Key Points
Dnmt3a-null hematopoietic stem cells (HSCs) cannot sustain long-term hematopoiesis.
Cooperating c-Kit mutations drive leukemic transformation of Dnmt3a-null HSCs.
Abstract
Genome sequencing studies of patient samples have implicated the involvement of various components of the epigenetic machinery in myeloid diseases, including the de novo DNA methyltransferase DNMT3A. We have recently shown that Dnmt3a is essential for hematopoietic stem cell differentiation. Here, we investigated the effect of loss of Dnmt3a on hematopoietic transformation by forcing the normally quiescent hematopoietic stem cells to divide in vivo. Mice transplanted with Dnmt3a-null bone marrow in the absence of wild-type support cells succumbed to bone marrow failure (median survival, 328 days) characteristic of myelodysplastic syndromes with symptoms including anemia, neutropenia, bone marrow hypercellularity, and splenomegaly with myeloid infiltration. Two out of 25 mice developed myeloid leukemia with >20% blasts in the blood and bone marrow. Four out of 25 primary mice succumbed to myeloproliferative disorders, some of which progressed to secondary leukemia after long latency. Exome sequencing identified cooperating c-Kit mutations found only in the leukemic samples. Ectopic introduction of c-Kit variants into a Dnmt3a-deficient background produced acute leukemia with a short latency (median survival, 67 days). Our data highlight crucial roles of Dnmt3a in normal and malignant hematopoiesis and suggest that a major role for this enzyme is to facilitate developmental progression of progenitor cells at multiple decision checkpoints.
Introduction
Hematopoietic stem cell (HSC) fate decisions are controlled by signaling pathways, cues from the niche, and the actions of cell-autonomous regulators such as transcription factors, but they are now also recognized to be influenced by a significant epigenetic component. DNA methylation is one of the major epigenetic modifications in the vertebrate genome and is important for development, stem cell differentiation, and oncogenesis.1-3 DNA methylation is catalyzed by the DNA methyltransferase enzymes Dnmt1, Dnmt3a, and Dnmt3b.4-6 Genome sequencing studies of myeloid malignancies have identified recurrent DNMT3A somatic mutations in approximately 22%,7,8 10%,9,10 and 8%11,12 of patients with acute myeloid leukemia (AML), myelodysplastic syndromes (MDS), and myeloproliferative neoplasms (MPN), respectively, and are associated with poor prognosis.13 The most prevalent DNMT3A mutation is an R882H variant that produces a protein that acts as a dominant negative.14,15 DNMT3A nonsense and frameshift mutations are all predicted to result in truncated proteins that eliminate or shorten the methyltransferase domain or are associated with nonsense-mediated decay, suggesting loss of function.8
We had previously studied the role of Dnmt3a in HSC function.16,17 Inducible conditional knockout mice were generated by crossing Dnmt3afl/fl mice18 with the Mx1-CRE driver, with deletion of floxed Dnmt3a alleles induced by sequential injections of polyinosinic-polycytidylic acid (pIpC; henceforth referred to as Dnmt3a-KO). Serial transfer of Dnmt3a-KO HSCs with fresh wild-type (WT) whole bone marrow (WBM) competitor revealed a decline in the differentiated cell output from Dnmt3a-KO over multiple rounds of transplantation on a per-HSC basis.16 Although the striking accumulation of phenotypically defined HSCs in the bone marrow of Dnmt3a-KO HSC recipient mice was reminiscent of a myeloid abnormality, these mice did not develop overt disease even when aged to over 14 months posttransplant.16
In these competitive transplants, the presence of WT WBM may have suppressed malignant transformation of the mutant HSCs. Dnmt3a-null HSCs were less proliferative than counterpart control HSCs,16 suggesting that the cellular turnover threshold necessary to generate additional genetic and/or epigenetic lesions required for leukemogenesis was not achieved. To further understand the contribution of Dnmt3a loss of function in hematopoiesis, we performed noncompetitive transplantation of Dnmt3a-KO bone marrow. This forces the mutant HSCs to divide in vivo to regenerate the hematopoietic system following lethal irradiation and should uncover any predispositions to transformation.
Methods
Mice and transplantation
Animal procedures were approved by the Institutional Animal Care and Use Committee and conducted in accordance with Washington University institutional guidelines. All mice were C57Bl/6 background, distinguished by CD45.1 or CD45.2 alleles. Dnmt3afl/fl mice were originally obtained from the Beaudet laboratory at Baylor College of Medicine (with the consent of En Li) and crossed to Mx1-CRE mice. Deletion of floxed alleles was mediated by 6 intraperitoneal injections (300 μg per mouse) of pIpC (Sigma) in phosphate-buffered saline every other day. For primary noncompetitive transplantation, WBM was harvested from donor mice 4 weeks after the last pIpC injection. Recipient mice were transplanted with 1 × 106 unfractionated WBM cells by retro-orbital injection following a split dose of 10.5 Gy irradiation. For secondary transplantation, 1 × 106 WBM or spleen cells from primary diseased mice were transplanted into sublethally irradiated (6.0 Gy) mice. Peripheral blood counts were performed with a Hemavet 950 (Drew Scientific). Peripheral blood smears and bone marrow and spleen cytospins were stained with the Hema 3 stat pack (Fisher Scientific) and images captured with a Nikon Eclipse E200 microscope equipped with an Infinity 2 color camera (Lumenera) controlled by Infinity Capture software (Lumenera).
Cell purification and flow cytometry
Antibody staining was performed as previously described.19 The following gating strategies were used: HSCs (CD150+ CD48− Lineage− Sca-1+ c-Kit+/Flk2− CD34− Lineage− Sca-1+ c-Kit+), common myeloid progenitors (CMPs) (Lineage− Il7rα− Sca-1low c-Kit+ CD34+ FCγr−), granulocyte-monocyte progenitors (GMPs) (Lineage− Il7rα− Sca-1low c-Kit+ CD34+ FCγr+), megakaryocyte-erythroid progenitors (MEPs) (Lineage− Il7rα− Sca-1low c-Kit+ CD34− FCγr−). Lineage marker cocktail consisted of Gr-1, Mac-1, B220, Ter119, CD4, and CD8. The following antibody (clones) were used (eBioscience or BioLegend): Gr-1 (RB6-8C5), Mac-1 (M1/70), B220 (RA3-6B2), Ter119 (TER119), CD4 (GK1.5), CD8 (53-6.7), Sca-1 (D7), c-Kit (2B8), CD34 (RAM34), Flk2 (A2F10.1), CD150 (TC15-12F12.2), CD48 (HM48-1), CD45.1 (A20), CD45.2 (104), CD71 (R17217), and FcεR1 (MAR-1). Proliferation analysis was performed with the FITC Mouse Anti-Human Ki-67 Set (BD Pharmingen). Apoptosis analysis was performed with the Annexin V Apoptosis Detection Kit APC (eBioscience). Cell sorting and analysis was performed at the Siteman Cancer Center flow cytometry core and the Department of Pathology and Immunology flow cytometry core.
Methocult serial replating
One hundred HSCs were sorted directly into each well of 6-well plates containing Methocult M3434 medium (Stem Cell Technologies) and cultured in vitro at 37°C. Colony-forming units (CFUs) were scored after 7 days, then cells were collected, pooled, and replated at a density of 5000 cells per well.
