Abstract
Ulva zoospores preferentially settle on N-acylhomoserine lactone (AHL) producing marine bacterial biofilms. To investigate whether AHL signal molecules also affect the success and rate of zoospore germination in addition to zoospore attraction, the epiphytic bacteria associated with mature Ulva linza were characterized and bacterial isolates representative of this community tested for the ability to produce AHLs. Two of these AHL-producing isolates, Sulfitobacter spp. 376 and Shewanella spp. 79, were transformed with plasmids expressing the Bacillus spp. AHL lactonase gene aiiA to generate AHL-deficient variants. The germination and growth of U. linza zoospores was studied in the presence of these AHL-deficient strains and their AHL-producing counterparts. This revealed that the AHLs produced by Sulfitobacter spp. and Shewanella spp. or the bacterial products they regulate have a negative impact on both zoospore germination and the early growth of the Ulva germling. Further experiments with Escherichia coli biofilms expressing recombinant AHL synthases and synthetic AHLs provide data to demonstrate that zoospores germinated and grown in the absence of AHLs were significantly longer than those germinated in the presence of AHLs. These results reveal an additional role for AHLs per se in the interactive relationships between marine bacteria and Ulva zoospores.
Introduction
The green seaweed Ulva is the most common macro-alga contributing to biofouling of man-made surfaces throughout the world, mainly due to its tolerance to diverse environmental conditions and antifouling surface coatings, and also to its enormous reproductive potential, with vast quantities of motile zoospores (or zygotes from the fusion of gametes) being released from each thallus (Callow et al., 1997). Once released from the thallus, the swimming zoospores select surfaces for attachment by ‘sensing’ a surface, and this is followed by temporary adhesion. If conditions are unsatisfactory, the zoospore detaches and continues to search for an optimum location. Once conditions are ‘sensed’ as satisfactory, the zoospore excretes a glycoprotein adhesive to form a permanent attachment, differentiates and grows to form a mature alga (Callow et al., 2000a). Several factors are thought to influence the surface selection of Ulva zoospores, including negative phototaxis, surface chemistry, wettability, surface topography and also the presence of a bacterial biofilm (Callow and Callow, 2000; Joint et al., 2000; Callow et al., 2000b; 2002). Zoospores were shown to preferentially settle on top of bacteria, suggesting a direct interaction between the bacteria and zoospores (Joint et al., 2000). Ulva zoospores can also utilize bacterial quorum-sensing (QS) signal molecules, specifically the N-acylhomoserine lactones (AHLs) as a cue for the selection of sites for attachment (Joint et al., 2002; Tait et al., 2005; 2009). QS molecules such as the AHLs are used by bacteria to coordinate their behaviour at the population level and by collectively controlling expression of multiple genes including those involved in secondary metabolism, virulence, motility and biofilm development (Williams et al., 2007). AHLs affect swimming behaviour of the zoospores through a process of chemokinesis (Wheeler et al., 2006), and the detection of AHLs by zoospores causes an increase in intracellular Ca2+ levels (Joint et al., 2007).
In addition to their effect on Ulva zoospore settlement, bacteria appear to have a profound impact on the morphology and growth of Ulva. When grown axenically, Ulva lactuca adopts an atypical ‘pin cushion’ undifferentiated morphology as opposed to the normal foliaceous growth developed when associated with marine bacteria (Provasoli and Pintner, 1980). Such changes in morphology, when grown axenically, have also been observed in Monostroma oxyspermum (Tatewaki et al., 1983; Matsuo et al., 2003), Ulva linza, Ulva pertusa and Ulva compressa (Nakanishi et al., 1996; Marshall et al., 2006). Adding back bacterial strains to axenic Ulva cultures has been shown to restore the foliaceous morphology (Nakanishi et al., 1996; Marshall et al., 2006). Of 1555 bacterial strains isolated from U. pertusa, 676 strains (41%) showed morphogenesis activity (Nakanishi et al., 1996). For U. linza, 13 out of 20 unique strains isolated from the algae induced morphological change when added to axenic cultures, and five of those also increased the relative growth rate of the alga (Marshall et al., 2006). However, phylogenetic analysis of these strains indicated that the bacterial effect on Ulva was independent of bacterial phylogeny: members of the Proteobacteria, Bacteroidetes and Gram-positive cocci all positively impacted Ulva growth (Marshall et al., 2006).
This suggests that there are either multiple cues produced by epiphytic bacteria that stimulate growth and morphogenesis of Ulva or that a common metabolite, produced by a wide range of bacteria, is responsible for these effects. There are reports of marine bacteria producing plant growth regulators and vitamins which may affect the morphological differentiation of algae (Maruyama et al., 1986; Croft et al., 2006). For example, an algal morphogenesis inducer, named thallusin, has been isolated from a Bacteroidetes strain. Very low concentrations of this compound strongly induced differentiation in M. oxyspermum (Matsuo et al., 2005). Additionally, other algal growth hormones such as cytokinin-type hormones, auxin-type hormones and indole-3-acetic acid have been shown to be produced by marine bacteria (Maruyama et al., 1986; Bradley, 1991). It has been hypothesized that the effect of bacteria on algal growth and morphology may be due to: (i) bacteria being responsible for supplying nitrogen to algae, a suggestion based upon an observation that bacterial strains isolated from green alga possess the nitrogenase gene nifH, and/or (ii) that some bacterial species metabolize plant hormones (Chisholm et al., 1996; Ashen and Goff, 2000).
