAGACGAGACAG | ||
| OPN1-M/LW | F: GGARGTGASSCGCATGGTGGTKGTGATG | e4–e5 |
| R: TCATGCAGGCGATACCGAGGACACAGATGAGAC | ||
| OPN2 | F: ATGAATGGCACAGAAGGCCCTAACTTCTACGTGCC | e1–e5 |
| R: TTAGGCCGGGGCCACCTGGCTCGTC | ||
| OPN3 | F: CGGCAACAACCTGCTGGTGCTCGTCCTCTAC | e1–e4 |
| R: CCTGTCCCCATCTTTCTGTGACATCACAATGG | ||
| F: GGTCATGGTCACCTGGTCACTCCAACAATATC | e3–e4 | |
| R: CCTGTCCCCATCTT TCTGTGACATCACAATGG | ||
| OPN5 | F: TTCAGATCCCTCGTGGTTCGAGAACAGA | e1–e3 |
| R: GCAAGCCGTTCACCATCATCTCTTGCTTTTGTCAC | ||
| F: GGTGCCAACCCTACTTGCAAAATCTGCAGC | e5–e6 | |
| R: GCAGAAGACTTCCTGACTGTGGTTACGGTGTGC |
F, forward primer; R, reverse primer.
For PCR amplification of full-length OPN2 transcripts we used 30–35 cycles of the following protocol: 95°C for 30 s, 58°C for 45 s and 72°C for 2 min. In some cases we obtained a faint ˜1 kb band that was purified and reamplified using the same protocol. PCR products were sequenced directly, or cloned into a plasmid vector (TOPO-TA, Invitrogen) before sequencing. For most reactions, we used the following touchdown PCR protocol: 94°C for 5 min, 5 cycles of 94°C for 15 s and 70°C for 4 min; 5 cycles of 94°C for 15 s and 68°C for 4 min; 20 cycles of 94°C for 15 s and 66°C for 3 min; followed by 35 regular PCR amplification cycles: 94°C for 30 s, 55°C for 45 s and 72°C for 1 min 30 s. For amplification of OPN3 we used the GC-Rich Polymerase kit (Invitrogen).
Quantitative PCR was used to determine the abundance of OPN1-SW, OPN2, OPN3 and OPN5 mRNA in HEMs or KERs, using the primers below:
| Gene | Primer (5′-3′) | Exon span |
|---|---|---|
| OPN1-SW | F: TGTGCCTCTCTCCCTCATCT | e3–e4 |
| R: GGCACGTAGCAGACACAGAA | ||
| OPN2 | F: GAGTCAGCCACCACACAGAA | e4 |
| R: CATGAAGATGGGACCGAAGTTGGAG | ||
| F: CGGAGGTCAACAACGAGTCT | e3–e4 | |
| R: TCTCTGCCTTCTGTGTGGTG | ||
| OPN3 | F: CAATCCAGTGATTTATGTCTTCATGATCAGAAAG | e3–e4 |
| R: GCATTTCACTTCCAGCTGCTGGTAGGT | ||
| F: CATTGCTATGGCCATATTCTATATTCCATTCG | e2–e3 | |
| R: ATCATTAAAAAGCACATTTTGGCCAGTTTC | ||
| OPN5 | F: CTAGACGAAAGAAGAAGCTGAGACC | e2–e3 |
| R: GCGGTGACAAAAGCAAGAGA |
F, forward primer; R, reverse primer.
Primer efficiency was tested by measuring the slope of the template dose–response curve with brain cDNA. Only primers with slopes within 10% of the theoretical slope were used for amplification.
Reactions were prepared according to manufacturer's instructions using Power SYBR Green (Applied Biosystems) and cycled on a 7500 Real-Time PCR System (Applied Biosystems). Actin was used as an internal control and all reactions were run in triplicate. mRNA levels were determined independently for four different HEM and four different KER donors. mRNA levels were determined by calculating  values for each opsin.
values for each opsin.
Light stimulation of cultured cells. Light stimulation was performed using a 200 W Hg-Xe arc lamp (Newport) with converging optics and filters consisting of a dichroic mirror (260–400 nm) combined with 280 nm long pass and 400 nm short pass filters. The light source comprises ˜90% UVA (320–400 nm) and ˜10% UVB (280–320 nm) and each 10 mJ cm−2 exposure equates to 10 s of solar UVR exposure on a day with a UV index ˜10. Filtered light was directed onto a microscope stage using a liquid light guide and a TTL-driven mechanical shutter (Newport). Power was measured using a hand-held silicon detector (Newport).
HEMs and KERs cultured in 35 mm Petri dishes were exposed to 500 mJ cm−2 UVR (4.5 mW cm−2 for 111 s) in extracellular buffer containing (in mm): 125 NaCl, 5 KCl, 1.5 MgCl2, 1.5 CaCl2, 20 HEPES, 10 glucose, pH 7.4. Cells were immediately rinsed and either used for RNA extraction (t = 0 h) or returned to their growth medium and incubated at 37°C and 5% CO2 until the 4, 16 or 24 h time points.
