Abstract
Background
MicroRNAs (miRNAs) have been reported to play crucial roles in regulating a variety of genes pivotal for tumor metastasis. MicroRNA-301a (miR-301a) is overexpressed and displays oncogenic activity in many cancers. However, little is known about the potential roles of miR-301a in colorectal cancer (CRC).
Methods
Taqman probe stem-loop real-time PCR was used to quantitatively measure the expression level of miR-301a in 48 cases of CRC tissues and the matched adjacent non-tumor mucosa as well as in CRC cell lines. miR-301a mimics and inhibitors were used to up-regulate and down-regulate miR-301a in CRC cells, respectively; lentivirus was used to construct miR-301a stably up- and down-regulated CRC cell lines. Metastasis ability was evaluated by transwell and wound healing assays while invasion was measured by transwell coated with matrix gel in vitro; in vivo metastasis was performed on nude mice model. The target of miR-301a was predicted by TargetScan software and validated by qRT-PCR, immunohistochemistry, western blot and luciferase reporter gene assay.
Results
The expression of miR-301a was significantly higher in lymph node metastasis positive CRC samples compared with negative ones. Downregulation of miR-301a significantly inhibited the migration and invasion both in vitro and in vivo while forced up-regulation of miR-301a promoted migration and invasion. TGFBR2 was identified to be the downstream target of miR-301a. Knockdown of TGFBR2 in cells treated by miR-301a inhibitor elevated the previously abrogated migration and invasion.
Conclusions
Our data indicated that miR-301a correlated with the metastatic and invasive ability in human colorectal cancers and miR-301a exerted its role as oncogene by targeting TGFBR2.
Keywords: CRC, miRNA-301a, Metastasis, Invasion, TGFBR2
Background
Colorectal cancer (CRC) is one of the most prevalent causes of cancer-related mortalities worldwide, with 610.000 deaths each year. Though efforts have been paid to the prevention of CRC, the incidence of CRC is still increasing in the past decades [1]. It has been reported that approximate 20% of CRC patients are diagnosed with metastases and 50% of early stage CRC patients treated will eventually develop metastases [2,3]. Despite advantages in early screening and development of new therapeutic strategies, the 5-year survival rate for patients with metastatic CRC is only about 10% [2]. Thus, an insight into the molecular mechanisms of CRC metastasis is important for developing new prognostic and therapeutic molecular markers to reduce CRC metastasis.
MicroRNAs (miRNAs) are a class of small non-coding RNAs (18–22 nt in length) which regulate the expression of target genes by binding to the 3’ un-translated regions (3’-UTR) [4,5]. Emerging evidences strongly suggest critical roles of miRNAs in the pathogenesis of cancer, and specially, the tumor metastasis [6,7]. The dysregulation of miRNAs in CRC has been reported using miRNA expression profiling studies with different miRNAs identified either as enhancers (miR-21, miR-31, miR-103, miR-107) or suppressors (miR-135, miR-145, miR-200c) in the initiation and evolution of tumor metastasis [8-13]. Considering the chemical stability, the ease of clinical detection, as well as the crucial roles of miRNA in tumor pathology, researches on the identification of potential miRNAs as metastatic biomarkers and therapeutic targets in CRC patients are rapidly increasing.
The gene encoding miR-301a is located in the human chromosome 17q22-17q23 and it has been previously reported to be overexpressed in many kinds of human cancers, including gastric cancer, liver cancer, breast cancer, etc. [14-20]. Besides, a series of recent functional studies demonstrated that miR-301a promoted cancer cell metastasis in breast, hepatocellular and gastric tumors through different target genes [17,21,22]. For CRCs, a miRNA array which analyzed the miRNA expression profiles of 44 pairs of CRCs revealed that the expression of miR-301a was elevated in tumor tissues of stage II patients compared to paired normal tissues [23]. However, until now, functional evidence of miR-301a in CRCs has not been documented and its role in CRC metastasis remains unclear. Thus, we wanted to know whether miR-301a was involved in the metastasis of CRC and we further wanted to evaluate the potential signalling pathways through which miR-301a regulated metastasis.
Methods
CRC samples
48 pairs of tumor samples and the paired adjacent non-tumor mucosa (at least 5 cm away from the tumor margin) were collected from CRC patients undergoing laparoscopic surgery from March 2011 to September 2011 at Shanghai Minimally Invasive Surgery Centre (Ruijin Hospital, Shanghai Jiao Tong University School of Medicine). The written informed consents were given by all patients prior to surgery and all procedures were approved by the ethics committee of Ruijin Hospital. None of the patients received preoperative chemoradiotherapy and all were diagnosed with resectable tumors by endoscopy and CT. All tumors were confirmed pathologically and classified according to the 7th edition of AJCC/UICC TNM system [24]. The clinicopathological information of included cases was prospectively collected during hospitalization. Samples were immediately excised and preserved in formalin for further immunohistochemical staining or snap-frozen in liquid nitrogen for future RNA extraction.
Cell culture
Five human CRC cell lines were obtained from American Type Culture Collection (Manassas, United States). HCT116 and HT29 were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, United States) with 10% fetal bovine serum (FBS). LOVO, SW480 and SW620 were maintained in RPMI 1640 (Sigma Aldrich, United States), supplemented with 10% FBS. All cells were maintained in a humidified incubator at 37°C with 5% CO2.
RNA isolation and miRNAs assays
Total RNA was isolated from cell lines and tissue samples using Trizol reagent (Invitrogen, United States) according to the manufacturer’s instructions. The expression levels of miR-301a and U6 small nuclear RNA (RNU6B, Applied Biosystems, United States) were assayed in triplicates by the TaqMan stem-loop RT-PCR method with a mirVana miRNA detection Kit and gene-specific primers (Applied Biosystems, United States). Expression levels of RNU6B were used for normalization. The relative expression of miR-301a in tissues and cell lines were calculated by the 2-Δct method.
SYBR green real-time RT-PCR
The mRNA level of TGBR2 was measured by quantitative real-time polymerase chain reaction (qRT-PCR) with Power SYBR Green PCR Master Mix (Applied Biosystems, United States) in an Applied Biosystems 7500 System. The Super Script III First-Strand Synthesis System (Invitrogen, United States) was used to prepare cDNA from total RNA. The primers for TGFBR2 gene were obtained from “Primerbank” (http://pga.mgh.harvard.edu/primerbank/). The relative amount of TGFBR2 mRNA was uniformly normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as endogenous control. Primer sequences are presented in Table 1.
Table 1.