Although AHLs affect Ulva zoospore settlement, their potential influence on the later stages of Ulva growth and development has never been investigated. Hence, the aim of the present study was to explore whether AHL signal molecules from marine bacteria could influence Ulva germination and growth. The bacterial population associated with U. linza was first phylogenetically characterized, bacteria representative of the epiphytic community isolated and their AHL profiles characterized using AHL bioreporters and liquid-chromatography tandem mass spectrometry (LC-MS/MS). The germination and growth response of Ulva zoospores exposed to bacterial biofilms composed of AHL-producing strains indigenous to the Ulva bacterial population was compared with the response to the same strains transformed with plasmids expressing the recombinant Bacillus sp. lactonase enzyme AiiA to create AHL-deficient variants (Dong et al., 2002). Additionally, Ulva germination and germling growth was measured in response biofilms of transgenic Escherichia coli which expressed various AHL synthase genes and with synthetic AHLs alone.
Results
Impact of AHL-producing biofilms from Ulva endogenous bacteria on zoospore germination
A phylogenetic tree resulting from the alignment and analysis of a clone library composed of 76 individual 16S rDNA clones at 97% sequence similarity shows the U. linza thallus epiphytic bacterial community to be dominated by Alphaproteobacteria, Gammaproteobacteria, Flavobacteria and Sphingobacteria (Fig. S1). Additionally, signal molecule characterization using both E. coli lux-based bioreporters and LC-MS/MS showed bacterial strains isolated from Ulva to produce a wide variety of AHLs (Table S1). The 16S rDNA clone library of bacteria associated with Ulva showed both Sulfitobacter (Alphaproteobacteria) and uncultured Alteromonadales (Gammaproteobacteria) to be abundant within the population. Based on these findings, two AHL-producing strains from these groups of bacteria were selected in order to investigate the zoospore response to Ulva's indigenous bacteria. To compare the impact on germination of these strains with their AHL-deficient counterparts, Sulfitobacter spp. 376 and Shewanella spp. 79 were transformed with pMT01, a plasmid harbouring the aiiA AHL lactonase gene from Bacillus sp. 240B1 within the broad host vector pBBRIMCS-5 (Kovach et al., 1995). Successful transformation with the pMT01 plasmid rendered the cognate AHLs of 376 and 79 biologically inactive as AiiA hydrolyses the homoserine lactone ring (Dong et al., 2000). Inactivation of AHL production by AiiA was confirmed by the lack of activation of appropriate lux-based E. coli AHL bioreporters with solvent extracts from cell-free supernatants of these strains (Fig. S2).
Ulva zoospores were settled onto Shewanella spp. 79 pBBRIMCS, Shewanella spp. 79 pMT01, Sulfitobacter spp. 376 pBBRIMCS and Sulfitobacter spp. 376 pMT01 biofilms of varying bacterial density. Zoospore germination and average germling length was measured after 48 h incubation (for representative images of germinated zoospores exposed to AHL-producing and non-producing marine bacterial biofilms, see Fig. S3). The varying volumes of inoculum used to grow biofilms of both strains produced biofilms with bacterial densities ranging from approximately 20–75% coverage, with no major difference in biofilm density observed between strains carrying the pBBRIMCS or pMT01 plasmids. After 48 h incubation, average germling length was greatly increased for zoospores settled on biofilms of both aiiA-expressing, AHL-deficient Shewanella spp. and Sulfitobacter spp. strains compared with the AHL-producing biofilms (Fig. 1). Additionally, successful zoospore germination was significantly increased on the AHL-deficient Shewanella spp. 79 pMT01 biofilms with an average percentage of germination of 77.9% (± 6.4%) in comparison with 53.4% (± 7.2%) on AHL-producing Shewanella spp. 79 pBBRIMCS biofilms. This trend was also observed in Sulfitobacter spp. 376 where the percentage of germination on Sulfitobacter spp. 376 pMT01 biofilms was 80.4% (± 2.8%) in comparison with 62.4% (± 6.3%) on Sulfitobacter spp. 376 pBBRIMCS biofilms.
Figure 1.