Western Blot analysis. For OPN2 expression in KERs, HA-tagged rhodopsin in pcDNA3.1 was used for transfections of HEK293 cells using Lipofectamine 2000 (Life Technologies). KERs and transfected HEK293 cells cultured in 10 cm Petri dishes were homogenized in ice-cold buffer containing: (in mm) 300 sucrose, 35 Tris (pH 7.4), 10 EDTA, 10 EGTA and protease inhibitor cocktail (Roche Applied Science). Samples were centrifuged at 7000 g for 5 min at 4°C using an OptimaMaxx Ultracentrifuge (Beckman Coulter), followed by two additional centrifugations at 140 000 g for 1 h at 4°C to enrich the pellet with membrane proteins. Total protein content was determined using the Pierce BCA Protein Assay Kit (Thermo Scientific). Equal amounts of protein were loaded onto each lane, separated by electrophoresis on NuPAGE Bis-Tris gels (Life Technologies), and then transferred to PVDF membranes (Roche Applied Science). Membranes were blocked at room temperature for 1 h and incubated overnight at 4°C with either mouse monoclonal antirhodopsin antibody clone 1D4 (1:1000, Sigma) or rat monoclonal anti-HA antibody clone 3F10 (1:500, Roche Applied Science), followed by 1 h at RT with HRP-conjugated goat anti-mouse IgG affinity purified antibody (1:2000, Invitrogen) or HRP-conjugated goat anti-rat IgG affinity purified antibody (1:5000, Chemicon). For OPN3 we used anti-Encephalopsin (1:500, Abcam) as primary antibody and anti-Integrin α5 (1:500, Santa Cruz Biotechnology) as loading control. Secondary antibodies were obtained from Santa Cruz and diluted 1:10 000. Immunoreactive protein bands were detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and visualized with CL-XPosure Film (Thermo Scientific).
Results and discussion
To investigate the ability of human skin to act as a sensory system for ultraviolet and visible radiation, we examined the expression pattern of opsin receptors in HEMs and KERs at the mRNA level. We used reverse transcription polymerase chain reaction (RT-PCR) to identify opsin mRNA transcripts expressed in HEMs and KERs, followed by quantitative PCR (qPCR) to determine their relative expression levels.
Functional opsins require a chromophore attached to a lysine residue homologous to K296 in bovine rhodopsin (Fig. 1A). Hence, we first investigated whether mRNA fragments encompassing the equivalent lysine residue of human OPN1-SW (NM_001708.2), OPN1-M/LW (NM_000513/020061), OPN2 (NM_000539), OPN3 (NM_014322), OPN4 (NM_033282) and OPN5 (NM_181744) are present in HEMs and KERs. Using cDNA obtained through reverse transcription (RT) of mRNA extracted from cultured HEMs or KERs, we PCR-amplified bands of the expected size using primers for OPN1-SW, OPN2, OPN3 and OPN5 from both cell types (Fig. 1B). These fragments were sequenced and found to be identical to the corresponding annotated opsin sequences (Fig. S1).
Figure 1.
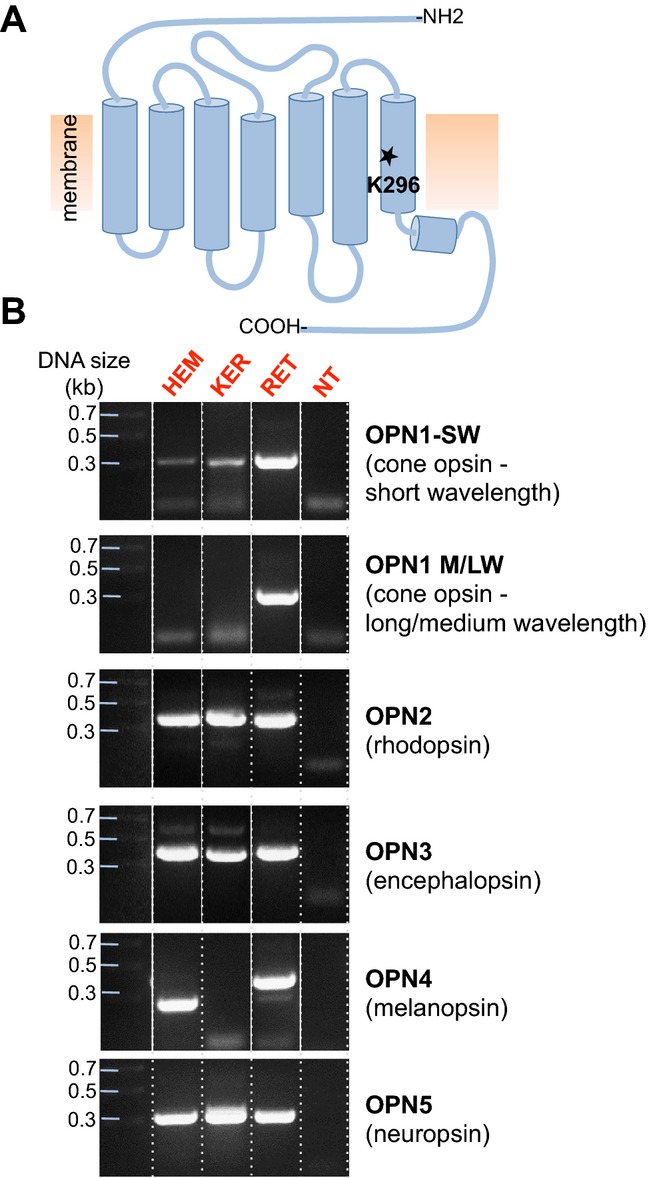
Identification of mRNA corresponding to the retinal-binding site of human opsins in HEMs and KERs. (A) Schematic representation of the topology of human opsins, seven transmembrane domain light-activated receptors that contain in the seventh transmembrane domain a conserved Lys residue (K296 in rhodopsin) required for binding retinal. (B) PCR screen of HEM, KER, retina (RET) or brain (BR) cDNA with primers encompassing the conserved lysine residue (Fig. S1) of cone opsin, short wavelength (OPN1-SW), medium and long wavelength (OPN1-M/LW), rhodopsin (OPN2), encephalopsin (OPN3), melanopsin (OPN4) and neuropsin (OPN5). DNA bands corresponding to OPN1-SW, OPN2, OPN3 and OPN5, but not OPN1-M/LW and OPN4, were amplified from HEMs and KERs. Retina cDNA was used as a positive control for OPN1-SW, OPN1-M/LW, OPN2 and OPN4, and brain cDNA for OPN3 and OPN5. NT, no template. Dotted lines separate the gel lanes, continuous white lines denote different gel lanes joined together.