Primer sequences used qRT-PCR
| Forward primer | Reverse primer | |
|---|---|---|
| TGFBR2 | 5′-GTAGCTCTGATGAGTGCAATGAC-3′ | 5′-CAGATATGGCAACTCCCAGTG-3′ |
| GAPDH | 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ | 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ |
Oligonucleotides transfection
miR-301a inhibitors (anti-miR-301a), miR-301a mimics and their corresponding negative controls (anti-miR-NC, miR-NC) were purchased from GenePharma (China, Table 2). siRNA of TGFBR2 was purchased from Dharmacon (D-003930, Dharmacon, United States). Cells were transfected with 100 nM oligonucleotides using Lipofectamine 2000 Reagent (Invitrogen, United States) according to the manufacturer’s instructions. 48 hours after transfection, cells were harvested and miR-301a expression level was monitored by qRT-PCR (U6 RNA was endogenous control), and the siRNA transfection efficiency was assessed by western blot using GAPDH as loading control.
Table 2.
The sequences of miR-301a inhibitor and mimics
| Sequences | |
|---|---|
| miR-301a-inhibitor | 5′-GCUUUGACAAUACUAUUGCACUG-3′ |
| Inhibitor NC | 5′-CAGUACUUUUGUGUAGUACAA-3′ |
| miR-301A-mimics | 5′-CAGUGCAAUAGUAUUGUCAAAGCUUUGACAAUACUAUUGCACUGUU-3′ |
| Mimics NC | 5′-UAGCUAAGAGACCCAGAGUUAUUCUUACAAUCUCUCUACUAUC-3′ |
Lentiviral transfection for stable expression clones
Plasmids LV3-pGLV-H1-GFP + Puro with hsa-miR-301a inhibitor or its negative control oligonucleotides, namely LV-anti-miR-301a and LV-anti-miR-301a-NC, were purchased from GenaPharma (China). The procedures to establish LV-anti-miR-301a stably expressing clones and the control clones in SW620 cells (SW620/LV-anti-miR-301a and SW620/LV-anti-miR-NC) were performed as previously described [25].
Migration and invasion assays
Cell migration was performed using transwell inserts (Corning, United States), and cell invasion assay was measured using matrigel-coated upper chambers (BD Bioscience, United States). Briefly, cells (2.5 × 105) in 200 μl serum-free medium were plated to the upper chambers and medium containing 10% serum was used as chemo attractant in lower chambers. After 24 hours incubation, non-migrating or non-invading cells were removed softly with cotton swabs and cells at the out surface of insert membranes were fixed with menthol for 30 min, stained with 1% crystal violet for another 30 min, and then photographed under 200 × magnification. Cells were counted from 5 random fields of view and means ± SD were calculated accordingly.
Wound-healing assay
Generally, cells were first plated in a six-well plate and then transfected with different oligonucleotides. Forty-eight hours later, cells were harvested and plated in a new six-well plate. When the cells grew to a confluence of 90–95%, the cell monolayer was scratched using a sterile 10 μl pipette tip. After washing and removing the detached cells, the plates were incubated at 37°C for 24 h and the wounds were photographed. Then the open wound area was quantified using “TScratch” as previously described [26]. At least 5 different wounds were performed, and the experiments were repeated 3 times independently.
In vivo metastasis assay
SW620/LV-anti-miR-301a and SW620/LV-anti-miR-NC cells (5 × 106 cells per mice) were implanted subcutaneously into the left flanks of 5-week-old male nude mice (10 mice per group). Local invasion and lung metastasis were examined 4 or 7 weeks after implantation respectively (5 mice per group for each). Tumors and lungs were harvested, fixed, embedded and stained with haematoxylin and eosin as described previously [27]. All animal experiments complied with protocols approved by the Animal Care and Use Committee of Shanghai Jiao Tong University School of Medicine.
Western blot analysis
Whole cell protein lysates were extracted using M-PER reagents and Halt Protease Inhibitor Cocktail kits (Pierce, United States). The protein concentrations were quantified with a Bicinchoninic Acid (BCA) protein assay kit (Pierce, United States). Western blotting for TGFBR2 was performed with established procedures as Ye et al. described previously [28]. The mouse monoclonal anti-TGFBR2 antibody (1:500, Abcam, United States) and anti-GAPDH antibody (1:2000, Kang Chen, China) were used as the primary antibodies. GAPDH served as loading control.
Immunohistochemistry analysis
Immunohistochemistry analysis of TGFBR2 was performed with an anti-TGFBR2 antibody (1:200, Abcam, United States). Tumor tissues were fixed, embedded and stained with haematoxylin and eosin (Sigma, United States). Immunohistochemistry staining was performed with established protocols [29]. Relative TGFBR2 expression were defined as positive (moderate or strong staining) and negative (no or weak staining) based on the intensity of TGFBR2 staining of the tumor cells.
Vector construction and luciferase reporter assay
miR-301a binding sites were predicted using TargetScan software (http://www.targetscan.org) and target genes which had the highest possibility and were also metastasis related were chosen for further validation. Bioinformatics analysis revealed two putative binding sites for miR-301a: a conserved 7mer-m8 at nt 266-272 of TGFBR2 3’-UTR and a poorly conserved 7mer-m8 at position 566-572 of TGFBR2 3’-UTR. Two mutant fragments of TGFBR2 3’-UTR (mut-266, mut-566) were designed using a Quick Change Site-Directed Mutagenesis kit (Stratagene, United States). Then the full length of miR-301a TGFBR2 3’-UTR including two wild-type binding sites and the two mutant fragments (mut-266, mut-566) were designed by and purchased from Sangon (Shanghai, China). After digestion by Sac I and Hind III, the fragments of wild-type and mutant were cloned into the Sac I and Hind III sites of pMIR-Report Luciferase Vector (Applied Biosystems, United States) and were named pMIR/TGFBR2-wt, pMIR/TGFBR2-mut-266 and pMIR/TGFBR2-mut-566, respectively. All constructs were verified by Sanger sequencing.
SW620 cells were co-transfected with 200 ng luciferase reporter gene construct, 2 ng pRL-TK vector (Promega, United States) containing Renilla luciferase, and 100 nM mimics or inhibitor in 24-well plates. Reporter assays were performed 48 h post-transfection using the Dual-luciferase assay system (Promega, United States). Firefly luciferase activity was normalized to renilla luciferase activity. All transfection experiments were conducted in triplicate and repeated 3 times independently.
Statistical methods
Data were expressed as means ± standard deviation (SD) and P < 0.05 was considered statistically significant. Student’s t test (two-tailed) was used to compare two groups unless indicated specifically (χ2 test was used for comparison of the rates of two groups). Correlation analysis was performed with spearman correlation analysis. All statistical analyses were performed using the SPSS 15.0 software (SPSS Inc, United States).