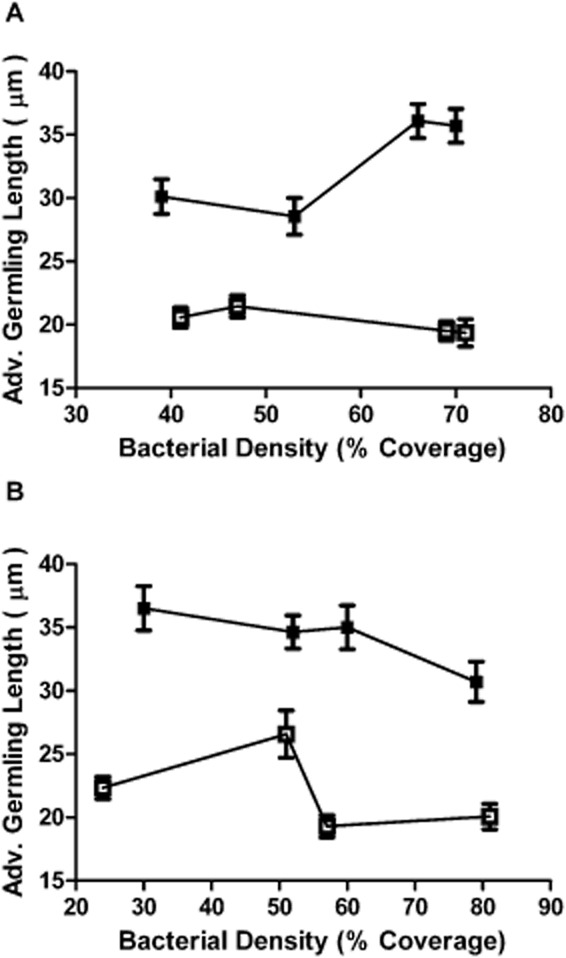
-
AAverage germling length of zoospores exposed to biofilms of AHL-expressing Shewanella sp. 79 pBBRIMCS (□) and non-AHL expressing Shewanella sp. 79 pMT01 (□) at 48 h incubation.
-
BAverage germling length of zoospores exposed to biofilms of AHL expressing Sulfitobacter sp. 376 pBBRIMCS (□) and non-AHL expressing Sulfitobacter sp. 376 pMT01 (□) at 48 incubation. Error bars represent 95% confidence intervals.
Ulva zoospore germination response when exposed to biofilms of E. coli expressing recombinant AHL synthases
The changes in zoospore germination observed following the expression of aiiA in the AHL-producing Sulfitobacter spp. 376 and Shewanella spp. 79 strains may be a consequence of either the direct action of AHLs on the zoospores or indirectly due to the loss of an AHL-dependent bacterial phenotype, or the action of the lactonase. To distinguish between these possibilities, Ulva zoospore germination was assayed using biofilms composed of transgenic E. coli strains that expressed different recombinant AHL synthase genes. These synthases were RhlI from Pseudomonas aeruginosa (producing C4-HSL), LuxI from Vibrio fischeri (producing 3-oxo-C6-HSL) and VanI from V. anguillarum (producing 3-oxo-C10-HSL) (Latifi et al., 1995; Milton et al., 1997; Joint et al., 2002). All recombinant AHL synthase genes were expressed from plasmids within the E. coli host and compared with E. coli harbouring the respective vector plasmids without recombinant AHL synthase genes (see Table S2). These strains constitutively produce AHLs, resulting in a more stable assay for monitoring the response of Ulva zoospores to AHLs (Joint et al., 2002). Owing to the detrimental effect of the osmotic pressure of seawater on the growth and survival of E. coli biofilms, zoospore slides were incubated for a reduced time of 24 h in 70% sterile filtered seawater prior to being fixed and stained. At 24 h incubation, there was a small yet significant decrease in average germling length when Ulva zoospores were settled and grown on biofilms of E. coli expressing rhlI and vanI. Although the response to E. coli expressing luxI was not found to be significant, a modest decrease in the presence of this biofilm was also apparent (Fig. 2). Reduction in successful zoospore germination was observed for all AHL-producing biofilms of E. coli strains at 24 h in comparison to their respective AHL-deficient vector control stains (33% for RhlI, 19% for LuxI and 33% VanI in comparison to 78%, 73% and 60% in the respective controls).
Figure 2.
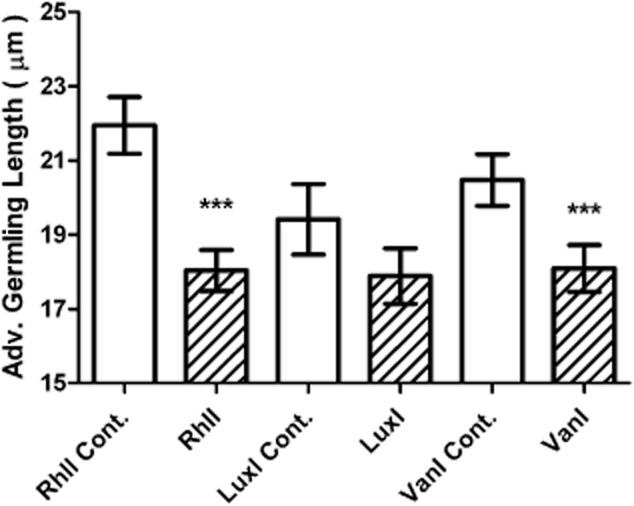
Effect of recombinant E. coli biofilms on Ulva germling growth. Average Ulva germling length when exposed to biofilms of E. coli producing AHLs from plasmids expressing recombinant AHL synthases, in comparison with those from their E. coli counterparts harbouring the corresponding control vectors without the AHL syntheses after 24 h incubation. The plasmids used in this experiment are listed in Table S2. Error bars represent 95% confidence intervals and asterisks show those values significantly different to the controls (one-way ANOVA * = P ≤ 0.001).