Primers specific for OPN1-M/LW did not amplify any transcripts from either HEMs or KERs, but produced a band of the expected size (˜300 bp) using cDNA obtained from retinal cells (RET). Primers specific to OPN4 amplified a cDNA fragment of the expected size (˜400 bp) from retina (RET), a smaller cDNA fragment from HEMs (˜250 bp) and no fragment from KERs (Fig. 1B). Sequencing of the two fragments from RET and HEMs indicated that the amplified retina cDNA corresponded to OPN4, whereas the fragment amplified from HEMs did not. Numerous attempts using different primers and amplification conditions for OPN4 did not yield a fragment corresponding to OPN4 in either HEMs or KERs. On the basis of these results, we next attempted to amplify full-length cDNAs for OPN1-SW, OPN2, OPN3 and OPN5 from HEMs and KERs.
OPN1-SW (short–wavelength-sensitive cone opsin)
OPN1-SW (short–wavelength-sensitive cone opsin) is one of the three types of cone opsins responsible for normal color vision, having a maximal absorption in the blue-violet range. OPN1-SW cDNA consists of five exons (e1–e5), encoding a 348 amino acid protein (Fig. 2B). Primers based on the coding sequence of OPN1-SW amplified DNA fragments of the expected size (˜1 kb) from both HEM and KER cDNA (Fig. 2A); these were sequenced and found to correspond to the full-length open reading frame of OPN1-SW.
Figure 2.
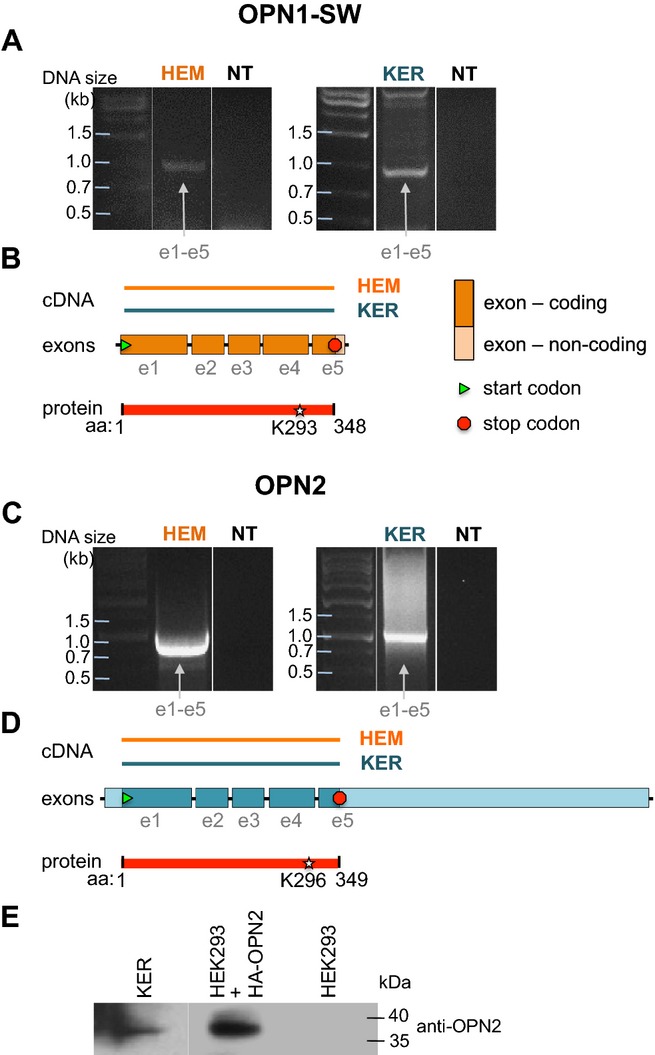
Identification of full-length OPN1-SW and OPN2 mRNA in HEMs and KERs. (A) RT-PCR using HEM or KER RNA and primers based on the first and last exon of OPN1-SW amplified a cDNA fragment (e1–e5) corresponding to the full-length coding sequence of OPN1-SW. No amplification occurred in the absence of HEM or KER cDNA (NT, no template). (B) The cDNA amplified from HEM or KER in (A) corresponds to a fragment of the coding sequence of OPN1-SW that encompasses all five exons and results in a 348 amino acids peptide containing the retinal binding residue K293. (C) RT-PCR using HEM or KER RNA and primers based on the first and last exon of OPN2 amplified a cDNA fragment (e1–e5) corresponding to the full-length coding sequence of OPN2. No amplification occurred in the absence of HEM or KER cDNA (NT, no template). (D) The cDNA amplified from HEM or KER in (C) corresponds to the coding sequence of OPN2 that encompasses all five exons and results in a 349 amino acids peptide containing the retinal binding residue K296. (E) Western blot analysis of HEM and HEK293 cell extracts probed with anti-OPN2 antibody shows a ˜ 36 kDa band in HEM and HEK293 cells expressing hemagglutinin (HA)-tagged OPN2, but not in untrasfected HEK293 cells. Representative of three independent experiments.