Results
miR-301a is up-regulated in lymph node metastatic CRC tissues and CRC cells lines
To understand the role of miR-301a in CRC progression, we examined the expression of miR-301a in 48 CRC tissues and their paired adjacent non-tumor tissues. qRT-PCR demonstrated that there was no significant difference of miR-301a expression between CRC and the adjacent non-tumor mucosa (Figure 1A, P > 0.05). However, the expression of miR-301a in tumors with lymph node metastases was significantly higher than that in tumors without metastases (Figure 1B, P < 0.05), which indicated that miR-301a might be involved in the metastasis of CRC. Importantly, the expression of miR-301a did not differ among patients with different gender, age, tumor differentiation, tumor location, tumor histopathology, tumor local invasion or distant metastasis (Table 3).
Figure 1.
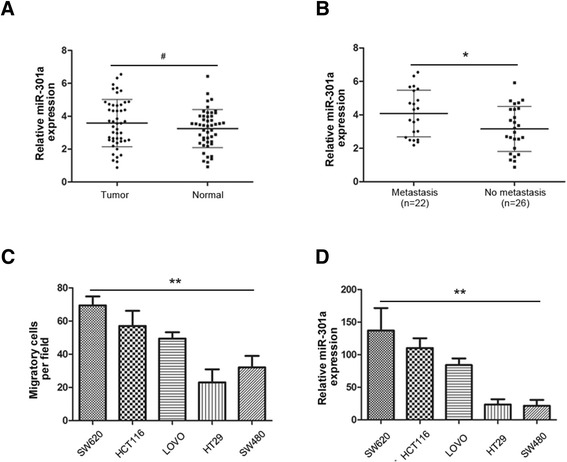
miR-301a was upregulated in lymph node metastatic CRC tissues and highly metastatic CRC cells. (A) qRT-PCR analysis of 48 pairs of CRC samples and matched adjacent normal mucosa; U6 small nuclear RNA was used as an internal control. There was no significant difference in miR-301a expression between CRC tissues and their adjacent non-tumor tissues. (B) miR-301a expression levels in primary CRCs with lymph node metastasis were much higher than that of CRCs without lymph node metastasis. (C) The migratory potential was examined by migration assays. (D) Relative expression of miR-301a in 5 CRC cell lines by qRT-PCR with U6 small nuclear RNA as an internal control. Data were representedas means ± SD from 3 independent experiments. #, P > 0.05; *, P < 0.05; **, P < 0.01.
Table 3.
Clinicopathological parameters
| Characteristics | No. of cases | miR-301 expression | P -value |
|---|---|---|---|
| Age(years) | |||
| <66 | 20 | 3.76 | |
| ≥66 | 28 | 3.63 | 0.79 |
| Sex | |||
| Male | 30 | 3.60 | |
| Female | 18 | 3.83 | 0.62 |
| Tumor location | |||
| Colon | 23 | 3.27 | |
| Rectum | 25 | 4.07 | 0.08 |
| Tumor invasion | |||
| T1, T2 | 10 | 3.03 | |
| T3, T4 | 38 | 3.86 | 0.14 |
| Lymphatic metastasis | |||
| Positive | 26 | 3.16 | |
| Negative | 22 | 4.08 | 0.03 |
| Distant metastasis | |||
| M0 | 43 | 3.72 | |
| M1 | 5 | 3.40 | 0.68 |
| Differentiation | |||
| Well and Moderate | 34 | 3.91 | |
| Poor | 14 | 3.14 | 0.13 |
Next we wanted to know whether miR-301a was expressed in CRC cell lines with differential metastatic potential and whether the expression was associated with the metastatic ability. We conducted the migration assay with five CRC cell lines and found SW620 and HCT116 cells exhibited high metastatic ability while LOVO being the moderate and HT29 and SW480 as less metastatic cell lines (Figure 1C). Subsequently, we evaluated the expression of miR-301a in these cells and as shown in Figure 1D, the expression of miR-301a was much higher in SW620 and HCT116 than that of LOVO, HT29, and SW480. Specifically, expression of miR-301a seemed to be correlated with the metastatic ability of CRC cells by spearman correlation analysis (r = 0.96, P < 0.01). Altogether, these data indicated that miR-301a may contribute to the metastasis of CRC.
miR-301a promotes CRC cell migration and invasion in vitro
To further determine whether miR-301a could affect CRC cell migration, we transfected SW620 cells, which showed a high level of miR-301a, with the anti-miR-301a in order to suppress miR-301a (i.e. SW620/anti-miR-301a cells). Meanwhile, anti-miR-control was transfected as negative control (i.e. SW620/anti-miR-NC cells). Transient transfection of anti-miR-301a in SW620 cells significantly suppressed the expression of miR-301a in SW620/anti-miR-301a cells (Figure 2A, P < 0.01). Next we conducted cell migration assay to evaluate whether miR-301a could influence metastasis directly. As shown in Figure 2C, SW620/anti-miR-301a cells showed a significant (P < 0.01) reduction of migratory ability, while, unexpectedly, the invasive capacity was also significantly (P < 0.01) decreased. To further support the role of miR-301a in migration, we performed the wound-healing assays to explore cell motility change. In consistent with the migratory change, SW620/anti-miR301a cells exhibited significant lower migratory potential compared with the control cells (Figure 2E, P < 0.01).
Figure 2.