Ulva zoospore germination response to synthetic AHLs
To ensure that the effect on zoospore germination observed by AHL-producing bacteria was not just due to phenotypic changes in these bacteria, the impact of synthetic AHLs on germination was tested. The alkaline pH of seawater is known to hydrolyse the homoserine lactone ring rendering AHLs biologically inactive, and this effect is particularly prevalent in short-chain AHLs and AHLs with 3-oxy and 3-hydroxy substitutions (Hmelo and Van Mooy, 2009). Longer chain AHLs such as C12-HSL are known to be more stable in seawater (Tait et al., 2005; Tait and Havenhand, 2005). As AHLs with acyl side chains of 12 carbons in length are produced by both Sulfitobacter spp. 376 and Shewanella sp. 79 (Table S1), synthetic C12-HSL was tested for its effect on zoospore germination and growth at concentrations ranging from 0.5 μM to 40 μM. After 48 h incubation, there was a significant reduction in average germling length in the presence of C12-HSL concentrations ranging between 5 μM and 40 μM in comparison to the absence of C12-HSL (Fig. 3). There was also a reduction in successful zoospore germination in the presence of C12-HSL at all concentrations tested (e.g. 76% at 5 μM C12-HSL) compared with the absence of this signal molecule (86%). Furthermore, after 48 h, Ulva germlings exposed to synthetic C4-HSL, (an AHL also shown to be produced by both Sulfitobacter spp.376 and Shewanella spp. 79), showed a significant reduction in size and percentage of germination in the presence (26.86 μm ± 1.3 μm and 61%) compared with the absence of 5 μM C4-HSL (39.65 μm ± 1.6 μm and 86%).
Figure 3.
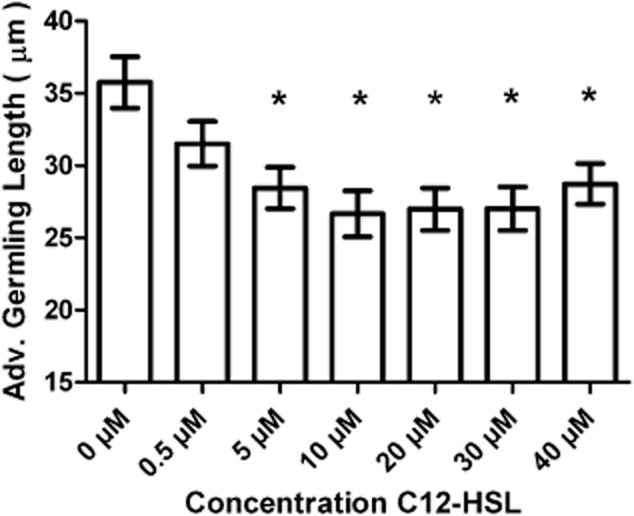
Effect of exogenously added synthetic C12-HSL on Ulva germling growth. Average Ulva germling length when exposed to a range of C12-HSL concentrations at 48 h incubation. Error bars represent 95% confidence intervals and asterisks show those values that differed significantly from that of 0 μM AHL (one-way ANOVA * = P ≤ 0.001).
Discussion
Ulva zoospores preferentially settle on AHL-producing bacterial biofilms and on agarose slides permeated with synthetic AHLs (Joint et al., 2002; Tait et al., 2005) and bacteria have been shown to affect the growth of Ulva (Matsuo et al., 2003). The present study has shown that many bacteria associated with the Ulva thallus are AHL producers, and hence, it would be a reasonable assumption that AHLs or the products of AHL-regulated bacterial genes may also enhance Ulva zoospore germination and germling growth. This is the case in higher plants where AHLs are known to promote post-embryonic root development in Arabidopsis thaliana (Ortíz-Castro et al., 2008; von Rad et al., 2008) and can accelerate auxin-dependent adventitious root formation in Vigna radiata (mung bean) seedlings (Bai et al., 2012). In contrast, this study has shown that U. linza zoospore germination and the early growth of the U. linza germling are reduced when an AHL-producing biofilm or a synthetic source of AHLs are present. This presents a paradox; why do AHLs promote zoospore settlement but reduce zoospore germination and growth? A study of estuarine waters has shown that slower growing algal species have the ability to outcompete fast-growing species in the long term as their slow growth imposes less of a metabolic burden when nutrient conditions become limiting (Pedersen and Borum, 1996). Previous research has also shown that Ulva spp. are particularly sensitive to levels of nitrates and phosphates, with both nutrients shown to significantly affect the early growth of Ulva (Sousa et al., 2007). Additionally, Ulva growth has been shown to be dependent on salinity and light with increased light levels and a salinity of 25 psu being optimum for early Ulva growth (Imchen, 2012). It may be the case that in its early growth stages Ulva utilizes AHL signal molecules as a cue to slow its growth rate. This may lessen the rate at which Ulva utilizes the nutrients available in the local environment and increase the time for Ulva to react to local physical conditions therefore helping to facilitate survival. Retarded growth, as a result of bacterial AHL production, may also allow Ulva to take advantage of any extracellular growth factors bacteria provide which lead to cellular differentiation and a healthy alga. Differences in algal morphology were not observed in Ulva zoospores germinated with or without the presence of AHLs, and the experiments presented here were restricted to investigating the effects of AHLs on both germination and the initial germling growth period. Therefore, further research investigating the latter stages of germling growth and differentiation is required to support the hypotheses that AHLs reduce Ulva growth either to mediate a better chance of long-term survival and/or the successful differentiation into an adult alga. Experiments into Ulva germination and early growth varying both physical conditions and the presence of AHL signal molecules would also allow further testing of this hypothesis.