OPN2 (rhodopsin)
OPN2 (rhodopsin) functions in dim-light photoreception and is highly expressed in retinal rod cells. We have previously shown that OPN2 mRNA and protein are expressed in HEMs (15) and Tsutsumi and colleagues detected rhodopsin immunoreactivity in the upper layer of human epidermis, which consists primarily of keratinocytes (18). To determine whether full-length OPN2 mRNA is expressed in KERs, we performed PCR using primers based on the coding sequence of OPN2, between its 5′ start (ATG) and 3′ stop codons, and amplified a transcript of the expected size (1047 bp) from both HEMs and KERs (Fig. 2C). The sequenced cDNA fragment corresponded to the five exons of OPN2 (Fig. 2D) and encodes a full-length protein (red bar). We thus conclude that both HEMs and KERs express full-length OPN2 mRNA. As we previously showed that HEMs express full-length OPN2 protein (15), we tested if KERs also contain full-length OPN2. Western blot analysis using an anti-OPN2 antibody revealed a protein band of the expected size (˜36 kDa) in KERs, similar in size to the band recognized by an anti-HA antibody in HEK293 cells expressing HA-OPN2, and which was absent in untransfected HEKs (Fig. 1E).
OPN3 (encephalopsin, panopsin)
OPN3 (encephalopsin, panopsin) is a nonvisual opsin of unknown function that is highly expressed in brain, testes and retina (9,10). Expression analysis of human OPN3 has shown that it is an ancestral-type opsin, highly conserved across vertebrates (19). Recently, the pufferfish teleost multiple tissue opsin (TMT opsin) and mosquito OPN3 were shown to have the ability to form a green–light activated photopigment (14). OPN3 expression or function in skin has not been previously detected.
On the basis of our isolation of an OPN3 fragment containing K299 (equivalent to bovine rhodopsin K296), we sought to amplify larger fragments of OPN3 containing all four coding exons. RT-PCR using a forward primer based on exon 1 and a reverse primer based on exon 4 resulted in two bands from both HEMs and KERs: a longer transcript of ˜900 bp and a smaller transcript ≥500 bp (Fig. 3A). Sequencing identified the longer transcript as a 878 bp sequence encompassing exons 1–4 (Fig. 3B), and the shorter transcript as a 558 bp sequence containing exons 1, 3 and 4 but lacking exon 2 (320 bp) (Fig. 3C). This novel splice variant encodes a protein containing a frameshift that causes truncation after exon 1, and resulting in a 143 amino acid peptide (Fig. S2B) that is presumably nonfunctional.
Figure 3.
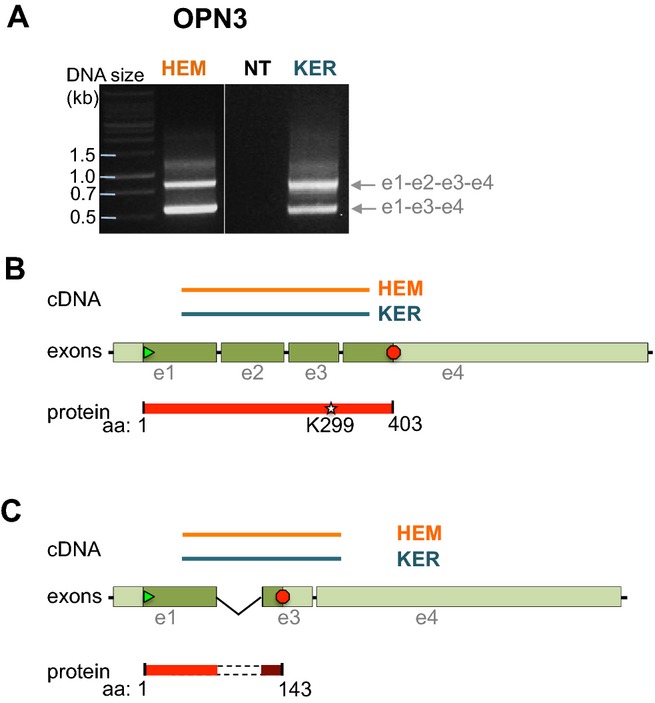
Identification of full length and truncated splice variant of OPN3 in HEMs and KERs. (A) RT-PCR using HEM or KER RNA and primers based on exon 1 and exon 4 of OPN3 amplified two bands that corresponded to exons 1–4 (e1-e2-e3-e4, upper band) and to a splice variant missing exon 2 (e1-e3-e4, lower band) (Fig. S2A). (B) The cDNA amplified in the upper band in (A) corresponds to a fragment of the coding sequence of OPN3 that encompasses all four exons and results in a 403 amino acids peptide containing the retinal binding residue K299. (C) The cDNA amplified in the lower band in (A) corresponds to exons 1, 3 and 4 of OPN4, coding for a 143 amino acid peptide that does not contain the K299 residue (Fig. S2B).
OPN5 (neuropsin)
OPN5 (neuropsin) is expressed in human brain, testes and retina (12). Recent studies suggest that OPN5 is expressed in mammalian skin (11,20), where it functions as a G protein-coupled UVR-sensitive receptor in humans (11) and birds (13,21). Together, these findings make OPN5 an attractive candidate for an epidermal UVR-sensitive photoreceptor.
