Abstract
Peptoniphilus spp. are Gram-positive anaerobic cocci (GPAC) that were formerly classified in the genus Peptostreptococcus. This study describes 15 cases of Peptoniphilus spp. bloodstream infection (BSI) diagnosed from 2007 to 2011 using 16S rDNA sequencing in patients with pneumonia, pre-term delivery, soft tissue infection or colon or bladder disease. Seven out of 15 (47%) of these cases had polymicrobial BSIs. One of the isolates was closely related to P. duerdenii (EU526290), while the other 14 isolates were most closely related to a Peptoniphilus sp. reference strain (ATCC 29743) and P. hareii (Y07839). Peptoniphilus is a rare but important cause of BSI.
Keywords: Anaerobic bacteria, bacteraemia, Gram-positive cocci, Peptoniphilus, 16S ribosomal DNA sequencing
Gram-positive anaerobic cocci (GPAC) are not routinely recovered or identified from many types of clinical specimens because they may be difficult to grow in culture and are frequently isolated as part of mixed polymicrobial infections 1. Peptoniphilus are commensals of the human vagina and gut that were formerly classified in the genus Peptostreptococcus 2. Recent implementation of non-biochemical methods including MALDI-TOF and 16S rDNA sequencing has allowed their accurate identification from a variety of clinical specimens. There are now more than 15 Peptoniphilus species within the genus, seven of which were discovered in 2012 3–10. Several new Peptoniphilus spp. were also recently discovered as part of a study of the human gut microbiome in which microbial culturomics was used to complement genetic analyses 11. To date, Peptoniphilus spp. have most commonly been associated with diabetic skin and soft tissue infections, bone and joint infections and surgical site infections 12–15. A recent study of pre-term labour and early neonatal sepsis also isolated Peptoniphilus spp. from amniotic fluid causing choramnionitis 16. To our knowledge, our study is the first case series of Peptoniphilus spp. causing bloodstream infection (BSI) either alone or as part of a polymicrobial infection.
A retrospective review of all adult patients (>17 years) with at least one blood culture positive for Peptoniphilus spp. was carried out. All cases were diagnosed by the regional centralized clinical microbiology laboratory, Calgary Laboratory Services (CLS), between 1 July 2007 and 31 December 2012. The study protocol was approved by the Conjoint Health Research Ethics Board at the University of Calgary.
All isolates were recovered from blood cultures. The regional protocol for adult blood culture draws routinely collects two sets, each consisting of both an aerobic (iFA) and anaerobic (iFN) bottle with incubation, culture and growth monitoring being performed by the BacT/Alert 3D System (bioMérieux, Laval, QC, Canada). Positive blood cultures were immediately pelleted, Gram stained and plated to standard aerobic and anaerobic culture media. Peptoniphilus spp. isolates demonstrated typical GPAC morphology on Gram stain from anaerobic Brucella blood agar. Prior to 2011, anaerobic GPAC isolates were preliminarily identified using antibiotic (KVC) disc testing and biochemical analysis with the Vitek 2 ANC card (bioMérieux), but subsequently, MALDI-TOF (Vitek MS; bioMérieux) has been used. Antibiotic susceptibility testing was performed using E-test strips (bioMérieux) according to CLSI guidelines and the manufacturer's instructions 17. All isolates were susceptible to penicillin and metronidazole, and all but one of the isolates (case #7) was also susceptible to clindamycin.
Preliminary GPAC identifications were confirmed by performing partial sequencing of the 16S rDNA gene using standard methods 2,10,18 (ABI Prism 3130 sequencer; Applied Biosystems, Foster City, CA, USA). The SmartGene Integrated Database Network System (IDNS) indicated the most closely related species (per cent identity >99%) as shown in Table1. A maximum likelihood phylogeny was produced illustrating the relation of the 15 clinical isolates with publicly available sequences 19,20. Genus-level identity (99–100%) was achieved for all 16S rDNA sequences in this study. Species level identification by the IDNS Smartgene database often resulted in multiple species with >99% sequence similarity. Independent phylogenetic analysis of the partial 16S rDNA sequences confirmed the similarity of CLS sequence KF963596 with P. duerdenii while the remaining 14 CLS sequences shared highest similarity with a Peptoniphilus spp. (ATCC 29743). These sequences were also highly similar (99.6%) to P. harei (Fig.1).
Table 1.
