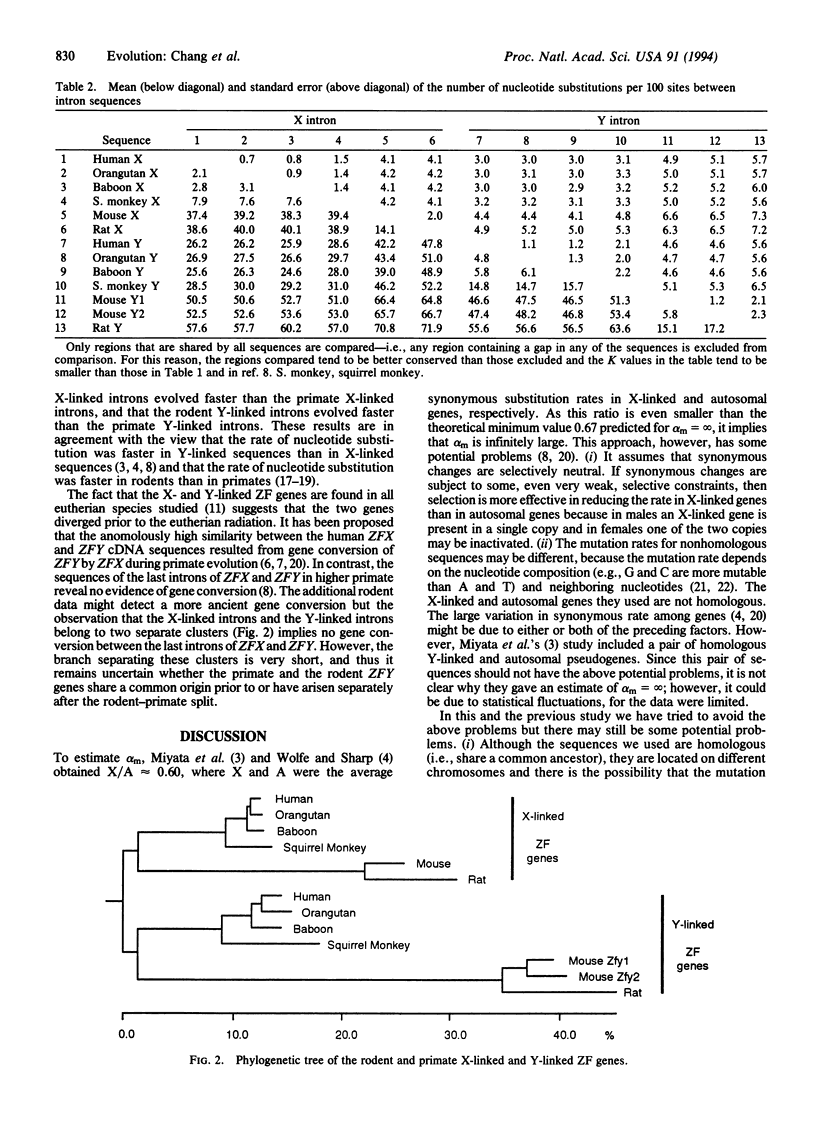
Abstract
In humans and rodents the male-to-female ratio of mutation rate (alpha m) has been suggested to be extremely large, so that the process of nucleotide substitution is almost completely male-driven. However, our sequence data from the last intron of the X chromosome-linked (Zfx) and Y chromosome-linked (Zfy) zinc finger protein genes suggest that alpha m is only approximately 2 in rodents with a 95% confidence interval from 1 to 3. Moreover, from published data on oogenesis and spermatogenesis we estimate the male-to-female ratio of the number of germ cell divisions per generation to be approximately 2 in rodents, confirming our estimate of alpha m and suggesting that errors in DNA replication are the primary source of mutation. As the estimated alpha m for rodents is only one-third of our previous estimate of approximately 6 for higher primates, there appear to be generation-time effects--i.e., alpha m decreases with decreasing generation time.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bulmer M. Neighboring base effects on substitution rates in pseudogenes. Mol Biol Evol. 1986 Jul;3(4):322–329. doi: 10.1093/oxfordjournals.molbev.a040401. [DOI] [PubMed] [Google Scholar]
- Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol Rev. 1972 Jan;52(1):198–236. doi: 10.1152/physrev.1972.52.1.198. [DOI] [PubMed] [Google Scholar]
- Ellsworth D. L., Rittenhouse K. D., Honeycutt R. L. Artifactual variation in randomly amplified polymorphic DNA banding patterns. Biotechniques. 1993 Feb;14(2):214–217. [PubMed] [Google Scholar]
- Hayashida H., Kuma K., Miyata T. Interchromosomal gene conversion as a possible mechanism for explaining divergence patterns of ZFY-related genes. J Mol Evol. 1992 Aug;35(2):181–183. doi: 10.1007/BF00183228. [DOI] [PubMed] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat Rec. 1971 Mar;169(3):533–557. doi: 10.1002/ar.1091690306. [DOI] [PubMed] [Google Scholar]
- Li W. H., Tanimura M., Sharp P. M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol. 1987;25(4):330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- Li W. H., Wu C. I., Luo C. C. Nonrandomness of point mutation as reflected in nucleotide substitutions in pseudogenes and its evolutionary implications. J Mol Evol. 1984;21(1):58–71. doi: 10.1007/BF02100628. [DOI] [PubMed] [Google Scholar]