We sought to amplify the full coding sequence of OPN5 (containing seven exons) from HEMs and KERs (Fig. 4A,B). Repeated attempts to amplify OPN5 with different primer pairs based on exon 1 and exon 7 did not detectably amplify transcripts from either HEM or KER cDNA, but did amplify bands of expected size using brain cDNA (data not shown). Using a forward primer based on the 5′ untranslated region (5′-UTR) and a reverse primer based on exon 3, we amplified two transcripts (319 and 408 bp) from both HEMs and KERs (Fig. 4C). Sequencing identified the shorter of these as the expected 319 bp transcript encompassing exons 1 through 3. The longer transcript corresponded to a sequence containing a novel 89 bp exon between exon 1 and 2 (exon 1a), resulting in a 408 bp splice variant (Fig. 4D), which truncates in exon 2 due to a frameshift introduced by exon 1a. Interestingly, the relative abundance of the two transcripts varies in HEMs vs KERs (Fig. 4A), suggesting that this short, and presumably nonfunctional, variant might function to regulate OPN5 expression levels.
Figure 4.
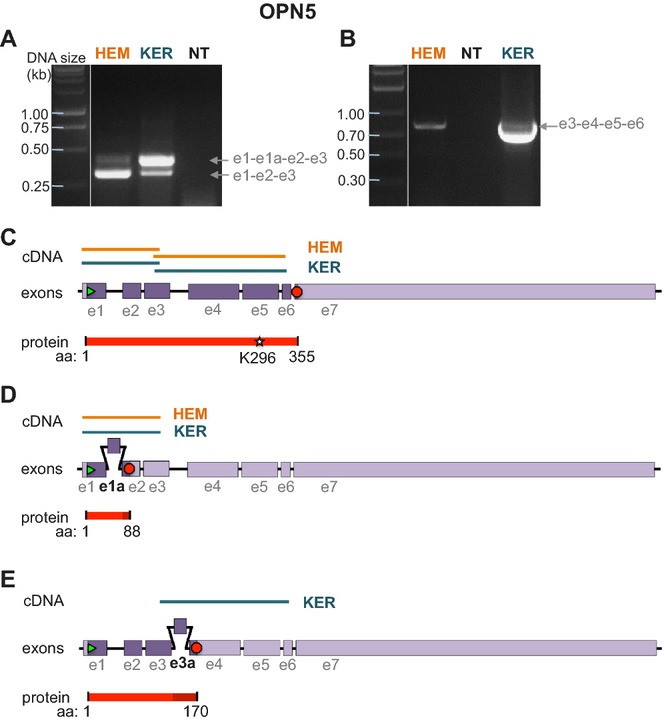
Identification of full length and two truncated splice variants of OPN5 in HEMs and KERs. (A) RT-PCR using HEM or KER RNA and primers based on exon 1 and 3 of OPN5 amplified two bands that corresponded to exons 1–3 (e1-e2-e3, lower band) and to a splice variant containing an additional exon e1a (e1-e1a-e2-e3, upper band). (B) RT-PCR using HEM or KER RNA and primers based on exon 3 and 6 of OPN5 amplified a band that corresponded to exons 3–6 (e3-e4-e5-e6). (C) The lower cDNA bands amplified in (A) and the bands amplified in (B) correspond to exons 1–6 of OPN5, coding for a 355 amino acid peptide containing the retinal binding site K296. (D) The upper cDNA band amplified in (A) contains an additional 89 bp exon (e1a) between exons 1 and 2. The coding sequence of this splice variant translates into an 88 amino acid peptide (Fig. S3A). (E) The cDNA band amplified in B from KERs comprises, in addition to the fragment shown in (C), a splice variant containing an additional 76 bp exon (e3a) between exons 3 and 4 and results in a 170 amino acid peptide (Fig. S3B).
Subsequent PCR reactions using primers designed to amplify OPN5 exons 3 through 6 produced a ˜750 bp band using KER cDNA (Fig. 4A, right). Sequencing analysis identified two distinct transcripts: the first corresponded to the expected 612 bp sequence encompassing exons 3–6, whereas the second corresponded to a 681 bp transcript containing a novel 69 bp exon between exon 3 and 4 (exon 3a), resulting in a variant truncated due to a frameshift introduced by exon 3a (Fig. 4E).
We next assessed the relative expression levels of OPN1-SW, OPN2, OPN3 and OPN5 mRNA in HEMs and KERs using qPCR. We used primer pairs designed to assess total mRNA levels, without discriminating between splice variants. When comparing the levels of OPN1-SW, OPN2, OPN3 and OPN5 to actin levels in HEMs and KERs, we found that OPN2 is expressed at higher levels in HEMs, whereas OPN5 is expressed at higher levels in KERs (Fig. 5). We measured opsin levels in at least three biological replicates. We observed significant variability (10–100-fold) between HEM and KER lines from different individuals, but did not detect any correlation between the expression levels of different opsins and the pigmentation phenotype of the donor. Surprisingly, we found that OPN3 is expressed in both HEMs and KERs at significantly higher levels than OPN1-SW or OPN5 (Fig. 5), suggesting that OPN3 might have an important functional role in human skin. We also found that OPN2, which we previously found to be involved in UVR phototransduction in melanocytes (15), is expressed in HEMs at levels comparable to OPN3 and significantly higher than OPN1-SW or OPN5 (Fig. 5).