Clinical characteristics and blood culture results of Peptoniphilus BSI cases
| Study ID# | Age & Sex | Significant past medical history | Discharge diagnoses | Blood culture –ID by Gram stain and either MALDI-TOF or Vitek2 ANC carda | ID by 16S sequence using Smartgene IDNS database (Genbank accession) | Sequence length (bp) | Days antibiotics (IV & PO) from day of positive blood draw | Outcome |
|---|---|---|---|---|---|---|---|---|
| 01 | 20F | Remote Guillan Barre Syndrome 2nd CMV | PROM (25 weeks), chorioamnionitis | MALDI: anaerobic Gram positive cocci | Peptoniphilus sp.DQ986463 (KF963587) | 505 | 1 | Discharged home, infant survived |
| 03 | 78F | COPD on home oxygen | Acute exacerbation of COPD | Vitek2: Peptoniphilus sp. | Peptoniphilus sp. DQ986463 (KF963593) | 518 | 11 | Discharged to rehabilitation facility |
| 04 | 84F | Remote stroke with deficits requiring total care, nutrition via gastrostomy tube | Pneumonia, sepsis | MALDI: anaerobic Gram-positive cocci | Peptoniphilus sp. DQ986463 (KF963588) | 488 | 17 | Deceased |
| 05 | 96M | Colon cancer diagnosed on admission with probably metastases | Urosepsis, colon cancer invading bladder | MALDI: polymicrobial (Peptoniphilus sp. Aerococcus urinae) | Peptoniphilus sp. DQ986463 (KF963585) | 488 | 18 | Discharged back to long-term care facility |
| 06 | 42F | Remote tubal ligation | Gastroenteritis | Vitek2: polymicrobial (Peptoniphilus sp., E. coli) | Peptoniphilus sp. DQ986463 (KF963595) | 508 | 4 | Discharged home |
| 07 | 54F | Type 2 diabetes mellitus | Diabetes mellitus extremity infection requiring above-knee amputation | MALDI: polymicrobial (Peptoniphilus sp., Group B Streptococcus) | Peptoniphilus sp. DQ986463 (KF963590) | 499 | 12 | Discharged to rehabilitation facility |
| 08 | 33F | None | Septic abortion (8 weeks) | MALDI: Peptoniphilus sp. | Peptoniphilus sp. DQ986463 (KF963583) | 496 | 18 | Discharged home |
| 09 | 66F | Remote bladder cancer with urostomy and ilieal conduit, smoker | Complicated urinary tract infection, bacteraemia | MALDI: Peptoniphilus sp. | Peptoniphilus sp. DQ986463 (KF963582) | 477 | 10 | Discharged home |
| 10 | 34F | End-stage renal disease on haemodialysis via fistula 2nd Goodpasture's disease, cardiac arrest 2004 | Recurrent pericarditis | Vitek2: Peptoniphilus spp. | Peptoniphilus duerdenii (KF963596) | 380 | 7 | Discharged home |
| 11 | 82M | Remote prostate cancer, Crohn's disease on prednisone | Ischaemic colitis with pelvic abscess, B-cell lymphoma of colon | Vitek2: polymicrobial (Peptoniphilus spp., Eggerthella lenta) | Peptoniphilus sp. DQ986463 (KF963592) | 518 | 21 | Deceased |
| 12 | 42M | Renal transplant, on sirolimus, tacrolimus and prednisone | Sepsis, deep vein thrombosis | MALDI: Peptoniphilus sp. | Peptoniphilus sp. DQ986463 (KF963591) | 494 | 13 | Discharged home |
| 13 | 32F | Unknown (paper chart unavailable) | Stillbirth (18 weeks), chorioamnionitis | Vitek2: Peptoniphilus spp. | Peptoniphilus sp. DQ986463 (KF963594) | 438 | 15 | Discharged home, foetal death |
| 14 | 80M | Metastatic prostate cancer with recent venous thromboembolism | Bladder tumour eroding into rectum | MALDI: polymicrobial (Peptoniphilus sp. B. fragilis, S. viridans) | Peptoniphilus sp. DQ986463 (KF963589) | 500 | 3 | Discharged to community hospital |
| 15 | 96M | COPD, pacemaker | Acute exacerbation COPD, aspiration pneumonia, stroke | MALDI: polymicrobial (anaerobic Gram-positive cocci, P. mirabilis,S. aureus) | Peptoniphilus sp. DQ986463 (KF963586) | 492 | 2 | Deceased |
| 16 | 87M | Type 2 diabetes mellitus | Acute renal failure secondary to obstruction, urinary tract infection | MALDI: polymicrobial (Peptoniphilus sp. E. faecalis) | Peptoniphilus sp. DQ986463 (KF963584) | 499 | 13 | Discharged home |
CMV, cytomegalovirus; PROM, premature rupture of membranes; COPD, chronic obstructive pulmonary disease.
For polymicrobial cultures identification was performed on all isolates individually.
Fig 1.