Plasmids and viral transduction
Mouse c-Kit and c-KitD814V complementary DNAs (cDNAs) were a kind gift of Dr Michael Tomasson (Washington University in St. Louis). The c-KitV750M variant was generated with the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent). All c-Kit variants were subcloned into the HIV-MND-IRES-GFP lentiviral vector as previously described.20 For lentiviral production, 293T cells were cotransfected with the packaging plasmids pMD.G, pRSV-Rev, and PMDLg plus the respective HIV-MND plasmid. Viral supernatant concentrated by centrifugation at 76 000g for 1.5 hours at 4°C. For lentiviral transduction, hematopoietic progenitors were enriched using CD117 microbeads (Miltenyi Biotec). The positive cell fraction was adjusted to 5 × 105 cells per mL in Stempro34 medium (Gibco) supplemented with l-glutamine (2 mM), murine stem cell factor (100 ng/mL), murine thrombopoietin (100 ng/mL), murine Flt3L (50 ng/mL), and murine interleukin-3 (5 ng/mL), and polybrene (4 μg/mL; Sigma), spin-infected with lentivirus at 250g for 2 hours, and transplanted into lethally irradiated mice (100 000 cells per mouse).
DNMT3A cDNA (Open Biosystems) was subcloned into MSCV-IRES-GFP (MIG) vector using Gateway recombination. MIG empty vector was used as a control in retroviral transduction experiments. The DNMT3AR882H variant was generated by site-directed mutagenesis as above. Retroviruses were packaged by cotransfection with pCL-Eco into 293T cells. For retroviral transduction, donor mice were treated with 5-fluorouracil (5-FU; 150 mg/kg; American Pharmaceutical Partners, Schaumburg, IL) 6 days prior to harvest. c-Kit+ cells were selected, spin-infected, and transplanted as above.
32D cell proliferation and western blot
32D cells transduced with lentiviruses were washed with Dulbecco’s modified Eagle’s medium (DMEM) to remove interleukin-3 (IL-3). A total of 50 000 cells were then plated into 24-well plates with DMEM containing 10% fetal bovine serum and 1× penicillin/streptomycin in the presence of either IL-3 or IL-3 and stem cell factor (SCF). Viable cells were counted using Cellometer (Nexcelom Biosciences).
For western blot, transduced 32D cells that were grown until the midexponential phase were washed with DMEM to remove IL-3 and SCF. These cells were resuspended with DMEM containing 10% fetal bovine serum in the absence of IL-3 and SCF for 12 hours. Using 20 µg of whole-cell extracts, western blot was performed to detect phosphorylated or nonphosphorylated Jnk1 and Jnk2 proteins.21
Quantitative real-time PCR
RNA was isolated using the RNAqueous kit (Ambion) and reverse transcribed with the SuperScript VILO kit (Life Technologies). cDNA input was standardized and real-time polymerase chain reaction (PCR) was performed with TaqMan master Mix (Applied Biosystems), 18 s-rRNA probe (VIC-MGB; Applied Biosystems), and a gene-specific probe (FAM-MGB; Applied Biosystems) on a StepOnePlus Real-Time PCR System (Life Technologies). Samples were normalized to 18S and fold change determined by the ∆∆Ct method.
Library construction, capture, and exome sequencing
Genomic DNA was prepared using a PureLink Genomic DNA Mini Kit (Invitrogen). Library construction, capture, and exome sequencing was performed by The Genome Institute at Washington University in St. Louis. Illumina paired-end small-insert multiplexed libraries were constructed according to the manufacturer’s recommendations (Illumina) with the following modifications: (1) 500 ng of native genomic DNA was fragmented using a Covaris E220 DNA Sonicator (Covaris) to range in size between 100 and 400 bp, (2) Illumina adapter-ligated library fragments were amplified in four 50-μL PCR reactions for 18 cycles, (3) solid-phase reversible immobilization bead cleanup was used for enzymatic purification throughout the library process, as well as final library size selection targeting 300-bp to 500-bp fragments. For capture hybridization, 10 indexed libraries were pooled prior to hybridization. Hybridization was performed with the Sure Select Mouse All Exon kit (Agilent). The capture target is 51 Mb. This pool of samples was sequenced on 2 lanes of Illumina HiSequation 2000 (Illumina). Sequence data were aligned to mouse reference sequence build mm9 using bwa version 0.5.9.22 Single-nucleotide variants were detected using the union of 3 callers: samtools version r963,23 VarScan 2.2.6, and Strelka v0.4.6.2.24 Indels were detected using the union of 4 callers: gatk-somatic-indel v5336,25 pindel v0.5,26 VarScan 2.2.6, and Strelka v0.4.6.2.24 Exome sequences are available at the National Center for Biotechnology Information’s sequence read archive under study accession SRP047158.
Statistics
Student t test and 1-way analysis of variance were used for statistical comparisons where appropriate. Significance is indicated on the figures using the following convention: *P < .05, **P < .01, and ***P < .001. Error bars on all graphs represent the standard error of the mean (SEM) unless indicated otherwise.
Results
Loss of Dnmt3a in HSCs leads to enhanced serial replating capacity
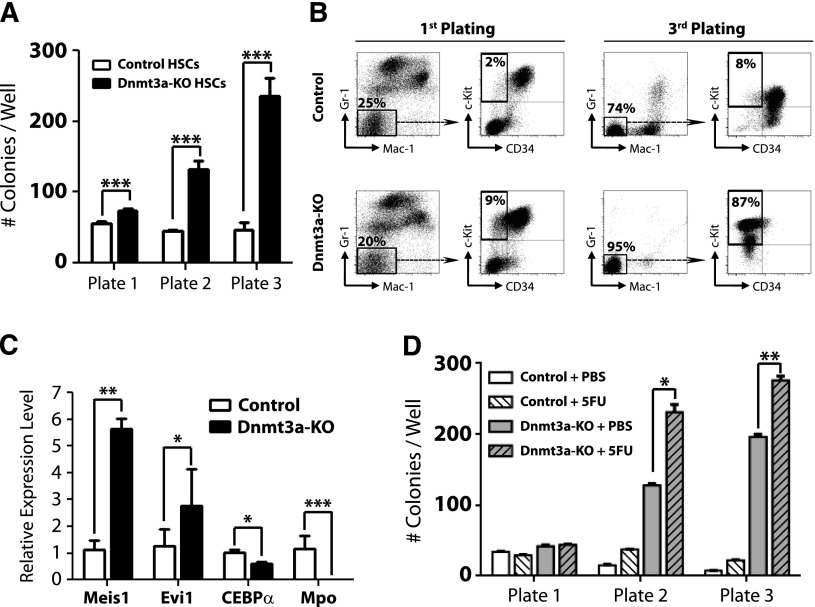
To further examine the kinetics of HSC regulation by Dnmt3a, we performed serial replating CFU assays. Four weeks after pIpC treatment, 100 control (Mx1-CRE:Dnmt3a+/+) or Dnmt3a-KO HSCs (Lineage− c-Kit+ Sca-1+ CD48− CD150+) were plated and scored for CFU after 7 days. A total of 5000 cells were then replated every 7 days. Dnmt3a-KO HSCs generated significantly more colonies after each successive round (Figure 1A), similar to their self-renewal advantage in vivo, which became more dramatic after serial transplantation. Fluorescence-activated cell sorter (FACS) analysis revealed that Dnmt3a-KO colonies possessed a more immature phenotype (Mac-1− Gr-1− c-Kit+ CD34−) after the third plating (Figure 1B). Third-plate Dnmt3a-KO colonies showed increased expression of the self-renewal regulators Meis1 and Evi1 and reduced expression of myeloid-specific factors such as CEBPα and Mpo (Figure 1C). These data suggested that cell turnover is required to enforce the molecular changes resulting from loss of Dnmt3a. In support of this, HSCs from Dnmt3a-KO mice given 2 prior injections of 5-FU 1 month apart (to force HSCs to divide in vivo) showed increased CFU ability compared with control Dnmt3a-KO mice (Figure 1D).
Figure 1.