Figure 5.
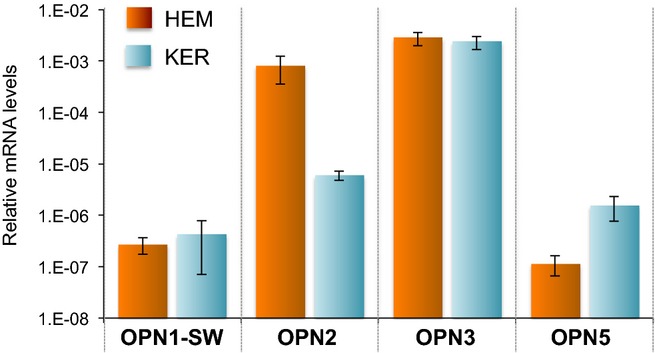
Relative expression levels of opsins mRNA in HEMs and KERs. qPCR analysis of the mRNA expression level of OPN1-SW, OPN2, OPN3 and OPN5 in HEM and KER, relative to actin. Bars represent the average of ≥4 experiments; ± SEM.
Because human epidermal skin is physiologically exposed to solar UVR, we tested if UVR stimulation leads to changes in the opsin expression levels in these cells. We exposed melanocytes and keratinocytes to 500 mJ cm−2 UVR (equivalent to ˜10 min of bright solar UVR) to minimize cell death and oxidative damage, and collected RNA at 0, 4, 16 and 24 h after the treatment. The cDNA obtained by RT was used to quantify the levels of each opsin by qPCR. We found that none of the opsins expressed changed their expression levels at any of the time points measured, in neither HEMs nor KERs (Fig. S4). In addition, we could not detect OPN4 at later time points after exposure (data not shown), suggesting that its expression is not UVR dependent. These results suggest that opsins are not transcriptionally regulated by UVR exposure, but might contribute to UVR signaling pathways in skin. Alternatively, their activation mechanism and function in skin could be light-independent.
As the levels of OPN3 mRNA measured in Fig. 5 consisted of both full-length coding sequence as well as a truncated isoform, we deemed it important to determine their relative expression levels. We designed primers based on exon 2 and 3 to detect full-length OPN3 exclusively, and primers based on exons 3 and 4 to detect both the full-length and truncated isoforms (Fig. S2A). We calculated the expression level of the truncated isoform by subtracting the expression of the full-length measured with the first primer pair from the expression of both isoforms measured with the second primer pair, and found that the ratio of full length to truncated OPN3 mRNA was approximately 1:1 both in HEMs and KERs (Fig. 6A), suggesting that at least one-half of the OPN3 mRNA detected in these cells (Fig. 5) encodes the full-length protein (Fig. S2B).
Figure 6.
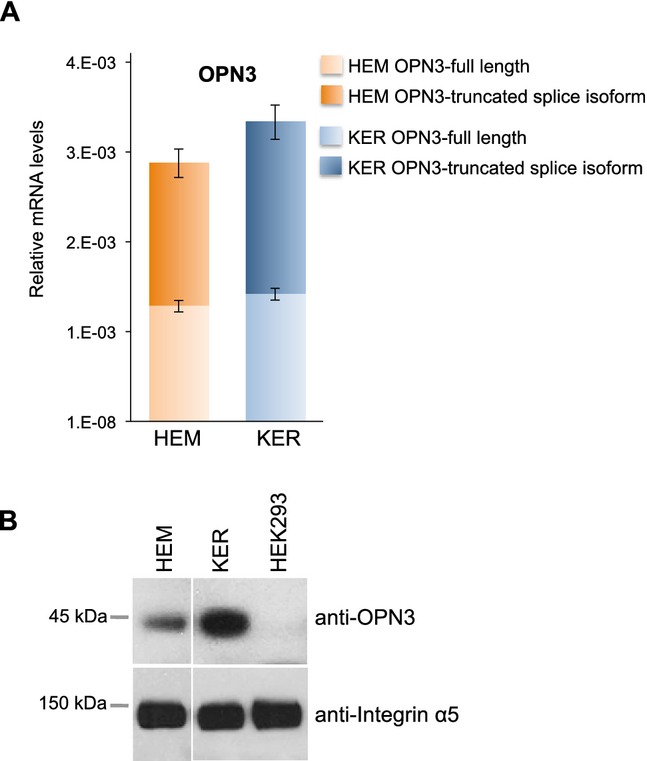
Relative OPN3 expression in HEMs and KERs. (A) Quantitative PCR analysis of the mRNA expression of full length (light color) and truncated splice variant (dark color) of OPN3 in HEM and KER, using the primers shown in Fig. S2A and calculated relative to actin levels. Bars represent the average of three experiments; ±SEM. (B) Western Blot analysis of OPN3 protein expression in HEM, KER and HEK293. A 45 kDa band was detected by anti-OPN3 antibodies in HEM and KER, but not in HEK293 cells. Integrin α5 was used as a positive control.
To test whether OPN3 mRNA is translated into a protein of the predicted size in HEMs and KERs, we performed western blot analysis using an anti-OPN3 antibody that recognizes the region between amino acids 181 and 210 of OPN3, present only in the full-length isoform. Anti-OPN3 detected a band of ˜45 kDa both in HEMs and KERs, but not in HEK293 cells (Fig. 6B), suggesting that OPN3 protein is expressed in epidermal skin cells.