This investigation and studies by Tait and colleagues (2005; 2009) have demonstrated that Ulva zoospore germlings are constantly exposed to a variety of AHL-producing bacteria in the natural environment. Tait and colleagues (2009) provide an estimate of the concentration of AHLs within natural rocky shore biofilms, which was around 6 μM. However, natural fluctuation in the concentration of AHLs, either due to the pH-dependent hydrolysis of the homoserine lactone ring, dilution by the action of waves on the rocky shore or the production of AHL inactivating enzymes by marine bacteria such as Shewanella sp. (Tait et al., 2009), would be expected to influence Ulva zoospore settlement, and may also affect the germination of Ulva zoospores and early germling growth. Interestingly, this study observed a lack of a dose response to increasing concentrations of C12-HSL. It is thought that Ulva responds to AHLs via some form of yet to be discovered receptor (Wheeler et al., 2006; Joint et al., 2007). Concentrations above 5 μM C12-HSL may be saturating this receptor therefore accounting for the lack of dose response to increasing, ecologically relevant, C12-HSL concentrations. To test this further repeated experiments would have to be carried out using AHL concentrations between 0.5 μM where no significant effect was observed and 5 μM where a significant reduction in zoospore germination and germling growth was observed. Indeed, further work mimicking natural conditions are required to fully appreciate the importance of AHL signalling to both Ulva zoospore settlement and successful germination and growth.
Many of the bacteria isolated from the Ulva thallus and/or representing the Ulva bacterial population are actively producing a range of AHL signal molecules (Table S1). AHL production by bacterial species isolated from the marine environment and specifically from Ulva has been reported previously (Gram et al., 2002; Mohamed et al., 2008; Tait et al., 2009). The strains assayed in this study were shown to predominantly produce AHLs with shorter fatty acid side chains. The production of AHLs with short to medium length fatty acid side chains differs from previous work that has investigated AHL production in bacterial strains, isolated from a variety of marine environments, which show a predominant bias to long-chain AHL production (Gram et al., 2002; Taylor et al., 2004; Wagner-Döbler et al., 2005; Tait et al., 2009). This study used an LC-MS/MS methodology better adapted for identifying shorter chain AHLs that may not have been used in previous studies investigating marine bacterial AHL production. This may account for why we observed more bacterial strains producing these short-chain AHLs. Our findings may also indicate that short-chain AHL production is more widespread in the marine environment than suggested in previous studies. One interesting finding was C4-HSL production in strains P13, UI13 and UI08 all of which were identified as being members of the Bacteroidetes phylum. Until recently, AHL production was thought to be limited to Proteobacterial species. However, Romero and colleagues (2010) have equivocally demonstrated that Tenacibaculum maritimum, also a member of the Bacteroidetes phylum, produces C4-HSL, and Sharif and colleagues (2008) have also shown AHL production in cyanobacterial strains (Sharif et al., 2008; Romero et al., 2010). C4-HSL production by Bacteroidetes strains shown here further extends the range of species known to be undertaking AHL signal molecule production, strengthening the evidence that AHL signal molecule production is not restricted to Proteobacterial species. This study has shown the Bacteroidetes to be a dominant group of bacteria within the Ulva thallus epiphytic population and that AHL signal molecule production by these bacteria could affect the Ulva growth process.
Further knowledge of how AHLs and Ulva's indigenous epiphytic bacterial community modulate algal growth may lead to improvements in the prevention of Ulva biofouling, enabling the marine industry to move away from the use of harmful antifouling agents, which can detrimentally impact the marine environment.
Experimental Procedures
Strains, plasmids and media
Non-marine bacterial strains and plasmids used in this study can be found in Tables S2 and S3. Post isolation marine bacterial strains were routinely cultured in Marine Broth (Difco, BD, Oxford, UK) at 30°C. E. coli strains were cultured in Luria broth (Bacto-tryptone 10 g l−1; Bacto yeast extract 5 g l−1; NaCl 10 g l−1) supplemented with appropriate antibiotics at 37°C (Table S3).