Loss of Dnmt3a in HSCs leads to enhanced serial replating capacity. (A) CFU assay of HSCs. A total of 100 control or Dnmt3a-KO HSCs were plated per well 4 weeks after the final pIpC injection. CFUs were scored every 7 days, and 5000 cells were replated each round in triplicate. Mean ± SEM values are shown from 3 independent experiments. (B) FACS analysis of CFU plates. Although control and Dnmt3a-KO colonies showed similar myeloid development after the first plating, by the third plating, most Dnmt3a-KO colonies were negative for mature myeloid markers (Gr-1− Mac-1−) and showed an immature c-Kit+ CD34− phenotype (red squares). (C) Real-time PCR at the third plating showed Dnmt3a-KO cells were characterized by increased expression of the self-renewal regulators Meis1 and Evi1 and decreased expression of the myeloid differentiation factors CEBPα and Mpo. (D) Serial replating of control or Dnmt3a-KO HSCs from mice previously injected with phosphate-buffered saline (PBS) or 5FU. Mean ± SEM values are shown from 3 independent experiments. *P < .05, **P < .01, ***P < .001.
Enforced differentiation of Dnmt3a-KO HSCs leads to bone marrow failure resembling MDS
As it appeared cell turnover was necessary to manifest the Dnmt3a-KO HSC phenotype, we hypothesized that transplantation of Dnmt3a-KO WBM in the absence of WT support WBM would elucidate any predisposition to transformation. We performed noncompetitive transplantation of Dnmt3a-KO WBM 4 weeks following pIpC treatment. Two transplant cohorts were established independently using different donor mice. Control mice in the first cohort were Mx1-CRE-:Dnmt3afl/fl and in the second were Mx1-CRE+:Dnmt3a+/+ to control for interferon-mediated effects, nonspecific deletion of floxed Dnmt3a alleles, and long-term CRE expression in HSCs. As there were no significant differences between the cohorts (supplemental Figure 1, available on the Blood Web site), the final data are compiled from both transplants.
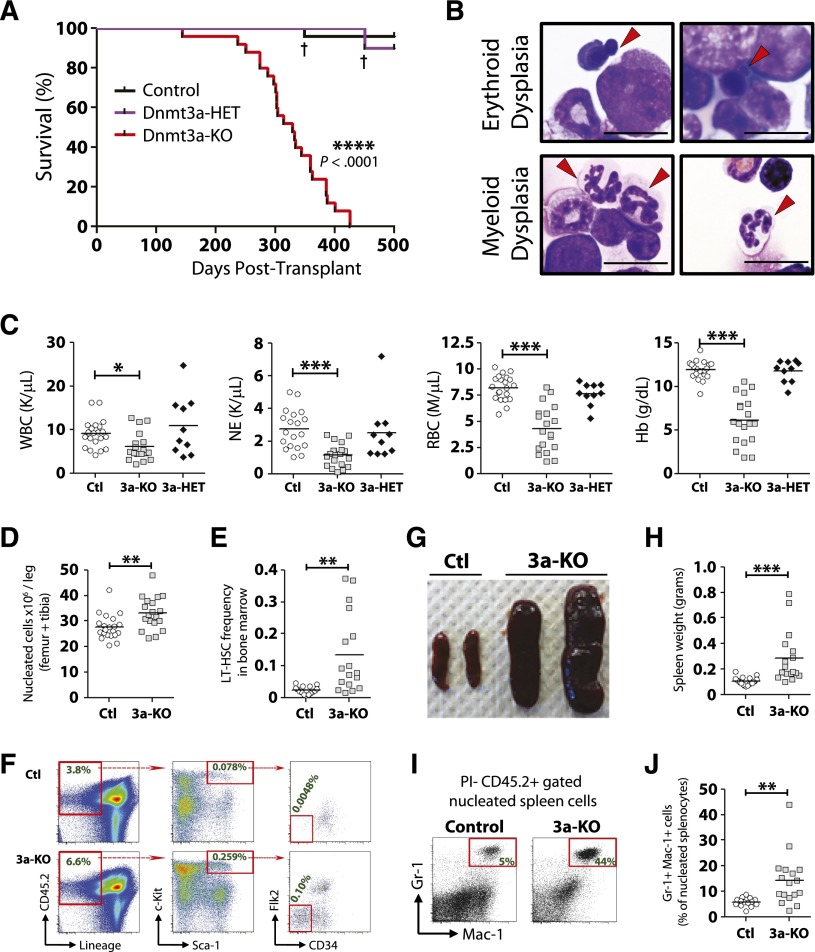
Mice transplanted with Dnmt3a-KO WBM succumbed to bone marrow failure (100% penetrance) ∼1 year posttransplant (median survival, 328 days). No hematopoietic-related mortality was observed from mice transplanted with control or Dnmt3a-heterozygous (Mx1-CRE+:Dnmt3afl/+ = Dnmt3a-HET) WBM when monitored out to 500 days posttransplant (Figure 2A). The majority of the moribund mice (19/25) demonstrated myeloid and erythroid pathologies most consistent with MDS27 (Figure 2B), including peripheral neutropenia and anemia (Figure 2C) and bone marrow hypercellularity (Figure 2D). These mice showed an accumulation of phenotypically defined HSCs in the marrow (Figure 2E-F), although not to the magnitude witnessed in competitive HSC transplants.16 These mice also developed splenomegaly (Figure 2G-H) with myeloid infiltration (Figure 2I-J). Two of the 25 mice developed AML with leukocytosis and >20% myeloid blasts in the blood and bone marrow, whereas 4 out of 25 mice developed diseases with intermediate peripheral anemia and mild leukocytosis with <5% blasts in the bone marrow, most consistent with a mixed MDS/MPN (supplemental Table 1).
Figure 2.
Enforced differentiation of Dnmt3a-KO HSCs leads to bone marrow failure resembling MDS. Noncompetitive transplantation of control (n = 25), Dnmt3a-HET (n = 10), and Dnmt3a-KO (n = 25) WBM. Compiled data are derived from 2 independent transplant cohorts. Control WBM in the first cohort was Mx1-CRE-:Dnmt3afl/fl (n = 10) and in the second cohort Mx1-CRE+:Dnmt3a+/+ (n = 15). Dnmt3a-HET WBM was Mx1-CRE+:Dnmt3afl/+ (n = 10). (A) Kaplan-Meier survival curve of all mice. † indicates death of a mouse from nonhematopoietic complications. The remainder of the figure shows data only for the Dnmt3a-KO (3a-KO) mice that were diagnosed with MDS at time of sacrifice. Data for control (Ctl) and Dnmt3a-HET (3a-HET) are from day 500 posttransplant. (B) Bone marrow pathology of the MDS phenotype. Erythroid dysplasia indicates orthochromic pronormoblasts with irregular nuclear contours and nuclear budding indicative of dyserythropoiesis. Myeloid dysplasia indicates lobated, hypersegmented neutrophils with pale blue cytoplasm indicative of dysmyelopoiesis. Red arrows indicate cells with described phenotypes. All photomicrographs taken at original magnification ×100. Scale bar represents 10 μm. (C) Peripheral blood counts show leukopenia, neutropenia, and anemia in recipients of Dnmt3a-KO WBM. Dnmt3a-KO MDS mice also showed bone marrow hypercellularity (D) and increased HSC (Lineage− CD45.2+ Sca-1+ c-Kit+ CD34− Flk2−) frequency (E). (F) FACS plots showing increase of phenotypically defined HSCs in Dnmt3a-KO recipient mice. Numbers represent percentage of that cell fraction in total bone marrow. Dnmt3a-KO MDS mice showed splenomegaly (G-H) with increased myeloid cell (Gr-1+ Mac-1+) cell infiltration (I-J). * P < .05, ** P < .01, *** P < .001, **** P < .0001.