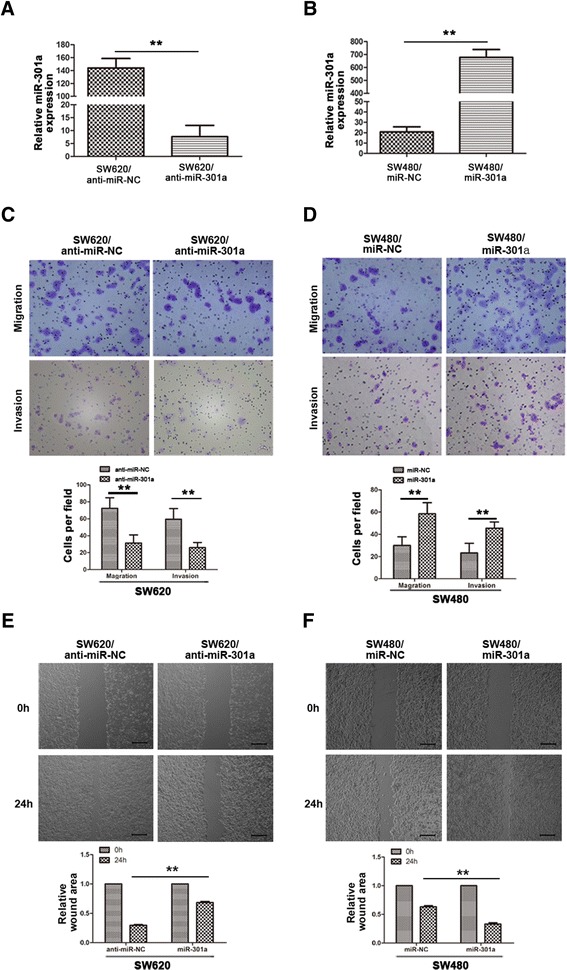
miR-301a promoted CRC cell migration and invasion in vitro . (A) Expression levels of miR-301a in SW620 cells after transfection with inhibitors of miR-301a, as determined by qRT-PCR. U6 small nuclear RNA was used as an internal control. (B) Expression levels of miR-301a in SW480 cells after transfection with synthetic miR-301a mimics, as determined by qRT-PCR. U6 small nuclear RNA was used as an internal control. (C) Downregulation of miR-301a with its inhibitors impeded cell migration and invasion of SW620 cells. (D) Overexpression of miR-301a significantly promoted cell migration and invasion in SW480 cells. (E) miR-301a inhibitors significantly inhibited SW620 cell motility as shown in wound healing assay. (F) miR-301a mimics significantly promoted SW480 cell migration in wound healing assay. Data were representedas means ± SD from 3 independent experiments. **, P < 0.01.
Additionally, the gain-of-function assay with miR-301a mimics transfected in miR-301a low-expressed SW480 cells was performed to further consolidate the role of miR-301a in cell migration and invasion. As shown in Figure 2B, exogenous expression of miR-301a in SW480 cells resulted in a significant increase of miR-301a by qRT-PCR (P < 0.01). Upregulation of miR-301a in SW480 cells led to a marked increase of both migratory and invasive abilities in contrast with the control cells (Figure 2D, P < 0.01). Consistently, SW480 cells transfected with miR-301a mimics migrated much faster than the control cells in the wound-healing assay (Figure 2F, P < 0.01). Conclusively, miR-301a promotes the migration and invasion ability of CRC cells in vitro.
miR-301a promotes tumor metastasis and cell invasion in vivo
The above mentioned results further led us to ask whether miR-301a functioned as a promoter of migration and invasion of CRC cells in vivo as well. To this end, we constructed miR-301a stably down-regulated and negative control SW620 cells with lentivirus particles LV-anti-miR-301a and LV-anti-miR-301a-NC, respectively. SW620/LV-anti-miR-301a cells and SW620/LV-anti-miR-NC cells were separately injected into nude mice subcutaneously and mice were sacrificed four weeks later. Tumors were processed and stained with H&E staining. As shown in Figure 3A, tumors grown of SW620/LV-anti-miR-301a cells were less-invasive as most tumors (4/5) confined within the fibrous capsules without breaking into the stromal (Figure 3A, right panel). In contrast, tumors of the control group had cancer nests formed by SW620/LV-anti-miR-NC cells which invaded the adjacent stromal tissues (Figure 3A, left panel).
Figure 3.
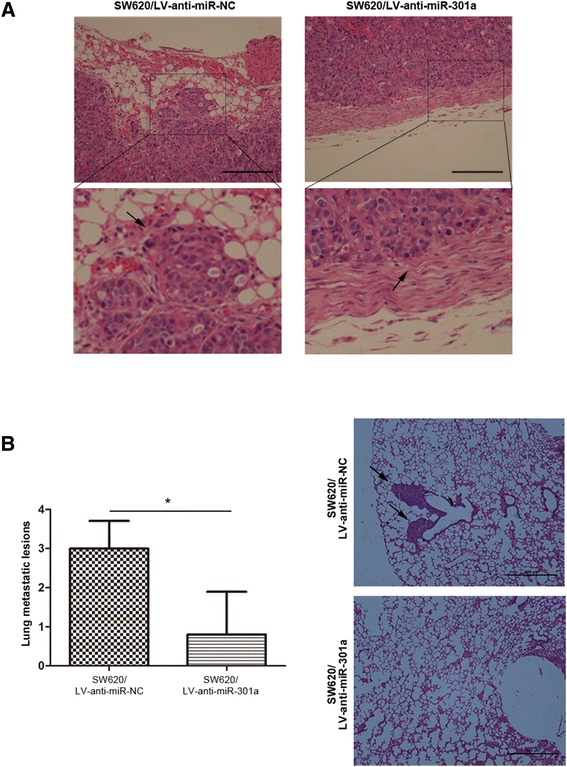
Downregualtion of miR-301a suppressed CRC cell invasion and lung metastases in vivo . (A) miR-301a inhibited CRC cell invasion. H&E staining of tumor acquied from nude mice bearing the SW620/LV-anti-miR-301a or SW620/LV-anti-miR-NC cells (indicated by arrows), 4 weeks post implantation. (B) Mean lung metastatic nodules (left) and histologic analysis (H&E staining) of lung (right) 7 weeks post implantation. Metastatic tumor lesions are indicated by arrows. Data were represented as means ± SD. Magnification, ×200. Scale bars, 100 μm. *, P < 0.05.
To validate the role of miR-301a in metastasis in vivo, we dissected the lung of nude mice and evaluated the metastatic sites both micro- and macroscopically. In line with the in vitro results, fewer metastatic lesions in the lung of nude mice injected with SW620/LV-anti-miR-301a were seen compared with that of SW620/LV-anti-miR-NC group (Figure 3B; P < 0.05), indicating the suppression of metastatic ability in SW620/LV-anti-miR-301a cells in vivo. Hence, miR-301a could promote tumor metastasis and invasion in vivo.
miR-301a represses TGFBR2 protein expression by binding to 3’-UTR
It’s well-established that miRNAs exert their biological functions by binding to the 3’-UTR and inhibiting the translation of their target genes. Thus we wanted to know the target through which miR-301a exerted its function in CRC cells. For this reason, we used bioinformatics method to predict the potential targets of miR-301a. By searching the online TargetScan and PicTar algorithms systems, we identified transforming growth factor receptor 2 (TGFBR2) as the target of miR-301a with the highest possibility.
TGFBR2 is a major trans-membrane receptor of transforming growth factor-β (TGF-β). Upon binding with its ligand TGF-β, TGFBR2 is phosphorylated at the serine and threonine residues within its GS box, which could activate the TGF-β signaling pathway and lead to acquisition of resistance to the anti-mitogenic effects of TGF-β and contribute to tumor development and progression [30,31].