Isolation and phylogenetic characterization of epiphytic bacteria
Marine bacteria were isolated from rocks colonized by Ulva, the Ulva holdfast–rock interface and from the thallus of wild Ulva. Ulva bacterial isolates were all obtained from either Wembury beach, Devon, UK (50°19'00' 'N 4°05'03' 'W), or Polzeath beach, Cornwall, UK (50°34'39' 'N 4°55'03' 'W). Bacterial isolation techniques were adapted from Patel and colleagues (2003) and Tait and colleagues (2009). Isolation of strains from the rocks colonized by Ulva was achieved by taking scrapings from the rocks, plating onto seawater agar (filtered seawater and 1.5% Oxoid number 1 agar) and incubating for 15 days at 15°C. Single colonies were then isolated on marine broth agar. Isolates from the Ulva thallus were obtained by vortexing Ulva tissue in phosphate-buffered saline (PBS) for approximately 3 min. The PBS supernatant was removed, serially diluted, plated onto either seawater agar, marine broth agar (Difco), Actinomycete Isolation Agar (Difco) or R2A agar (Difco) and cultured for up to 3 weeks, before being isolated on marine broth agar (Difco). Strains isolated from the Ulva holdfast–rock interface were obtained by swabbing the interface onto marine broth agar (Difco) and incubating for 72 h at 22°C. Single colonies were picked, streaked on marine broth agar (Difco) and incubated at 22°C (Patel et al., 2003; Tait et al., 2009). Phylogenetic typing of marine bacterial strains was carried out by amplifying 16S rDNA using primers 96bfm (5'-GAGTTTGATYHTGGCTCAG-3') and 1152uR (5'-ACGGHTACCTTGTTACGACTT-3') (Muhling et al., 2008) via colony polymerase chain reaction (PCR). PCR amplification was carried out using GoTaq DNA Polymerase (Promega, Southampton, UK). Reactions were made as per manufactures instructions to a total volume of 50 μl using 1 μl of boiled bacterial colony suspended in d.H2O has a template. Amplification was carried out using an initial step of 96°C for 2 min followed by 35 cycles of 95°C for 1 min, 53°C for 30 s and 72°C for 2 min. Amplified DNA was subsequently sequenced bidirectional using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Warrington, UK) with the same primers. All sequences were submitted to GenBank (see Table S1 for accession numbers).
DNA extraction, clone library construction and sequencing
Epiphytic bacteria were obtained from the surfaces of Ulva thallus material collected from Wembury beach, UK (50°19'00' 'N 4°05'03' 'W) by prolonged vortexing in sterile PBS. Bacteria were pelleted by centrifugation and DNA extracted using a Wizard DNA Purification Kit (Promega) as per manufacturer's instructions. 16S rDNA corresponding to nucleotides 341–926 of the E. coli 16S rDNA sequence was amplified via PCR from this chromosomal DNA using primers 341F (5'-CCTACGGGAGGCAGCAG-3') and 907R (5'-CCGTCAATTCMTTTGAGTTT-3'). PCR was carried out using the same condition described previously with the exception of the extension time which was 1 min. The amplified 16S rDNA was cloned into the pGEM T easy vector (Promega) and transformed into E. coli DH5a via electroporation according to the manufacturer's instructions. A total of 96 clones were selected for sequencing using the M13F and M13R primers. Sequencing was bidirectional using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Phylogenetic analysis and tree topology was carried out using the MEGA programme (Tamura et al., 2011).
AHL extraction, detection and characterization assays
AHLs were extracted from 20 ml of cell-free culture supernatants acidified with 2 M HCl to pH 2 using dichloromethane in accordance with the method described by Yates and colleagues (2002). AHL detection assays were carried in accordance with the method described by Tait and colleagues (2009) using bioreporters E. coli JM109 pSB536, pSB401 and pSB1075 (Swift et al., 1997; Winson et al., 1998). A black clear bottomed 96-well microtitre plate (Greiner Bio One, Gloucestershire, UK) was used for all assays with 100 μl of cell-free supernatant extract dried to each well assayed. After 3 h coincubation with bioreporters at 37°C, the resulting luminescence was measured using a Berthold MITHRAS microtitre plate reader (Berthold, Bad Wildbad, Germany). Intensity of bioluminescence was calculated in relative light units (Tait et al., 2009). AHLs from extracted planktonic cultures were further characterized by LC-MS/MS using a methodology previously described by Yates and colleagues (2002). Solvent extracts were separated by reverse phase high performance liquid chromatography using and Ascentis Express C18 2.7 μm column (150 × 2.1 mm) coupled to a tandem mass spectrometer (Bruker HCT Plus ion trap; Bruker Daltoniks, Bremen, Germany). Extracted ion chromatograms of precursor ion m/z 102.1 (homoserine lactone ring) were produced from the mass of the parent molecular ion [M + H] of each AHL screened for, (for fragment ion m/z of each AHL, see Yates et al., 2002), and retention times and peak spectra were matched to 1 μM standards of each AHL (Yates et al., 2002; Ortori et al., 2007; 2011).