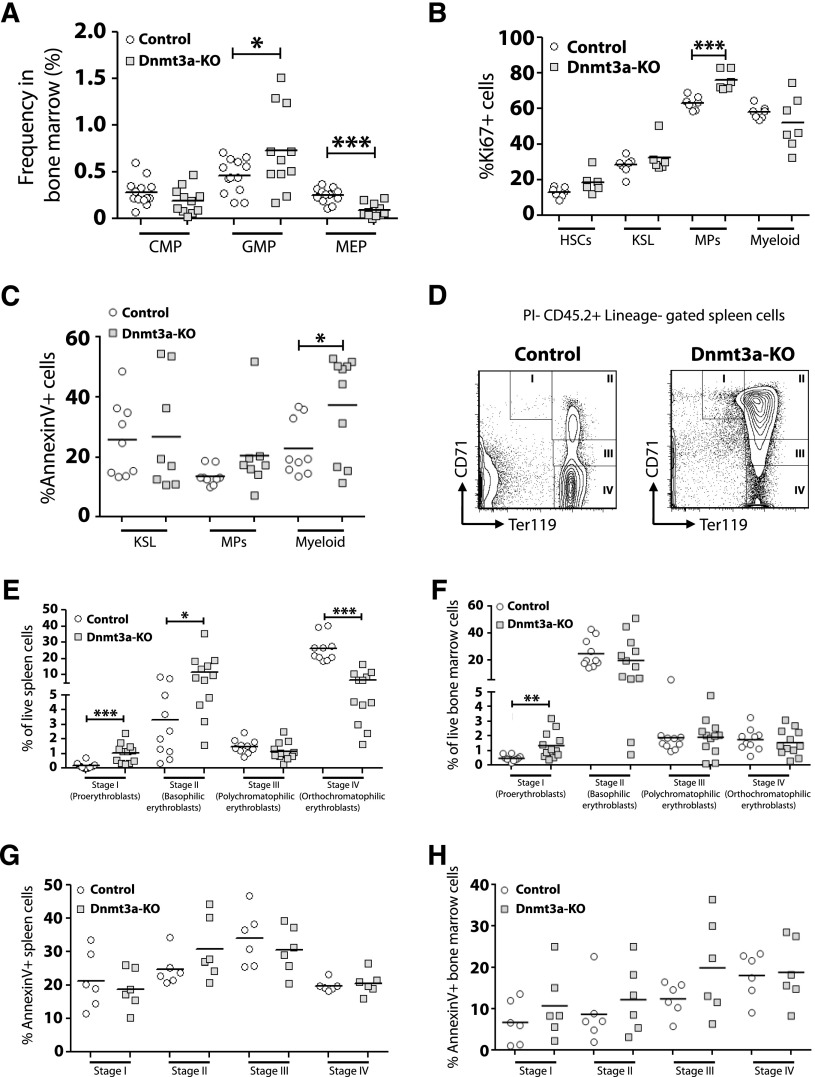
We performed a detailed analysis of myeloid cell development in the bone marrow of MDS mice. Although the frequency of CMPs was not different, Dnmt3a-KO mice had increased GMPs and reduced MEPs (Figure 3A). To account for these differences, analysis of proliferation and apoptosis was performed (supplemental Figure 2A). Dnmt3a-KO mice showed increased Ki67+ myeloid progenitors (MPs), but not HSCs, KSL (c-Kit+ Sca-1+ Lineage−) progenitors, or mature (Gr-1+ Mac-1+) myeloid cells (Figure 3B). The frequency of cells undergoing apoptosis within each myeloid cell fraction was determined by Annexin V staining (supplemental Figure 2B). No differences were observed for the proportion of Annexin V+ cells within Dnmt3a-KO KSL or MP populations, but mature myeloid cells from Dnmt3a-KO bone marrow showed significantly higher rates of apoptosis compared with control cells (Figure 3C). Peripheral cytopenias with bone marrow hypercellularity, increased apoptosis, and increased cell proliferation are consistent with an MDS.28
Figure 3.
Dysfunctional myeloid and erythroid cell development in the absence of Dnmt3a. (A) Increased frequency of GMPs but decreased frequency of MEPs in the bone marrow of Dnmt3a-KO MDS mice (n = 11) compared with control mice (n = 13). (B) Quantification of Ki67+ cells within bone marrow populations from Dnmt3a-KO MDS (n = 6-7) and control (n = 7) mice showed increased proliferation of Dnmt3a-KO myeloid progenitors (MPs). (C) Quantification of Annexin V+ cells within bone marrow populations from Dnmt3a-KO MDS (n = 8-10) and control (n = 9) mice showed increased apoptosis in Dnmt3a-KO mature myeloid cells (Gr-1+ Mac-1+) but not progenitors. (D) FACS analysis showing accumulation of immature erythroid progenitors (stage I and stage II) in the spleen of Dnmt3a-KO MDS mice. (E) Quantification of erythroid progenitors in the spleens of Dnmt3a-KO MDS (n = 12) and control (n = 10) mice showed arrest of Dnmt3a-KO progenitors at stage I (proerythroblast) and stage II (basophilic erythroblast) of erythroid development, leading to a subsequent decrease in the more mature stage IV (orthochromatophilic erythroblast) cells. (F) Quantification of erythroid progenitors in the bone marrow of Dnmt3a-KO MDS (n = 12) and control (n = 10) mice. (G) Apoptosis analysis of erythroid progenitors in the spleens of Dnmt3a-KO MDS (n = 6) and control (n = 6) revealed no differences in percentages of Annexin V+ cells, suggesting the accumulation of Dnmt3a-KO early progenitors arose from a block in developmental progression. (H) Apoptosis analysis of erythroid progenitors in the bone marrow of Dnmt3a-KO MDS (n = 6) and control (n = 6) mice. * P < .05, ** P < .01, *** P < .001.
To explore the mechanism leading to anemia, a comparison of erythroid progenitor development between control and Dnmt3a-KO MDS mice was performed. The maturation of erythroid progenitors can be discriminated based on staining for CD71 and Ter119.29 These are from least to most differentiated: proerythroblasts (CD71high Ter119med, stage I), basophilic erythroblasts (CD71high Ter119high, stage II), polychromatophilic erythroblasts (CD71med Ter119high, stage III), and orthochromatophilic erythroblasts (CD71low Ter119high, stage IV). Analysis revealed an accumulation of stage I and stage II progenitors in the spleens of Dnmt3a-KO MDS mice and a significant decrease in the numbers of stage IV progenitors (Figure 3E). Stage I progenitors were also increased in the bone marrow of Dnmt3a-KO MDS mice (Figure 3F). Annexin V staining did not reveal any significant differences in the proportion of apoptotic cells within each cell fraction between control and Dnmt3a-KO mice (Figure 3G-H). This suggests that the accumulation of early erythroid progenitors in the spleens of Dnmt3a-KO MDS mice represents a developmental arrest and not an increase to compensate for abortive loss of later progenitors.
The DNMT3AR882H variant drives myeloproliferation
In myeloid neoplasms, patients with DNMT3A mutations exist in the heterozygous state. The most common mutation occurs at amino acid 882, with over 60% of DNMT3A mutations in AML being the DNMT3AR882H variant.8 The resultant mutant protein acts as a dominant negative by inhibiting oligomerization of higher-order complexes and thereby blocking the ability of DNMT3A to form active tetramers.15 To explore the biological functions of the DNMT3AR882H variant in a genetically relevant system, we transduced Dnmt3a-HET bone marrow with retroviruses carrying green fluorescent protein (GFP)-only control (MIG), WT DNMT3A (MIG-DNMT3A), and DNMT3AR882H (MIG-R882H). To exacerbate the phenotype, we transduced posttransplant Dnmt3a-HET bone marrow.