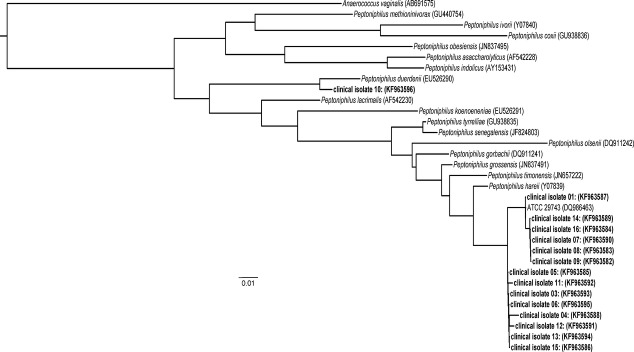
Maximum likelihood phylogeny of partial 16S rDNA (variable regions V1–V3) sequences from the Peptoniphilus BSI isolates (GenBank accessions KF963582–KF96396) in relation to publicly available Peptoniphilus reference strains (Genbank accession in parentheses). Fasttree 2 software was used to create the tree using the generalized time-reversible nucleotide substitution model 20.
Patient characteristics and blood culture results are summarized in Table1. During the study period, 15 hospitalized patients were diagnosed with Peptoniphilus BSI; nine women and six men. The mean age was 62 years (range, 20–96 years; median, 66 years) and the mean length of stay was 18 days (range, 4–62 days; median, 11 days). BSI due to P. duerdenii was confirmed in a 34-year-old female with end-stage renal disease secondary to Goodpasture's disease and recurrent pericarditis. All of the other patients with Peptoniphilus spp. BSI were due to septic abortion with choramnionitis (n = 3), acute exacerbation of COPD and/or pneumonia (n = 3), skin and soft tissue infection (n = 2) or underlying bowel and/or bladder disease (n = 6) (Table1). Seven (50%) patients had polymicrobial BSIs with other bacteria besides Peptoniphilus spp., including (i) Aerococcus urinae, (ii) Escherichia coli, (iii) Group B Streptococcus, (iv) Eggerthella lenta, (v) Bacteroides fragilis and viridians streptococcus group, (vi) Proteus mirabilis and Staphylococcus aureus, and (vii) Enterococcus faecalis. Most polymicrobial BSIs originated from a bowel or bladder source, except for one patient with diabetes mellitus and severe necrotizing infection of the leg, and one elderly patient with aspiration pneumonia.
Only one of the three women with Peptoniphilus BSI secondary to septic abortion/choramnionitis was discharged home with a live infant. Three of 15 (20%) of the patients died, and all of them were elderly with significant underlying co-morbidities. All of the other patients with Peptoniphilus BSI were successfully treated with antibiotics and discharged from hospital.
Peptoniphilus spp. would typically be recovered as a component of mixed bacterial flora in complex abscesses in the abdomen or pelvis. We would therefore expect to recover Peptoniphilus spp. as part of a polymicrobial BSI infection in patients with underlying bowel or bladder disease as occurred in our study. The recovery of Peptoniphilus alone indicates that this organism is a rare but important cause of BSI as a primary pathogen in certain clinical settings (i.e. septic abortion, soft tissue infection in immunocompromised patients and pneumonia).
Sequencing of the 16S rDNA gene is a valuable tool for definitive molecular identification of important clinical isolates that cannot be readily identified by phenotypic methods or MALDI-TOF 21. Peptoniphilus spp. are often misidentified using biochemical methods 22, and MALDI-TOF databases currently do not include most species. Most of our isolates had highly similar sequences to a reference Peptoniphilus spp. strain that has been called P. assacharolyticus ATCC 29743 (GenBank DQ986463). However, it is clear from our phylogenetic analysis (Fig.1) that this reference strain is divergent from the other typed P. assacharolyticus ATCC 14963 (AF542228), and most closely related to P. harei (Y07839). Our results support a previous report of 89.5% 16S rDNA similarity between P. asaccharolyticus strains ATCC 29743 and ATCC 14963 while the ATCC 29743 shares 99.4% similarity with P. harei (Y07839) 22. We have therefore chosen for now to designate most of our isolates as Peptoniphilus spp.
In summary, clinical microbiology laboratories should be aware that Peptoniphilus are rare but important causes of BSI infection either as the primary pathogen or as part of a polymicrobial infection. Isolates meeting the preliminary phenotypic characteristics for GPAC should be referred for definitive identification using partial 16S rDNA sequencing. Delineation of the clinical and epidemiological significance and pathogenic potential of Peptoniphilus spp. in humans is dependent upon further isolates from clinical samples and full phenotypic and genotypic characterizations.
Nucleotide Sequence Accession Numbers
The sequences of the Peptoniphilus strains described here have been deposited in GenBank under accession numbers KF963582–KF963596.
Author Contributions
Kristen Brown: study design, patient chart reviews and manuscript preparation. Tarah Lynch: composed phylogenetic tree and manuscript preparation. Deirdre Church and Daniel Gregson: study design, 16S rDNA sequencing and manuscript preparation.
Transparency Declaration
The authors declare that they have no conflicts of interest.