- Mardon G., Luoh S. W., Simpson E. M., Gill G., Brown L. G., Page D. C. Mouse Zfx protein is similar to Zfy-2: each contains an acidic activating domain and 13 zinc fingers. Mol Cell Biol. 1990 Feb;10(2):681–688. doi: 10.1128/mcb.10.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon G., Mosher R., Disteche C. M., Nishioka Y., McLaren A., Page D. C. Duplication, deletion, and polymorphism in the sex-determining region of the mouse Y chromosome. Science. 1989 Jan 6;243(4887):78–80. doi: 10.1126/science.2563173. [DOI] [PubMed] [Google Scholar]
- Mardon G., Page D. C. The sex-determining region of the mouse Y chromosome encodes a protein with a highly acidic domain and 13 zinc fingers. Cell. 1989 Mar 10;56(5):765–770. doi: 10.1016/0092-8674(89)90680-6. [DOI] [PubMed] [Google Scholar]
- Miyata T., Hayashida H., Kuma K., Mitsuyasu K., Yasunaga T. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb Symp Quant Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- Pamilo P., Bianchi N. O. Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol Biol Evol. 1993 Mar;10(2):271–281. doi: 10.1093/oxfordjournals.molbev.a040003. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmin L. C., Chang B. H., Hewett-Emmett D., Li W. H. Potential problems in estimating the male-to-female mutation rate ratio from DNA sequence data. J Mol Evol. 1993 Aug;37(2):160–166. doi: 10.1007/BF02407351. [DOI] [PubMed] [Google Scholar]
- Shimmin L. C., Chang B. H., Li W. H. Male-driven evolution of DNA sequences. Nature. 1993 Apr 22;362(6422):745–747. doi: 10.1038/362745a0. [DOI] [PubMed] [Google Scholar]
- Tajima F., Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984 Apr;1(3):269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- Vogel F., Rathenberg R. Spontaneous mutation in man. Adv Hum Genet. 1975;5:223–318. doi: 10.1007/978-1-4615-9068-2_4. [DOI] [PubMed] [Google Scholar]
- Winter R. M., Tuddenham E. G., Goldman E., Matthews K. B. A maximum likelihood estimate of the sex ratio of mutation rates in haemophilia A. Hum Genet. 1983;64(2):156–159. doi: 10.1007/BF00327115. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Sharp P. M., Li W. H. Mutation rates differ among regions of the mammalian genome. Nature. 1989 Jan 19;337(6204):283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- Wu C. I., Li W. H. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]