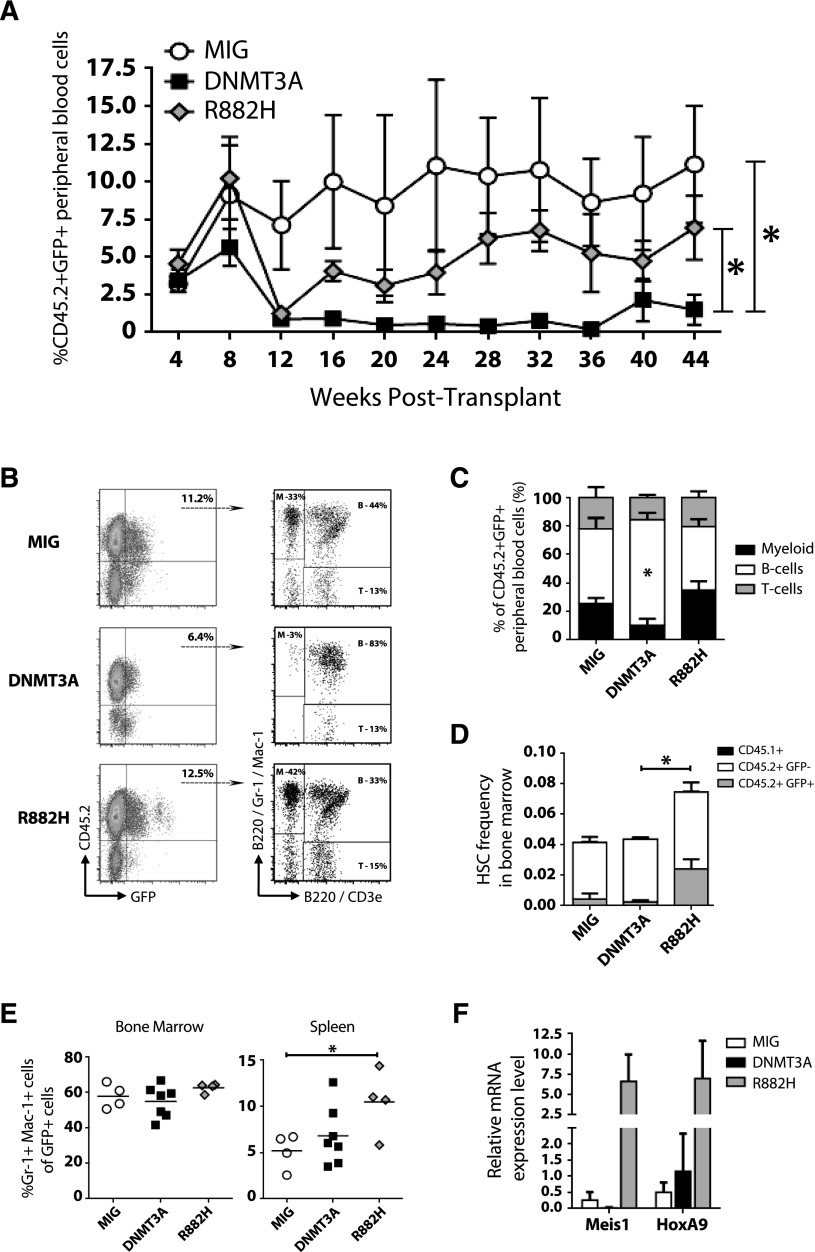
Ectopic expression of WT DNMT3A had a negative effect on blood differentiation. Expression of DNMT3AR882H produced levels of GFP+ blood chimerism similar to those of control MIG cells (Figure 4A). However, DNMT3AR882H did have a significant effect on the lineage distribution of engrafted cells (Figure 4B), causing a shift toward myeloid differentiation (Figure 4C). Bone marrow analysis 1 year posttransplant showed an expansion of phenotypically defined HSCs in these mice (Figure 4D) and increased numbers of Gr-1+ Mac-1+ cells (Figure 4E), but no overt leukemia. Gene expression analysis confirmed upregulation of Meis1 and HoxA9 induced by DNMT3AR882H30 (Figure 4F). Together, these data suggest DNMT3AR882H drives myeloproliferation and may act as a dominant negative in vivo due to the phenocopy of Dnmt3a loss-of-function alleles.
Figure 4.
The DNMT3AR882H variant drives myeloproliferation. Transduction of Dnmt3a-HET bone marrow posttransplant with control vector (MIG, n = 4), WT DNMT3A (DNMT3A, n = 7), and DNMT3AR882H (R882H, n = 4) followed by bone marrow transplant. (A) Peripheral blood engraftment of transduced donor cells (CD45.2+ GFP+) sampled at monthly intervals posttransplant. (B) Representative flow cytometry plots of engraftment and lineage distribution of transduced donor cells in peripheral blood (B, B cells; M, myeloid; T, T cells). (C) Compiled lineage distribution of donor peripheral blood cells. (D) Frequency of phenotypically defined HSCs (Lineage− c-Kit+ Sca-1+ CD48− CD150+) in the bone marrow of recipient mice 1 year posttransplant, showing contribution of recipient-derived (CD45.1+), donor-derived untransduced (CD45.2+ GFP−), and donor-derived transduced cells (CD45.2+ GFP+). (E) Percentage of mature myeloid cells (Gr-1+ Mac-1+) in the bone marrow and spleen of recipient mice 1 year posttransplant. (F) Gene expression analysis of CD45.2+ GFP+ Mac-1+ bone marrow cells 1 year posttransplant confirmed upregulation of Meis1 and HoxA9 reported by overexpression of DNMT3AR882H in WT mouse bone marrow cells.
The dominant clone establishes disease upon secondary transfer
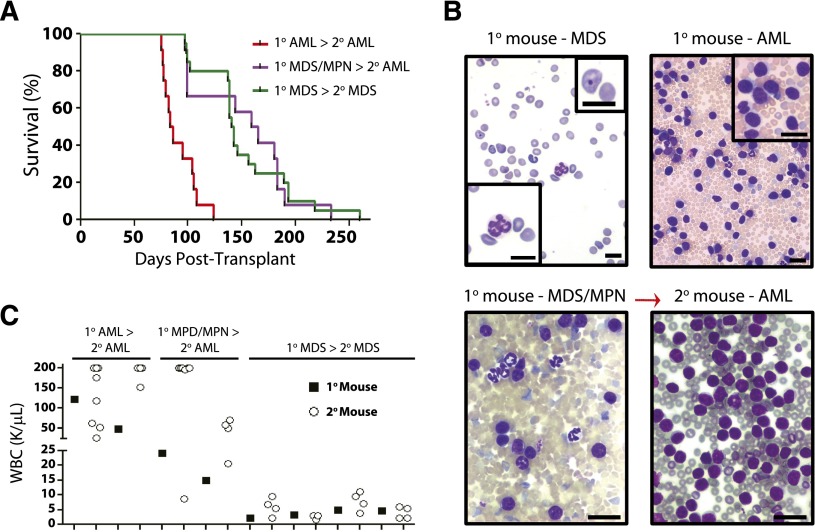
Secondary transplantation of diseased Dnmt3a-KO bone marrow and/or spleen into sublethally irradiated mice was performed to determine if a dominant clone would prevail upon disease reconstitution (and to confirm the phenotypes were cell intrinsic). Secondary transplantation of primary AML quickly recapitulated AML (Figure 5A; median survival, 84 days). Transplantation of primary MDS/MPN revealed that in most secondary mice, AML evolved, albeit with long latency (Figure 5B; median survival 162 days). For Dnmt3a-KO MDS, secondary transfer largely recapitulated the primary disease (median survival, 142 days). Peripheral blood counts of the secondary recipients of individual tumors were strikingly similar to the original tumor they were transplanted with at the time of sacrifice (Figure 5C).