Growth of biofilm material
Bacterial biofilms were grown from varying volumes of 0.5 ml, 1 ml, 2 ml and 4 ml initial inoculum in accordance with methods described by Tait and colleagues (2005). Biofilm inoculum was prepared using stationary phase cultures which were pelleted, washed in sterile filtered seawater and adjusted to an OD600 of 1.00 before inoculation. Marine biofilms were grown for 72 h at 22°C. Biofilms of E. coli strains were grown in 70% filtered seawater for 24 h in order to reduce osmotic stress on the bacteria by salinity (Tait et al., 2005). Biofilm density was determined as per the method described by Joint and colleagues (2002) with microscope image analysis at 400× magnification, using a Reichert Jung Polyvar microscope (Leica microsystems, Milton Keynes, UK) and an Optronics Magna Fire SP camera (Milton Keynes, UK) (Joint et al., 2002).
Ulva zoospore germination assays
Ulva zoospore release was carried out in accordance with methodology of Callow and colleagues (1997). All Ulva zoospores used in this study were released from fertile Ulva thalli collected from Wembury beach, UK (50°19'00' 'N 4°05'03' 'W) (Callow et al., 1997). Ulva zoospores were settled onto biofilms or sterile glass slides using the method described by Tait and colleagues (2005); in all cases, 15 ml of the final Ulva zoospore suspension adjusted to an OD600 of 0.5 was used; additionally, all experimental treatments were carried out on three separate slides. Post settlement zoospore slides were transferred to sterile 60 mm Petri dishes and submerged in 10 ml sterile filtered seawater. For zoospore assays using E. coli biofilms, 70% sterile filtered seawater was used. For the assays using synthetic AHLs, C12-HSL was added to the separate dishes post zoospore settlement at final concentrations ranging from 0.5 μM and 40 μM and at 5 μM for C4-HSL. Zoospore slides were incubated at 18°C in proximity to a light source with a 16 h light, 8 h dark cycle for 48 h (24 h for assay using E. coli biofilms). After incubation, the zoospore slides were fixed with 2% (v/v) glutaraldehyde and stained with dilute carbol fuchsin.
Image and statistical analysis
For all Ulva zoospore germination assays, the three separate zoospore slides for each experimental treatment were viewed at 100× magnification using a Reichert Jung Polyvar microscope with attached Optronics Magna Fire SP camera. The zoospore slides from each experimental condition were imaged 10 times in randomly selected locations. The lengths of 300 individual Ulva zoospores and germlings were measured from randomly selected images of each experimental condition using Image ProPlus Version 5 imaging software (Media Cybernetics, Rockville, MD, USA). Zoospores over 15 μm were defined as successfully germinated, as such percentage zoospore germination was calculated by dividing the number of successfully germinated zoospores by the total number of settled zoospores. Statistical analysis of Ulva zoospore germination assays was carried out by performing analysis of variance (ANOVA) tests using Minitab 16 (Minitab).
Acknowledgments
This work was supported by the UK Natural Environment Research Council (Grant NE/F012365/1). We thank Mary Bruce and Alex Truman for their technical contributions.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site
Fig. S1. Phylogenetic tree of the Ulva thallus bacterial population. The tree resulted from a sequence alignment of 16S rDNA from bacterial clones and isolates obtained from the Ulva thallus at 97% sequence similarity. The reference strains were taken from GenBank. In brackets is the number of clones in each OTU. The tree topology is based on neighbour- joining and bootstrap analysis was performed with 1000 replications.
Fig. S2. AHL bioreporter activation assay.
A. Cell-free supernatant extracts from Shewanella spp. 79 pBBRIMCS and pMT01 were assayed with lux-based AHL bioreporters E. coli pSB536 (clear bars) and pSB1075 (cross-hatched bars).
B. Cell-free supernatant extracts from Sulfitobacter spp. 376 pBBRIMCS and pMT01 were assayed with lux-based AHL bioreporter E. coli pSB401. All strains harbouring pMT01 and therefore expressing the aiiA lactonase failed to activate the bioreporters showing that their cognate AHLs were being deactivated.
Fig. S3. AHL germination assays slides. Example images of Ulva zoospores germinated on (A) Sulfitobacter sp. pMT01 biofilms (AHL-deficient) and (B) Sulfitobacter sp. pBBRIMCS biofilms (AHL-producing). Bar = 50 μm.
Table S1. Phylogenetic identification and AHL signal molecule production in Ulva-associated marine bacteria. The ability of each strain to produce AHLs was assayed using lux-based AHL bioreporters, a positive result is indicated by + and a negative result by –. AHL production and identity was confirmed by LC-MS/MS.
Table S2. Non-marine bacterial strains used in this study.
Table S3. Plasmids used in this study.