References
- Veloo AC, Erhard M, Welker M, Welling GW, Degener JE. Identification of Gram-positive anaerobic cocci by MALDI-TOF mass spectrometry. Syst Appl Microbiol. 2011;34:58–62. doi: 10.1016/j.syapm.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L, Shu S. Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol. 2001;51:1521–1528. doi: 10.1099/00207713-51-4-1521. [DOI] [PubMed] [Google Scholar]
- Cho E, Park SN, Kim HK, et al. Draft genome sequence of the novel Peptoniphilus sp. strain ChDC B134, isolated from a human periapical abscess lesion. Genome Announc. 2013;1(pii):e00822–13. doi: 10.1128/genomeA.00822-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron DM, Tyrrell KL, Goldstein EJ. Peptoniphilus coxii sp. nov. and Peptoniphilus tyrrelliae sp. nov. isolated from human clinical infections. Anaerobe. 2012;18:244–248. doi: 10.1016/j.anaerobe.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Kim DS, Jung MY, Kang A, et al. Genome sequence of Peptoniphilus rhinitidis 1-13T, an anaerobic coccus strain isolated from clinical specimens. J Bacteriol. 2012;194:2405–2406. doi: 10.1128/JB.00192-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Hugon P, Lagier JC, et al. Non contiguous-finished genome sequence and description of Peptoniphilus obesi sp. nov. Stand Genomic Sci. 2013;7:357–369. doi: 10.4056/sigs.3276687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Hugon P, Robert C, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Peptoniphilus grossensis sp. nov. Stand Genomic Sci. 2012;7:320–330. doi: 10.4056/sigs.3076460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney AP, Swezey JL, Pukall R, Schumann P, Spring S. Peptoniphilus methioninivorax sp. nov., a Gram-positive anaerobic coccus isolated from retail ground beef. Int J Syst Evol Microbiol. 2011;61:1962–1967. doi: 10.1099/ijs.0.024232-0. [DOI] [PubMed] [Google Scholar]
- Song Y, Liu C, Finegold SM. Peptoniphilus gorbachii sp. nov., Peptoniphilus olsenii sp. nov., and Anaerococcus murdochii sp. nov. isolated from clinical specimens of human origin. J Clin Microbiol. 2007;45:1746–1752. doi: 10.1128/JCM.00213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulger-Toprak N, Lawson PA, Summanen P, O'Neal L, Finegold SM. Peptoniphilus duerdenii sp. nov. and Peptoniphilus koenoeneniae sp. nov., isolated from human clinical specimens. Int J Syst Evol Microbiol. 2012;62:2336–2341. doi: 10.1099/ijs.0.031997-0. [DOI] [PubMed] [Google Scholar]
- Lagier JC, Armougom F, Million M, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS One. 2008;3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolcott RD, Gontcharova V, Sun Y, Dowd SE. Evaluation of the bacterial diversity among and within individual venous leg ulcers using bacterial tag-encoded FLX and titanium amplicon pyrosequencing and metagenomic approaches. BMC Microbiol. 2009;9:226. doi: 10.1186/1471-2180-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Snow DE, Rees E, et al. Evaluation of the bacterial diversity of pressure ulcers using bTEFAP pyrosequencing. BMC Med Genomics. 2010;3:41. doi: 10.1186/1755-8794-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter G, Vernier M, Pinelli PO, et al. Bone and joint infections due to anaerobic bacteria: an analysis of 61 cases and review of the literature. Eur J Clin Microbiol Infect Dis. 2014 doi: 10.1007/s10096-014-2073-3. ; [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Wang X, Buhimschi CS, Temoin S, Bhandari V, Han YW, Buhimschi IA. Comparative microbial analysis of paired amniotic fluid and cord blood from pregnancies complicated by preterm birth and early-onset neonatal sepsis. PLoS One. 2013;8:e56131. doi: 10.1371/journal.pone.0056131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute (CLSI) Methods for antimicrobial susceptibility testing of anaerobic bacteia; approved standards. Wayne, PA: CLSI; 2012. Eighth Edition MM11-A8. [PubMed] [Google Scholar]
- Gontcharova V, Youn E, Sun Y, Wolcott RD, Dowd SE. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol J. 2010;4:8–19. doi: 10.2174/1874285801004010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute (CLSI) Interpretive criteria for identification of Bacteria and Fungi by DNA target sequencing; approved guideline. Wayne, PA: CLSI; 2008. MM18-A, Vol. 28 No. 12. [Google Scholar]
- Veloo AC, Welling GW, Degener JE. Mistaken identity of Peptoniphilus asaccharolyticus. J Clin Microbiol. 2011;49:1189. doi: 10.1128/JCM.00043-11. [DOI] [PMC free article] [PubMed] [Google Scholar]