Figure 5.
The dominant clone establishes disease upon secondary transfer. Secondary transplantation of 1 × 106 primary bone marrow and/or spleen cells from Dnmt3a-KO primary diseased mice. (A) Kaplan-Meier survival curve of secondary recipient mice. All secondary recipients of bone marrow (n = 4) and spleen (n = 4, 4) cells from primary Dnmt3a-KO AML developed fatal AML within 124 days. Transplantation of bone marrow from 5 primary Dnmt3a-KO MDS samples also generated MDS in secondary mice (n = 4 recipients per tumor). Transplantation of bone marrow and/or spleen from 2 mixed MDS/AML primary samples generated AML in secondary recipients with long latency (n = 4, 8). (B) Peripheral blood smears of an individual Dnmt3a-KO tumor at time of sacrifice in primary and secondary recipients. Sample showed MDS characteristics in the blood of primary recipients (insets: lower left shows Howell-Jolly body, and upper right shows abnormal nucleated neutrophil), but blood of secondary recipient was packed with myeloid blasts (upper right inset; white blood cell [WBC] count >200 K/μL). Scale bar represents 10 μm. (C) WBC counts at time of sacrifice of individual tumors in primary and secondary recipients. For each paired sample, the black square is WBC count of primary tumor used as the donor for secondary transplant. Open circles are WBC count at time of sacrifice in secondary mice. Dnmt3a-KO MDS samples produced the same disease in primary and secondary mice.
Exome sequencing identifies c-Kit mutations in leukemic transformation
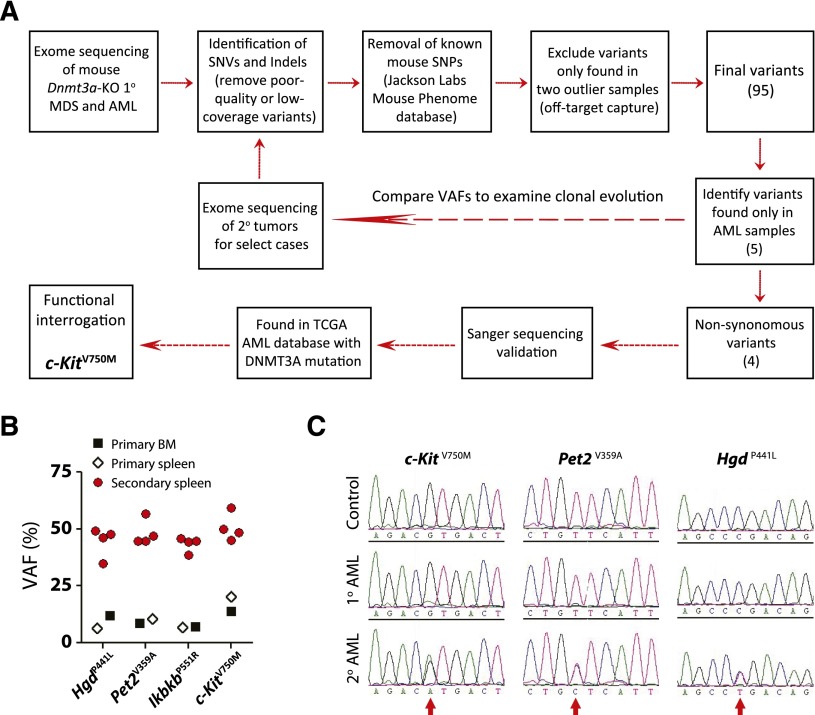
The long latency suggested that acquisition of secondary genetic and/or epigenetic lesions subsequent to inactivation of Dnmt3a were required to produce disease. As we have previously performed extensive DNA methylation profiling,16 here we performed whole-exome sequencing to uncover potential cooperating genetic mutations (Figure 6A). We chose 5 representative primary MDS samples (bone marrow), 1 primary MDS/MPN that progressed to AML in secondary recipients (bone marrow and spleen), and 1 primary AML sample (bone marrow and spleen) for analysis. Diseased spleen cells in secondary mice were also sequenced for select cases to examine clonal evolution. As banked germline DNA from the original bone marrow donor mice was either not available or of insufficient quality/quantity for sequencing to definitively assign the somatic status of prospective mutations, the comparator DNAs for this analysis were derived from pooled bone marrow of recipient mice transplanted with control (Mx1-Cre-:Dnmt3afl/fl) bone marrow from littermate mice. Following capture hybridization, high-throughput sequencing of exome DNA was performed on an Illumina platform.
Figure 6.
Exome sequencing identifies c-Kit mutation in leukemic transformation. (A) Schematic overview of workflow for exome sequencing. (B) Comparison of variant allele frequencies (VAFs) of four non-synonymous AML-specific variants in primary and secondary disease. The VAFs approximate 50% in secondary recipients, indicative of a clonal disease. (C) Independent Sanger sequencing validation of AML-specific mutations in primary and secondary disease. The mutations are identifiable (but not dominant) in the sequencing traces of the primary disease, whereas they are found in equal proportion to the WT allele in the secondary mice.
Initial analysis of primary disease samples identified 55 to 69 single-nucleotide variants and indels for 4 of the 5 MDS samples and 65 and 74 variants for the AML bone marrow and spleen samples, respectively, compared with the pooled reference bone marrow. As paired control DNA was not available to confirm the somatic basis of these mutations, we filtered out any known mouse SNPs identified in the Mouse Phenome Database (Jackson Laboratories) to exclude known strain polymorphisms. One of the 5 primary MDS bone marrow samples and the MDS/MPN spleen sample generated substantially more variant calls (1770 and 2405 respectively) than any other exome, suggesting some off-target capture. Any variants that were exclusively found only in these 2 samples were filtered out. These additional criteria resulted in identification of 16 to 31 high-quality coding variants per sample (supplemental Table 2), a number consistent with other mouse hematopoietic tumor sequencing studies.31 Comparison of these variants to The Cancer Genome Atlas AML database32 highlighted several genes that are also recurrently mutated in AML patients such as Fam5c and Smg1.
We were particularly interested in identifying mutations that were associated with leukemic transformation (present in Dnmt3a-KO AML, but not MDS). Five variants were exclusive to the AML sample, of which 4 were nonsynonymous: HgdP441L (amino acid change 441 P > L), Pet2V359A, IkbkbP551R, and c-KitV750M. Although the variant allele frequencies (VAFs) for these AML-specific variants (VAF range = 6.9% to 20.3%) suggested the pathogenic clone had not achieved complete clonal dominance in the primary sample, the VAFs approximated 50% in the secondary leukemic spleen cells (Figure 6B and supplemental Table 3). Exome sequencing results were confirmed for these variants by Sanger sequencing (Figure 6C). Of particular interest was the c-Kit variant given that AML and mastocytosis patients with DNMT3A mutations can also harbor KIT mutations,32,33 although we could not find evidence for c-Kit mutations in the other de novo AML sample or secondary AML derived from primary MDS/MPN.