Thus, we attempted to figure out whether TGFBR2 was a downstream target of miR-301a or not. qRT-PCR and western blot analysis were first employed and somehow unexpectedly, there was no significant difference of TGFBR2 mRNA levels between SW620/anti-miR-301a and SW620/anti-miR-NC cells or SW480/miR-301a and SW480/miR-NC cells (Figure 4A). However, western blot demonstrated that expression of TGFBR2 protein was markedly increased in miR-301a downregulated SW620 cells and significantly reduced in miR-301a overexpressed SW480 cells (Figure 4B). This suggested that miR-301a suppressed TGFBR2 by post-transcriptional mechanism rather than directly inhibiting the transcription. Besides, suppression of TGBFR2 by miR-301a was also validated in vivo: in the xenograft tumors from SW620/LV-anti-miR-301a nude mice, staining intensity of TGFBR2 elevated remarkably compared to that of the SW620/LV-anti-miR-NC tumors (Figure 4C). Combined together, these results suggested that miR-301a suppressed TGFBR2 in CRC cells.
Figure 4.
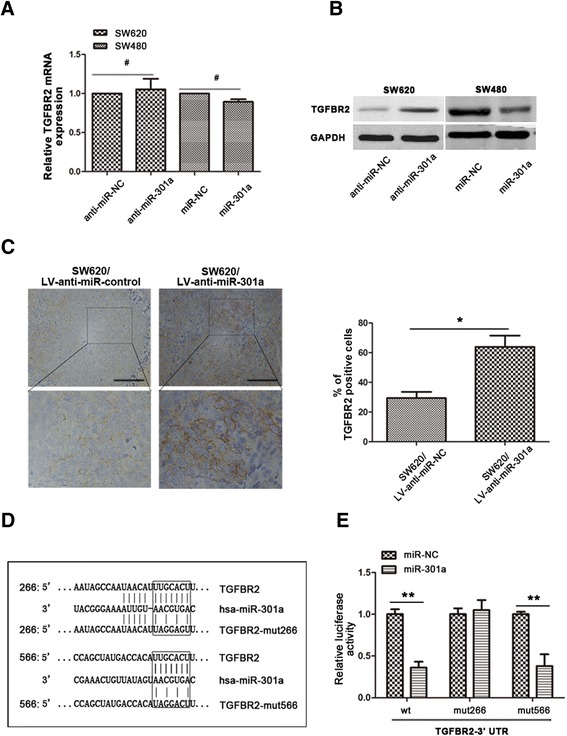
TGFBR2 was a direct target gene of miR-301a. (A) qRT-PCR showed that neither miR-301a mimics nor anti-miR-301a changed the mRNA level of TGFBR2. (B) anti-miR-301a in SW620 cells suppressed, while miR-301a mimics in SW480 cells increased the protein expression of TGFBR2. (C) Immunohistochemical staining of TGFBR2 in tumor sections. Expression of TGFBR2 was evaluated in tumors formed by SW620/LV-anti-miR-301a cells. (D) Schematic picture of two miR-301a binding sites and the corresponding mutants on TGFBR2 3’-UTR. (E) Luciferase activity assay identified the binding sites of miR-301a. Data were representedas means ± SD from 3 independent experiments. Magnification, ×200. Scale bars, 100 μm. *, P < 0.05; **, P < 0.01.
We then wanted to know the mechanism through which miR-301a regulated TGFBR2. In the 3’-UTR of TGRBR2 mRNA there existed two putative binding sites for miR-301a: a highly conserved 7mer-m8 at nt 266-272 and a poorly conserved 7mer-m8 at position 566-572 (Figure 4D). To clarify whether miR-301a could interact with the 3’-UTR of TGFBR2, we performed luciferase reporter assays. Wild type and two mutant fragments of 3’-UTR were co-transfected with miR-301a mimics or NC, respectively. As shown in Figure 4E, compared to the miR-NC, miR-301a mimics significantly (P < 0.01) reduced the relative luciferase activity of pMIR/TGFBR2-wt and pMIR/TGFBR2-mut566 of TGFBR2. However, miR-301a did not affect the relative luciferase activity of the pMIR/TGFBR2-mut266, suggesting that miR-301a specifically bind to the conserved 7mer-m8 at nt 266-272 on 3’-UTR of TGFBR2. These data demonstrated unambiguously that miR-301a suppressed TGFBR2 protein expression via targeting its specific RNA binding site.
TGFBR2 is a functional target through which miR-301a regulated metastasis and invasion in CRC cells
The results presented above confirmed TGFBR2 as one downstream target of miR-301a; however, it remained unclear whether miR-301a regulated metastasis and invasion in CRC cells through modulating TGFBR2. To this end, we focused on whether artificial expression or suppression of TGFBR2 protein could offset the effects caused by miR-301a mimics or anti-miR-301a, respectively. First, siRNA targeting TGFBR2 was transiently transfected into SW480 cells and as shown in Figure 5A, expression of TGFBR2 was markedly suppressed and this resulted in enhanced migration and invasion in SW480 cells (Figure 5B, P < 0.01), suggesting the involvement of TGFBR2 in CRC cell metastasis and invasion.
Figure 5.
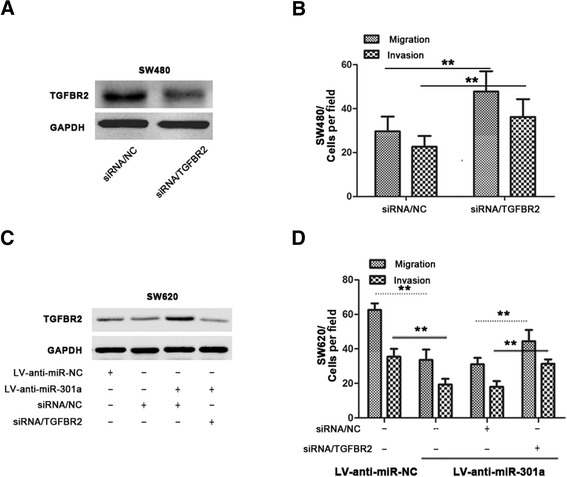
miR-301a regulated metastasis and invasion through suppressing TGFBR2 in CRC cells. (A) Western bloting showed TGFBR2 was downregulated in SW480 cells transiently transfected with TGFBR2 siRNA. (B) The cell invasion and migration ability were improved in TGFBR2 downregulated SW480 cells. (C) Expression of TGFBR2 protein in SW620/LV-anti-miR-301a or SW620/LV-anti-miR-NC cells transiently transfected with TGFBR2 siRNA or siRNA/NC. (D) Cell invasion and migration assays of SW620/LV-anti-miR-301a or SW620/LV-anti-miR-NC cells transiently transfected with TGFBR2 siRNA or siRNA/NC. Data were representedas means ± SD from 3 independent experiments. **, P < 0.01.