The detection of mRNA for four different opsins in the two main types of human epidermal cells made us wonder whether other cells express opsin mRNA. We first tested cDNA obtained by RT from HEK293 cells with the primers used in Fig. 1. Unexpectedly, we amplified bands that, when sequenced, were found to correspond to each of the opsins tested (Fig. S5A). However, because no protein product was detected for OPN2 (Fig. 2E) or OPN3 (Fig. 6B) in HEK293 cells, the significance of opsin mRNA expression in these cells remains unclear. As HEK293 cells are a polyploidy cell line that might exhibit aberrant mRNA expression, we tested human lung fibroblasts and found that the same primers also amplified from lung fibroblasts cDNA bands that were sequenced and found to correspond to the respective opsins (Fig. S5B).
Intrigued by this finding, we tested by qPCR the levels of a representative opsin, OPN3, in cell lines and primary cells derived from diverse tissues: human lung fibroblasts, primary human neutrophils, buffy coat cells, the leukemia cell line HL60 and human umbilical vein endothelial cells (HUVEC). Compared to HEMs and KERs, OPN3 mRNA was expressed at levels two- to 40-fold lower in other tested cells (Fig. S6A). Interestingly, western blot with anti-OPN3 antibodies detected a band in HEMs and KERs (as also shown in Fig. 6B), in fibroblasts, and in buffy coat cells, a weak band in HUVEC and no band in neutrophils or HL60 cells (Fig. S6B). The discrepancy between mRNA and protein expression could be due to specific protein degradation pathways used by different cells. Nevertheless, in addition to expression in cells physiologically exposed to light (HEM and KER), the presence of opsins in primary cells that are not exposed to light raises the question of their activation mechanism and functionality in these cells.
In conclusion, we find that multiple opsin receptors are expressed in human epidermal cells. We demonstrate that OPN1-SW, a cone opsin activated by blue-violet light, is expressed in epidermal skin. In addition, we confirm our previous observation that OPN2 is present in HEMs (15) and show that it is also expressed in KERs. Our findings also agree with previous reports suggesting that OPN5 is expressed in human epidermis (11) and, in addition, identify two novel OPN5 splice variants. The truncated proteins produced by alternative splicing of OPN3 and OPN5 may serve to regulate the expression levels and function of these opsins in skin. We found that OPN3 mRNA and protein is expressed at higher levels that the other opsins in the two major human epidermal cell types and identified a novel splice variant. The expression of opsins in skin suggests that they may function as epidermal photoreceptors, prompting further studies that might uncover their physiological roles. The expression of opsins in other cell types suggests that in the absence of light they could be activated by alternative mechanisms and serve other functions than phototransduction.
Acknowledgments
We thank members of the Oancea lab for technical assistance and critical reading of the manuscript. We also thank Dr. Anita Zimmerman for critical discussions and reading of the manuscript and Dr. Jonathan Reichner for providing the blood immune cells, as well as HUVEC and HL60 cell line. This work was supported by Grants from Brown University to E.O. Additional support: National Institute of General Medical Sciences of the National Institutes of Health Award No. R25GM083270 to K.D.H., and NSF Graduate Research Fellowship Award No. DGE-1058262 to K.D.H., grant from The Suna and Inac Kirac Foundation to R.N.O., Natural Sciences and Engineering Research Council (NSERC) to N.L.W. and NIH Predoctoral Fellowship to J.A.N.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Alignment of the human opsins cDNA region surrounding the conserved retinal-binding lysine residue.
Figure S2. Alignment of OPN3 full length and splice isoform.
Figure S3. Alignment of OPN5 full length and splice isoforms.
Figure S4. Opsin expression levels in HEMs and KERs stimulated with UVR. (A) qPCR analysis of OPN1-SW, OPN2, OPN3 and OPN5 in HEMs exposed to 500 mJ cm−2 UVR (see methods) and collected at 0, 4, 6 and 24 h after exposure. Bars represent the average of ≥3 experiments; ±SEM. (B) qPCR analysis of OPN1-SW, OPN2, OPN3 and OPN5 in KERs exposed to 500 mJ cm−2 UVR (see methods) and collected at 0, 4, 6 and 24 h after exposure. Bars represent the average of ≥2 experiments; ±SEM.
Figure S5. RT-PCR amplification with primers encompassing the region surrounding the retinal-binding site of the respective human opsins in HEK293 cells and human lung fibroblasts.
Figure S6. qPCR and western blot analysis of OPN3 expression in different human cells.