c-Kit mutations functionally cooperate with loss of Dnmt3a in vivo
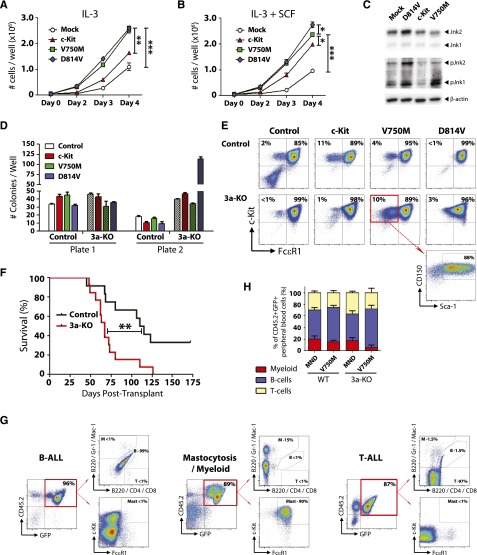
The functional significance of c-KitV750M (identified in our exome sequencing studies) and c-KitD814V (mouse homolog of KITD816V, most common variant found in DNMT3A-mutant AML and mastocytosis) were investigated using lentiviral expression models. Ectopic expression of c-Kit variants in the 32D mouse MP cell line lead to increased proliferation, both at baseline (Figure 7A) and in the presence of the c-Kit ligand SCF (Figure 7B). Analysis of these cells starved of IL-3 and SCF for 12 hours showed increased phosphorylation of Jnk1 and Jnk2 in 32D cells expressing c-KitV750M and c-KitD814V, indicative of ligand-independent activation of c-Kit signaling (Figure 7C). To determine the functional effects of c-Kit variants in primary bone marrow cells, hematopoietic progenitors were isolated and transduced with lentiviruses. Two days posttransduction, 100 GFP+ c-Kit+ Sca-1+ CD150+ cells were sorted and plated per well for CFU assay. Expression of c-KitD814V in a Dnmt3a-deficient background lead to explosive colony generation in the second plate (Figure 7D). c-KitD814V drove mast cell proliferation (Gr-1− Mac-1− c-Kit+ FcεR1+) in both genotypes, but the c-KitV750M variant seemed to have a specific effect in maintaining a more immature phenotype (Gr-1− Mac-1− c-Kit+ FcεR1− Sca-1+ CD150+) specifically in Dnmt3a-KO cells (Figure 7E).
Figure 7.
c-Kit mutations functionally cooperate with loss of Dnmt3a in vivo. (A) Growth rate of mouse 32D cells transduced with WT c-Kit, c-KitV750M, or c-KitD814V. Data represent mean ± SEM of 3 independent experiments. (B) Growth rate of mouse 32D cells transduced with WT c-Kit, c-KitV750M, or c-KitD814V in the presence of the c-Kit ligand SCF (10 ng/mL). Data represent mean ± SEM of 3 independent experiments. (C) Western blot to assess c-Kit signaling transducers Jnk1 and Jnk2 in 32D cells in the absence of IL-3 and SCF. Constitutive phosphorylation of Jnk1 and Jnk2 in c-KitV750M and c-KitD814V cells indicates ligand-independent signaling indicative of gain-of-function mutations. (D) Methocult serial replating of control and Dnmt3a-KO bone marrow progenitors transduced with c-Kit variants. In a Dnmt3a-KO background, c-KitD814V produced significantly more colonies in the second plate. Data represent mean ± SEM of 3 independent experiments. (E) Flow cytometry profiles of secondary Methocult colonies showing Gr-1− Mac-1− gated cells. c-KitD814V drives mast cell (Gr-1− Mac-1− c-Kit+ FcεR1+) proliferation, whereas c-KitV750M maintains an immature progenitor phenotype (Gr-1− Mac-1− c-Kit+ FcεR1− Sca-1+ CD150+) in a Dnmt3a-KO background. (F) Survival curve of mice transplanted with control or Dnmt3a-KO bone marrow progenitors transduced with c-KitD814V. (G) Bone marrow flow cytometry plots of mice transplanted with Dnmt3a-KO c-KitD814V reveal 3 distinct pathologies: B-ALL, T-ALL, and mastocytosis with myeloid blasts. (H) Peripheral blood lineage distribution of surviving mice (6 months posttransplant) transplanted with control or Dnmt3a-KO c-KitV750M showing expansion of B cells only in mice receiving Dnmt3a-KO c-KitV750M cells. Transplant data are compiled from 2 independent cohorts. * P < .05, ** P < .01, *** P < .001.
To determine if these 2 pathways could cooperate in vivo, we transduced hematopoietic progenitor cells with c-Kit variants and transplanted into lethally irradiated mice. As previously reported, expression of c-KitD814V in WT cells lead to development of B-cell acute lymphoblastic leukemia (B-ALL).34 However, c-KitD814V in a Dnmt3a-KO background led to explosive leukemia with a much shorter latency (Figure 7F). Moreover, although many of the mice transplanted with Dnmt3a-KO c-KitD814V cells also succumbed to a B-ALL, 4 out of 13 (31%) developed mastocytosis with involvement of myeloid blasts, and 4 out of 13 (31%) mice developed a T-cell acute lymphoblastic leukemia (T-ALL; Figure 7G). Expression of the c-KitV750M mutant lead to a selective advantage seen only in a Dnmt3a-deficient background, with 2 out of 8 mice (25%) succumbing to B-ALL within 6 months posttransplant, while all mice transplanted with WT cells transduced with c-KitV750M (8/8) were alive and healthy at this time point. Of the surviving mice, analysis of the lineage distribution of transduced donor-derived (CD45.2+ GFP+) peripheral blood cells showed a relative increase in the proportion of B cells in Dnmt3a-KO c-KitV750M cells (Figure 7H). These data suggest that this particular mutation may only be competent for transformation in a Dnmt3a-deficient background.
Discussion
Our data highlight crucial roles of Dnmt3a in normal and malignant hematopoiesis. Loss of Dnmt3a leads to developmental arrest of erythroid progenitors, enhanced proliferation of MPs, and increased apoptosis of mature myeloid cells. Although virtually all of the reported DNMT3A mutations in myeloid neoplasms are heterozygous,8,9,32 we did not observe a phenotype from Dnmt3a-heterozygous bone marrow. The most prevalent mutation in AML is the DNMT3AR882H variant, which acts as a dominant negative,14,15 making the patients effectively null for DNMT3A, akin to our genetic mouse model. The remainder of patients with heterozygous DNMT3A mutations typically present at an advanced age,8,32,35 suggesting the time to transformation may be dosage dependent in DNMT3A haploinsufficiency. The timeframe of our mouse experiments may not have been sufficient to observe this, and it is possible a phenotype for Dnmt3a-heterozygous bone marrow could be unveiled by extended transplantation or subjection to other stressors of HSC turnover such as serial 5-FU injection.
Activating mutations of KIT are associated with AML,36 germ cell tumors,37 and systemic mastocytosis.38 Exome sequencing identified acquisition of a c-Kit mutation (c-KitV750M) that was exclusively identified during leukemic transformation of Dnmt3a-KO cells. Characterization of this variant suggests it acts as a gain-of-function mutation, as it conferred a similar growth advantage and ligand-independent activation of downstream signaling pathways as the prototypical c-KitD814V variant. Functional studies showed synergism between Dnmt3a loss of function and c-Kit gain of function in generating an explosive leukemia with a much shorter latency than expression of c-KitD814V in WT bone marrow. Although DNMT3A and KIT have been shown to be concurrently mutated in mastocytosis33 and AML,32 Dnmt3a-KO c-KitV750M transplanted mice developed B-ALL. This is likely due to ectopic introduction of c-KitV750M into a lineage-biased HSC or progenitor cell. Similar to the BCR-ABL oncogene,39 the type of malignancy generated is likely dependent on the nature of the initiating cell that successfully integrates the virus, as evidenced by the spectrum of different diseases generated from c-KitD814V in Dnmt3a-KO transplanted mice (B-ALL, T-ALL, mastocytosis). To fully understand the relationship between Dnmt3a and activating c-Kit mutations in myeloid leukemogenesis, a genetic mouse model of regulatable stem cell–specific induction of c-Kit activation in Dnmt3a-KO HSCs will be required. However, here we show for the first time that these pathways can cooperate to accelerate transformation in vivo. This Dnmt3a/c-Kit disease model resembles the classical “two-hit” model of leukemogenesis40 in which one mutation inhibits differentiation (Dnmt3a loss-of-function), while another drives proliferation (c-Kit gain-of-function). Although we did not identify other mutations typically associated with DNMT3A-mutant AML patients (eg, FLT3-ITD, NPM1c), only approximately 10% of primary mice (2/25) developed de novo AML, and sequencing of many more samples would be required to fully ascribe to repertoire of cooperating mutations in this mouse model. As cooperating DNMT3A and KIT mutations are relatively rare in AML, the observation that this Dnmt3a mouse model selected for this combination in vivo likely represents stochastic selection and evolution at the level of the HSC.