To further determine whether TGFBR2 is responsible for the migration and invasion caused by miR-301a, we transfected TGFBR2 siRNA into SW620/LV-anti-miR-301a cells transiently. As shown in Figure 5C, the enhanced TGFBR2 level by LV-anti-miR-301a was depleted upon TGFBR2 siRNA transfection. Remarkably, compared with the SW620/LV-anti-miR-301a cells treated with siRNA/NC, SW620/LV-anti-miR-301a cells transfected with TGFBR2 siRNA regained the previously abrogated migration and invasion ability (Figure 5D, P < 0.01). Additionally, we evaluated the expression of TGFBR2 in CRCs. The archived tumors of the included 48 cases were retrieved and stained. As shown in Figure 6, higher TGFBR2 staining positivity was seen in tumors with low miR-301a expression; furthermore, we quantified the TGFBR2 staining intensity and made a correlation analysis between TGFBR2 and miR-301a. Expression of TGFBR2 was significantly and inversely associated with the expression of miR-301a (r = -0.72, P < 0.001). Conclusively, these results suggest that downregulation of TGFBR2 is involved in miR-301a-induced metastasis and invasion; TGFBR2 is a functional target of miR-301a.
Figure 6.
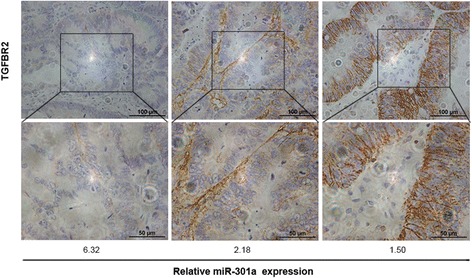
Immunohistochemical staining of TGFBR2 in CRCs. Higher expression of TGFBR2 was observed mainly in CRCs with low miR-301a level (r = -0.72, P < 0.001). Magnification, ×200. Scale bars, 100 μm.
Discussion
Cancer metastasis is a complex, multi-step process which includes intricate cascades of molecular events. It’s well-established that miRNAs are involved in CRC tumorigenesis and many of them can serve as activators or suppressors of CRC metastasis [8-10,13]. miR-301a, one micro-RNA reported to be expressed at different levels in a variety of cancers, has been demonstrated to be important in promoting cancer metastasis [17,21,22]. However, the expression of miR-301a in CRCs and its role in CRC metastasis remain unclear. In the current study, we reported for the first time that miR-301a was up-regulated in lymph node metastatic CRC tissues and promoted CRC metastasis both in vitro and in vivo. Furthermore, we identified TGFBR2 to be one downstream target of miR-301a and also the effector of miR-301a in the regulation of metastasis in CRC cells.
miR-301a has been discovered up-regulated in various types of human cancers and proved to be an oncogene in gastric cancer [22], pancreatic cancer [32], hepatocellular cancer [21], and esophageal cancer [33]. In this study, we investigated the expression of miR-301a in CRC specimens for the first time. Unlike its expressions in the cancers mentioned above, there is no statistical difference of miR-301a expression between CRC tissues and their adjacent non-tumor tissues, nevertheless, increased miR-301a expression was observed in tumors with lymph node metastasis. This was in consistent with its role as an oncogene in aforementioned cancers and might indicate a metastasis promotion role rather than a tumor initiation role of miR-301a in CRCs. In supporting this, we found CRC cell lines with high metastasis capacity expressed higher miR-301a than those with low metastasis capacity and we went a step further by showing that miR-301a expression was correlated positively with the cell migratory ability in these CRC cells. These data led us to ask whether miR-301a could regulate metastasis in CRCs or not.
Actually, previous studies have shown that miR-301a acted as a metastatic promoter in different kinds of human cancers. Shi et al first reported the expression of miR-301a in human cancer: miR-301a overexpression has been implicated as a negative prognostic indicator in lymph node negative invasive ductal breast cancer and strongly associated with tumor recurrence. Besides knockdown of miR-301a reduced migration and invasion of both MCF-7 and MDA-MB-231 breast cancer cell lines in vitro [17]. Likewise, a recent study described that the expression level of miR-301a was significantly elevated in the primary tumors of 10 breast cancer patients with distant metastasis, as compared with that in primary tumors from 10 patients without detectable distant metastasis [34]. Additionally, overexpression of miR-301a in breast cancer cell lines could promote cell migration and invasion in vitro and lung metastases in mice models. Moreover, downregulation of miR-301a in hepatocellular carcinoma (HCC) cells suppressed invasion and migration of HCC cells in vitro [21]. In gastric cancer cells, ectopic expression of miR-301a led to significantly enhanced cell migration and invasion, whereas knockdown of miR-301a also inhibited cell metastasis [22].
In line with these findings, we demonstrated that overexpression of miR-301a in less aggressive SW480 cells led to a significant increase of cell migration whereas downregulation of miR-301a in more aggressive SW620 cells resulted in decreased migratic ability, indicating the metastasis promoter role of miR-301a in CRC. Furthermore, this effect was also prominent in vivo: knockdown of miR-301a significantly reduced xenograft tumor invasion and lung metastases in nude mice. These results indicated that miR-301a is a metastatic promoter in CRC.
miRNAs exert their biological functions by binding with the 3’-UTR of their target genes. Thus, identifying the downstream target genes could help understand miRNA physiological and pathological roles. Though several approaches have been developed to search miRNA targets in recent years, the computational approach, a method based on the principle that all known miRNA targets have conserved perfect or near-perfect complementary sites for miRNAs, was a powerful and prevalently used strategy [35-37]. Thus, we used the online TargetScan and PicTar algorithms systems to search for potential targets of miR-301a. As a result, TGFBR2, a gene previously reported involved in metastasis, was identified to be the target with highest possibility.