References
- 1.Yau KW, Hardie RC. Phototransduction motifs and variations. Cell. 2009;139(2):246–264. doi: 10.1016/j.cell.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nickle B, Robinson PR. The opsins of the vertebrate retina: Insights from structural, biochemical, and evolutionary studies. Cell. Mol. Life Sci. 2007;64(22):2917–2932. doi: 10.1007/s00018-007-7253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palczewski K. G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumbalasiri T, Provencio I. Melanopsin and other novel mammalian opsins. Exp. Eye Res. 2005;81(4):368–375. doi: 10.1016/j.exer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: Architecture, projections, and intrinsic photosensitivity. Science. 2002;295(5557):1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Provencio I, Rodriguez IR, Jiang G, Hayes WP, Moreira EF, Rollag MD. A novel human opsin in the inner retina. J. Neurosci. 2000;20(2):600–605. doi: 10.1523/JNEUROSCI.20-02-00600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panda S, Sato TK, Castrucci AM, Rollag MD, DeGrip WJ, Hogenesch JB, Provencio I, Kay SA. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science. 2002;298(5601):2213–2216. doi: 10.1126/science.1076848. [DOI] [PubMed] [Google Scholar]
- 8.Ruby NF, Brennan TJ, Xie X, Cao V, Franken P, Heller HC, O'Hara BF. Role of melanopsin in circadian responses to light. Science. 2002;298(5601):2211–2213. doi: 10.1126/science.1076701. [DOI] [PubMed] [Google Scholar]
- 9.Halford S, Freedman MS, Bellingham J, Inglis SL, Poopalasundaram S, Soni BG, Foster RG, Hunt DM. Characterization of a novel human opsin gene with wide tissue expression and identification of embedded and flanking genes on chromosome 1q43. Genomics. 2001;72(2):203–208. doi: 10.1006/geno.2001.6469. [DOI] [PubMed] [Google Scholar]
- 10.Blackshaw S, Snyder SH. Encephalopsin: A novel mammalian extraretinal opsin discretely localized in the brain. J. Neurosci. 1999;19(10):3681–3690. doi: 10.1523/JNEUROSCI.19-10-03681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kojima D, Mori S, Torii M, Wada A, Morishita R, Fukada Y. UV-sensitive photoreceptor protein OPN5 in humans and mice. PLoS ONE. 2011;6(10):e26388. doi: 10.1371/journal.pone.0026388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarttelin EE, Bellingham J, Hankins MW, Foster RG, Lucas RJ. Neuropsin (Opn5): A novel opsin identified in mammalian neural tissue. FEBS Lett. 2003;554(3):410–416. doi: 10.1016/s0014-5793(03)01212-2. [DOI] [PubMed] [Google Scholar]
- 13.Yamashita T, Ohuchi H, Tomonari S, Ikeda K, Sakai K, Shichida Y. Opn5 is a UV-sensitive bistable pigment that couples with Gi subtype of G protein. Proc. Natl Acad. Sci. USA. 2010;107(51):22084–22089. doi: 10.1073/pnas.1012498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koyanagi M, Takada E, Nagata T, Tsukamoto H, Terakita A. Homologs of vertebrate Opn3 potentially serve as a light sensor in nonphotoreceptive tissue. Proc. Natl Acad. Sci. USA. 2013;110(13):4998–5003. doi: 10.1073/pnas.1219416110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wicks NL, Chan JW, Najera JA, Ciriello JM, Oancea E. UVA phototransduction drives early melanin synthesis in human melanocytes. Curr. Biol. 2011;21(22):1906–1911. doi: 10.1016/j.cub.2011.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellono NW, Kammel LG, Zimmerman AL, Oancea E. UV light phototransduction activates transient receptor potential A1 ion channels in human melanocytes. Proc. Natl Acad. Sci. USA. 2013;110(6):2383–2388. doi: 10.1073/pnas.1215555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellono NW, Najera JA, Oancea E. UV light activates a Galphaq/11-coupled phototransduction pathway in human melanocytes. J. Gen. Physiol. 2014;143(2):203–214. doi: 10.1085/jgp.201311094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsutsumi M, Ikeyama K, Denda S, Nakanishi J, Fuziwara S, Aoki H, Denda M. Expressions of rod and cone photoreceptor-like proteins in human epidermis. Exp. Dermatol. 2009;18(6):567–570. doi: 10.1111/j.1600-0625.2009.00851.x. [DOI] [PubMed] [Google Scholar]
- 19.Fischer RM, Fontinha BM, Kirchmaier S, Steger J, Bloch S, Inoue D, Panda S, Rumpel S, Tessmar-Raible K. Co-expression of VAL- and TMT-opsins uncovers ancient photosensory interneurons and motorneurons in the vertebrate brain. PLoS Biol. 2013;11(6):e1001585. doi: 10.1371/journal.pbio.1001585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denda M, Fuziwara S. Visible radiation affects epidermal permeability barrier recovery: Selective effects of red and blue light. J. Invest. Dermatol. 2008;128(5):1335–1336. doi: 10.1038/sj.jid.5701168. [DOI] [PubMed] [Google Scholar]
- 21.Nakane Y, Ikegami K, Ono H, Yamamoto N, Yoshida S, Hirunagi K, Ebihara S, Kubo Y, Yoshimura T. A mammalian neural tissue opsin (Opsin 5) is a deep brain photoreceptor in birds. Proc. Natl Acad. Sci. USA. 2010;107(34):15264–15268. doi: 10.1073/pnas.1006393107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the human opsins cDNA region surrounding the conserved retinal-binding lysine residue.
Figure S2. Alignment of OPN3 full length and splice isoform.
Figure S3. Alignment of OPN5 full length and splice isoforms.
Figure S4. Opsin expression levels in HEMs and KERs stimulated with UVR. (A) qPCR analysis of OPN1-SW, OPN2, OPN3 and OPN5 in HEMs exposed to 500 mJ cm−2 UVR (see methods) and collected at 0, 4, 6 and 24 h after exposure. Bars represent the average of ≥3 experiments; ±SEM. (B) qPCR analysis of OPN1-SW, OPN2, OPN3 and OPN5 in KERs exposed to 500 mJ cm−2 UVR (see methods) and collected at 0, 4, 6 and 24 h after exposure. Bars represent the average of ≥2 experiments; ±SEM.
Figure S5. RT-PCR amplification with primers encompassing the region surrounding the retinal-binding site of the respective human opsins in HEK293 cells and human lung fibroblasts.
Figure S6. qPCR and western blot analysis of OPN3 expression in different human cells.