We show that enforced differentiation of Dnmt3a-KO HSCs by noncompetitive transplantation leads to bone marrow failure resembling MDS. This is markedly distinct from our previous competitive transplantation outcomes and shows the critical requirement of cell division to acquire secondary genetic and epigenetic lesions necessary to promote disease. Such systems present a unique opportunity to study the sequence of early events leading to HSC transformation following Dnmt3a loss of function.
Acknowledgments
The authors thank Dr Michael Tomasson (Washington University School of Medicine [WUSM]) for c-Kit plasmids, Dr Timothy Ley and the Genome Institute (WUSM) for exome sequencing, Dr Eric Duncavage (WUSM) for pathology imaging, and the Alvin J. Siteman Cancer Center at WUSM for the use of the Siteman flow cytometry core, which provided cell sorting and analysis.
The Siteman Cancer Center is supported in part by the National Institutes of Health, National Cancer Institute Cancer Center Support Grant CA91842. This work was supported by the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (DK084259), the Children’s Discovery Institute, the Mallinckrodt Foundation, Alex's Lemonade Stand Foundation, and an American Society of Hematology Scholar award (all to G.A.C.).
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.C., C.M., A.K., E.L.O., E.E., A.M., and G.A.C. designed and performed experiments; H.C., C.A.M., J.H., J.M.K., and G.A.C. analyzed data; and G.A.C. wrote and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for J.M.K. is St. Jude Children’s Research Hospital, Memphis, TN, 38105.
Correspondence: Grant A. Challen, Washington University School of Medicine, 660 Euclid Ave, St. Louis, MO 63110; e-mail: gchallen@DOM.wustl.edu.
References
- 1.Jaenisch R, Jähner D. Methylation, expression and chromosomal position of genes in mammals. Biochim Biophys Acta. 1984;782(1):1–9. doi: 10.1016/0167-4781(84)90099-x. [DOI] [PubMed] [Google Scholar]
- 2.Ng HH, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9(2):158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 3.Surani MA. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93(3):309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- 4.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23(16):5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 6.Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 7.Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter MJ, Ding L, Shen D, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25(7):1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewalt M, Galili NG, Mumtaz M, et al. DNMT3a mutations in high-risk myelodysplastic syndrome parallel those found in acute myeloid leukemia. Blood Cancer J. 2011;1(3):e9. doi: 10.1038/bcj.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdel-Wahab O, Pardanani A, Rampal R, Lasho TL, Levine RL, Tefferi A. DNMT3A mutational analysis in primary myelofibrosis, chronic myelomonocytic leukemia and advanced phases of myeloproliferative neoplasms. Leukemia. 2011;25(7):1219–1220. doi: 10.1038/leu.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stegelmann F, Bullinger L, Schlenk RF, et al. DNMT3A mutations in myeloproliferative neoplasms. Leukemia. 2011;25(7):1217–1219. doi: 10.1038/leu.2011.77. [DOI] [PubMed] [Google Scholar]
- 13.Ribeiro AF, Pratcorona M, Erpelinck-Verschueren C, et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119(24):5824–5831. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]
- 14.Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013;122(25):4086–4089. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russler-Germain DA, Spencer DH, Young MA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25(4):442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Challen GA, Sun D, Mayle A, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 2014;15(3):350–364. doi: 10.1016/j.stem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaneda M, Okano M, Hata K, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 19.Challen GA, Boles N, Lin KK, Goodell MA. Mouse hematopoietic stem cell identification and analysis. Cytometry A. 2009;75(1):14–24. doi: 10.1002/cyto.a.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuettpelz LG, Gopalan PK, Giuste FO, Romine MP, van Os R, Link DC. Kruppel-like factor 7 overexpression suppresses hematopoietic stem and progenitor cell function. Blood. 2012;120(15):2981–2989. doi: 10.1182/blood-2012-02-409839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chian R, Young S, Danilkovitch-Miagkova A, et al. Phosphatidylinositol 3 kinase contributes to the transformation of hematopoietic cells by the D816V c-Kit mutant. Blood. 2001;98(5):1365–1373. doi: 10.1182/blood.v98.5.1365. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28(14):1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 25.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25(21):2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kogan SC, Ward JM, Anver MR, et al. Hematopathology subcommittee of the Mouse Models of Human Cancers Consortium. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100(1):238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 28.Nimer SD. Myelodysplastic syndromes. Blood. 2008;111(10):4841–4851. doi: 10.1182/blood-2007-08-078139. [DOI] [PubMed] [Google Scholar]
- 29.Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. Ineffective erythropoiesis in Stat5a(-/-)5b(-/-) mice due to decreased survival of early erythroblasts. Blood. 2001;98(12):3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 30.Xu J, Wang YY, Dai YJ, et al. DNMT3A Arg882 mutation drives chronic myelomonocytic leukemia through disturbing gene expression/DNA methylation in hematopoietic cells. Proc Natl Acad Sci USA. 2014;111(7):2620–2625. doi: 10.1073/pnas.1400150111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wartman LD, Larson DE, Xiang Z, et al. Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J Clin Invest. 2011;121(4):1445–1455. doi: 10.1172/JCI45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Traina F, Visconte V, Jankowska AM, et al. Single nucleotide polymorphism array lesions, TET2, DNMT3A, ASXL1 and CBL mutations are present in systemic mastocytosis. PLoS ONE. 2012;7(8):e43090. doi: 10.1371/journal.pone.0043090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiang Z, Kreisel F, Cain J, Colson A, Tomasson MH. Neoplasia driven by mutant c-KIT is mediated by intracellular, not plasma membrane, receptor signaling. Mol Cell Biol. 2007;27(1):267–282. doi: 10.1128/MCB.01153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan XJ, Xu J, Gu ZH, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43(4):309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 36.Beghini A, Larizza L, Cairoli R, Morra E. c-kit activating mutations and mast cell proliferation in human leukemia. Blood. 1998;92(2):701–702. [PubMed] [Google Scholar]
- 37.Nakai Y, Nonomura N, Oka D, et al. KIT (c-kit oncogene product) pathway is constitutively activated in human testicular germ cell tumors. Biochem Biophys Res Commun. 2005;337(1):289–296. doi: 10.1016/j.bbrc.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 38.Longley BJ, Tyrrell L, Lu SZ, et al. Somatic c-KIT activating mutation in urticaria pigmentosa and aggressive mastocytosis: establishment of clonality in a human mast cell neoplasm. Nat Genet. 1996;12(3):312–314. doi: 10.1038/ng0396-312. [DOI] [PubMed] [Google Scholar]
- 39.Wong S, Witte ON. Modeling Philadelphia chromosome positive leukemias. Oncogene. 2001;20(40):5644–5659. doi: 10.1038/sj.onc.1204638. [DOI] [PubMed] [Google Scholar]
- 40.Gilliland DG, Tallman MS. Focus on acute leukemias. Cancer Cell. 2002;1(5):417–420. doi: 10.1016/s1535-6108(02)00081-8. [DOI] [PubMed] [Google Scholar]