TGFBR2, a trans-membrane serine-threonine kinase, is a major member of TGF-β signaling and often found to be altered in CRCs [38]. The TGF-β signaling pathway plays a complex role in several CRC biological processes, including cell differentiation, proliferation, apoptosis, and motility [39]. It is commonly speculated that CRCs acquire partial TGF-β resistance largely because of TGFBR2 inactivation. Importantly, TGFBR2 is a metastatic suppressor in CRC: previous research showed that downregulation of TGFBR2 increased migration and invasion of HT29 cells in vitro [27] whereas evidences from in vivo studies suggested that loss of TGFBR2 contributed to colon cancer development and metastasis in cooperation with mutant K-ras or in the context of PTEN loss [40,41]. Indeed we found TGFBR2 was one target of miR-301a and the luciferase reporter gene assay successfully identified the binding site for miR-301a on the 3’-UTR of TGFBR2 gene. In consistent with this, we found knockdown of miR-301a in CRC cells upregulated the protein level of TGFBR2 while ectopically overexpression of miR-301a suppressed its expression. Supporting this, we observed an inverse correlation between miR-301a and TGFBR2 protein expression in CRC tumor tissues. Functionally, we found TGFBR2 acted as a metastasis suppressor and also the effector of miR-301a in CRC cells: knockdown of TGFBR2 induced a significant increase in motility and invasiveness of SW480 cells whereas downregulation of TGFBR2 also elevated the previously abrogated migration and invasion ability initiated by LV-anti-miR-301a in SW620 cells. Taken together, TGFBR2 played an important role in miR-301a mediated tumor migration and invasion in CRC.
Interestingly, we found miR-301a regulated TGFBR2 expression by some translational suppression mechanisms: the mRNA level of TGFBR2 did not change significantly upon transfection with miR-301a inhibitor or mimics while the protein level showed remarkable differences. It’s proposed that miRNAs exert their functions in two ways: when an miRNA perfectly or near-perfectly pairs with its target mRNAs, it was thought that mRNA cleavage is the primary mechanism for miRNA mediated gene regulation; in case of a miRNA imperfectly pairs with its target mRNAs, translational suppression is thought to happen with protein expression downregulated while mRNA level remained stable [42,43]. In this study, though we found miR-301a indeed bound with TGFBR2, they might not perfectly match in sequence since we observed marked downregulation of TGFBR2 protein while the mRNA of TGFBR2 remained unchanged. Thus we suggest that miR-301a imperfectly bind with the 3’-UTR of TGFBR2 and regulate the expression of TGFBR2 via translational suppression.
Conclusions
In summary, our study presented convincing evidences to show that miR-301a exerted its role as oncogene in CRC metastasis, and for the first time we identified TGFBR2 as a functional target involved in miR-301a modulated CRC cell metastasis. miR-301a could be regarded as a new target for CRC prevention and therapy.
Acknowledgements
This work was supported by grants from the Scientific Research Foundation of science and Technology Commission of Shanghai Municipality (No. 14ZR1426300).
Abbreviations
- miRNAs
microRNAs
- miR-301a
microRNA-301a
- CRC
Colorectal cancer
- TGFBR2
Transforming growth factor receptor 2
- 3’-UTR
3’ un-translated regions
- qRT-PCR
Quantitative real-time polymerase chain reaction
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- ATCC
American Type Culture Collection
- cDNA
Complementary DNA
- nt
Nucleotide
- FBS
Fetal Bovine Serum
- NC
Negative control
- HE
Hematoxylin-eosin
- FBS
Fetal bovine serum
- DMEM
Dulbecco’s modified Eagle’s medium
- RNU6B
U6 small nuclear RNA
- HCC
Hepatocellular carcinoma
- PTEN
Phosphatase and tensin homologue on chromosome 10
Footnotes
Wenpeng Zhang and Tao Zhang contributed equally
Competing interests
The authors declare no competing financial interests.
Authors' contributions
MLW conceived and designed the experiments. WPZ, TZ, HCZ, JH and RSJ performed the experiments. WPZ, BF and LZ analyzed the data. WPZ wrote the paper. MLW and MHZ supervised the whole experimental work and revised the manuscript. All authors read and approved the manuscript.
Authors’ information
Tao Zhang co-first author.
Contributor Information
Wenpeng Zhang, Email: darvid0610@sina.com.
Tao Zhang, Email: woodyhom@yahoo.com.
Runsen Jin, Email: nkvincent@163.com.
Hongchao Zhao, Email: zhaohongchao9037@163.com.
Jin Hu, Email: vincyhu16@hotmail.com.
Bo Feng, Email: fengbo2022@163.com.
Lu Zang, Email: zanglu@yeah.net.
Minhua Zheng, Email: zmhtiger@yeah.net.
Mingliang Wang, Email: mingliang-99@hotmail.com.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Surveillance Epidemiology and End Results (SEER), Cancer Statistics [http://seer.cancer.gov/statfacts/html/colorect.html] [DOI] [PubMed]
- 3.Kindler HL, Shulman KL. Metastatic colorectal cancer. Curr Treat Options Oncol. 2001;2:459–471. doi: 10.1007/s11864-001-0068-7. [DOI] [PubMed] [Google Scholar]
- 4.Ma R, Jiang T, Kang X. Circulating microRNAs in cancer: origin, function and application. J Exp Clin Cancer Res. 2012;31:38. doi: 10.1186/1756-9966-31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Chen X, Yu X, Tao Y, Bode AM, Dong Z, Cao Y. Regulation of microRNAs by epigenetics and their interplay involved in cancer. J Exp Clin Cancer Res. 2013;32:96. doi: 10.1186/1756-9966-32-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo L, Bai H, Zou D, Hong T, Liu J, Huang J, He P, Zhou Q, He J. The role of microRNA-133b and its target gene FSCN1 in gastric cancer. J Exp Clin Cancer Res. 2014;33:99. doi: 10.1186/s13046-014-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang CJ, Stratmann J, Zhou ZG, Sun XF. Suppression of microRNA-31 increases sensitivity to 5-FU at an early stage, and affects cell migration and invasion in HCT-116 colon cancer cells. BMC Cancer. 2010;10:616. doi: 10.1186/1471-2407-10-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faltejskova P, Besse A, Sevcikova S, Kubiczkova L, Svoboda M, Smarda J, Kiss I, Vyzula R, Slaby O. Clinical correlations of miR-21 expression in colorectal cancer patients and effects of its inhibition on DLD1 colon cancer cells. Int J Colorectal Dis. 2012;27:1401–1408. doi: 10.1007/s00384-012-1461-3. [DOI] [PubMed] [Google Scholar]
- 10.Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, Wang WC, Lin FM, Chen Z, Huang HD, Shyy JY, Liang JT, Chen RH: miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4.Cancer Res 2012, 72:3631–3641. [DOI] [PubMed]
- 11.Tokarz P, Blasiak J. The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment. Acta Biochim Pol. 2012;59:467–474. [PubMed] [Google Scholar]
- 12.Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, Eisbacher M, Zhang C, Tran N, Fan H, Retzlaff K, Bittner A, Raponi M: Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer.BMC Cancer 2009, 9:374. [DOI] [PMC free article] [PubMed]
- 13.Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR, Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315–1326. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z, Li Y, Takwi A, Li B, Zhang J, Conklin DJ, Young KH, Martin R. miR-301a as an NF-kappaB activator in pancreatic cancer cells. EMBO J. 2011;30:57–67. doi: 10.1038/emboj.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi W, Gerster K, Alajez NM, Tsang J, Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, McCready D, Fyles A, Liu FF: MicroRNA-301 mediates proliferation and invasion in human breast cancer.Cancer Res 2011, 71:2926–2937. [DOI] [PubMed]
- 18.Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia W, Li J, Li S, Chen L, Ding H, Zhao Q, Fan M, Shen B, Shao N: Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways.Biochem Biophys Res Commun 2010, 396:978–982. [DOI] [PubMed]
- 19.Liu C, Kelnar K, Vlassov AV, Brown D, Wang J, Tang DG. Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res. 2012;72:3393–3404. doi: 10.1158/0008-5472.CAN-11-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu YC, Chen YJ, Wang HM, Tsai CY, Chen WH, Huang YC, Fan KH, Tsai CN, Huang SF, Kang CJ, Chang JT, Cheng AJ: Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling.Cancer Prev Res (Phila) 2012, 5:665–674. [DOI] [PubMed]
- 21.Zhou P, Jiang W, Wu L, Chang R, Wu K, Wang Z. miR-301a is a candidate oncogene that targets the homeobox gene Gax in human hepatocellular carcinoma. Dig Dis Sci. 2012;57:1171–1180. doi: 10.1007/s10620-012-2099-2. [DOI] [PubMed] [Google Scholar]
- 22.Wang M, Li C, Yu B, Su L, Li J, Ju J, Yu Y, Gu Q, Zhu Z, Liu B. Overexpressed miR-301a promotes cell proliferation and invasion by targeting RUNX3 in gastric cancer. J Gastroenterol. 2013;48:1023–1033. doi: 10.1007/s00535-012-0733-6. [DOI] [PubMed] [Google Scholar]
- 23.Monzo M, Navarro A, Bandres E, Artells R, Moreno I, Gel B, Ibeas R, Moreno J, Martinez F, Diaz T, Martinez A, Balagué O, Garcia-Foncillas J: Overlapping expression of microRNAs in human embryonic colon and colorectal cancer.Cell Res 2008, 18:823–833. [DOI] [PubMed]
- 24.Hu H, Krasinskas A, Willis J. Perspectives on current tumor-node-metastasis (TNM) staging of cancers of the colon and rectum. Semin Oncol. 2011;38:500–510. doi: 10.1053/j.seminoncol.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Zhao H, Dong T, Zhou H, Wang L, Huang A, Feng B, Quan Y, Jin R, Zhang W, Sun J, Zhang D, Zheng M: miR-320a suppresses colorectal cancer progression by targeting Rac1.Carcinogenesis 2014, 35:886–895. [DOI] [PubMed]
- 26.Geback T, Schulz MM, Koumoutsakos P, Detmar M. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques. 2009;46:265–274. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]
- 27.Feng B, Dong TT, Wang LL, Zhou HM, Zhao HC, Dong F, Zheng MH. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS One. 2012;7:e43452. doi: 10.1371/journal.pone.0043452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye Q, Feng B, Peng YF, Chen XH, Cai Q, Yu BQ, Li LH, Qiu MY, Liu BY, Zheng MH. Expression of gamma-synuclein in colorectal cancer tissues and its role on colorectal cancer cell line HCT116. World J Gastroenterol. 2009;15:5035–5043. doi: 10.3748/wjg.15.5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao Z, Sun J, Feng B, Ma J, Zang L, Dong F, Zhang D, Zheng M. The metastasis suppressor, N-myc downregulated gene 1 (NDRG1), is a prognostic biomarker for human colorectal cancer. PLoS One. 2013;8:e68206. doi: 10.1371/journal.pone.0068206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gobbi H, Arteaga CL, Jensen RA, Simpson JF, Dupont WD, Olson SJ, Schuyler PA, Plummer WD, Jr, Page DL. Loss of expression of transforming growth factor beta type II receptor correlates with high tumour grade in human breast in-situ and invasive carcinomas. Histopathology. 2000;36:168–177. doi: 10.1046/j.1365-2559.2000.00841.x. [DOI] [PubMed] [Google Scholar]
- 31.Gobbi H, Dupont WD, Simpson JF, Plummer WD, Jr, Schuyler PA, Olson SJ, Arteaga CL, Page DL. Transforming growth factor-beta and breast cancer risk in women with mammary epithelial hyperplasia. J Natl Cancer Inst. 1999;91:2096–2101. doi: 10.1093/jnci/91.24.2096. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z, Chen LY, Dai HY, Wang P, Gao S, Wang K. miR-301a promotes pancreatic cancer cell proliferation by directly inhibiting Bim expression. J Cell Biochem. 2012;113:3229–3235. doi: 10.1002/jcb.24200. [DOI] [PubMed] [Google Scholar]
- 33.Su H, Jin X, Zhang X, Xue S, Deng X, Shen L, Fang Y, Xie C. Identification of microRNAs involved in the radioresistance of esophageal cancer cells. Cell Biol Int. 2014;38:318–325. doi: 10.1002/cbin.10202. [DOI] [PubMed] [Google Scholar]
- 34.Ma F, Zhang J, Zhong L, Wang L, Liu Y, Wang Y, Peng L, Guo B. Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt/beta-catenin signaling. Gene. 2014;535:191–197. doi: 10.1016/j.gene.2013.11.035. [DOI] [PubMed] [Google Scholar]
- 35.Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- 36.Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 37.Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA::target interaction. Chem Biol. 2004;11:1619–1623. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Pasche B. TGF-beta signaling alterations and susceptibility to colorectal cancer. Hum Mol Genet. 2007;16 Spec No 1:R14–20. doi: 10.1093/hmg/ddl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 40.Trobridge P, Knoblaugh S, Washington MK, Munoz NM, Tsuchiya KD, Rojas A, Song X, Ulrich CM, Sasazuki T, Shirasawa S, Grady WM. TGF-beta receptor inactivation and mutant Kras induce intestinal neoplasms in mice via a beta-catenin-independent pathway. Gastroenterology. 2009;136:1680–1688 e1687. doi: 10.1053/j.gastro.2009.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu M, Trobridge P, Wang Y, Kanngurn S, Morris SM, Knoblaugh S, Grady WM. Inactivation of TGF-beta signaling and loss of PTEN cooperate to induce colon cancer in vivo. Oncogene. 2014;33:1538–1547. doi: 10.1038/onc.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 43.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]