References
- Ashen JB. Goff LJ. Molecular and ecological evidence for species specificity and coevolution in a group of marine algal-bacterial symbioses. Appl Environ Microbiol. 2000;66:3024–3030. doi: 10.1128/aem.66.7.3024-3030.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Todd CD, Desikan R, Yang Y. Hu X. N-3-oxo-decanoyl-l-homoserine-lactone activates auxin-induced adventitious root formation via hydrogen peroxide- and nitric oxide-dependent cyclic GMP signaling in mung bean. Plant Physiol. 2012;158:725–736. doi: 10.1104/pp.111.185769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PM. Plant hormones do have a role in controlling growth and development of algae. J Phycol. 1991;27:317–321. [Google Scholar]
- Callow JA, Stanley MS, Wetherbee R. Callow ME. Cellular and molecular approaches to understanding primary adhesion in Enteromorpha: an overview. Biofouling. 2000a;16:141–150. [Google Scholar]
- Callow ME. Callow JA. Substratum location and zoospore behaviour in the fouling alga Enteromorpha. Biofouling. 2000;15:49–56. doi: 10.1080/08927010009386297. [DOI] [PubMed] [Google Scholar]
- Callow ME, Callow JA, Pickett-Heaps JD. Wetherbee R. Primary adhesion of EnteromorphaChlorophytaUlvales) propagules: quantitative settlement studies and video microscopy. J Phycol. 1997;33:938–947. [Google Scholar]
- Callow ME, Callow JA, Ista LK, Coleman SE, Nolasco AC. Lopez GP. Use of self-assembled monolayers of different wettabilities to study surface selection and primary adhesion processes of green algal (Enteromorpha) zoospores. Appl Environ Microbiol. 2000b;66:3249–3254. doi: 10.1128/aem.66.8.3249-3254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow ME, Jennings AR, Brennan AB, Seegert CE, Gibson A, Wilson L, et al. Microtopographic cues for settlement of zoospores of the green fouling alga Enteromorpha. Biofouling. 2002;18:229–236. [Google Scholar]
- Chisholm JRM, Dauga C, Ageron E, Grimont PAD. Jaubert JM. Roots in mixotrophic algae. Nature. 1996;381:382. [Google Scholar]
- Croft MT, Warren MJ. Smith AG. Algae need their vitamins. Eukaryot Cell. 2006;5:1175–1183. doi: 10.1128/EC.00097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Xu JL, Li XZ. Zhang LH. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci USA. 2000;97:3526–3531. doi: 10.1073/pnas.060023897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Gusti AR, Zhang Q, Xu JL. Zhang LH. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl Environ Microbiol. 2002;68:1754–1759. doi: 10.1128/AEM.68.4.1754-1759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gram L, Grossart HP, Schlingloff A. Kiorboe T. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl Environ Microbiol. 2002;68:4111–4116. doi: 10.1128/AEM.68.8.4111-4116.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hmelo L. Van Mooy BAS. Kinetic constraints on acylated homoserine lactone-based quorum sensing in marine environments. Aquat Microb Ecol. 2009;54:127–133. [Google Scholar]
- Imchen T. Recruitment potential of a green alga ulva flexusa Wulfen dark preserved zoospore and its development. PloS ONE. 2012;7:e32651. doi: 10.1371/journal.pone.0032651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joint I, Callow ME, Callow JA. Clarke KR. The attachment of Enteromorpha zoospores to a bacterial biofilm assemblage. Biofouling. 2000;16:151–158. [Google Scholar]
- Joint I, Tait K, Callow ME, Callow JA, Milton D, Williams P. Camara M. Cell-to-cell communication across the prokaryote-eukaryote boundary. Science. 2002;298:1207–1207. doi: 10.1126/science.1077075. [DOI] [PubMed] [Google Scholar]
- Joint I, Tait K. Wheeler G. Cross-kingdom signalling: exploitation of bacterial quorum sensing molecules by the green seaweed Ulva. Philos Trans R Soc Lond B Biol Sci. 2007;362:1223–1233. doi: 10.1098/rstb.2007.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM., 2nd Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Latifi A, Winson MK, Foglino M, Bycroft BW, Stewart GS, Lazdunski A. Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- Marshall K, Joint I, Callow ME. Callow JA. Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga Ulva linza. Microb Ecol. 2006;52:302–310. doi: 10.1007/s00248-006-9060-x. [DOI] [PubMed] [Google Scholar]
- Maruyama A, Maeda M. Simidu U. Occurrence of plant hormone (cytokinin)-producing bacteria in the sea. J Appl Microbiol. 1986;61:569–574. [Google Scholar]
- Matsuo Y, Suzuki M, Kasai H, Shizuri Y. Harayama S. Isolation and phylogenetic characterization of bacteria capable of inducing differentiation in the green alga Monostroma oxyspermum. Environ Microbiol. 2003;5:25–35. doi: 10.1046/j.1462-2920.2003.00382.x. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Imagawa H, Nishizawa M. Shizuri Y. Isolation of an algal morphogenesis inducer from a marine bacterium. Science. 2005;307:1598. doi: 10.1126/science.1105486. [DOI] [PubMed] [Google Scholar]
- Milton DL, Hardman A, Camara M, Chhabra SR, Bycroft BW, Stewart GS. Williams P. Quorum sensing in Vibrio anguillarum: characterization of the vanIvanR locus and identification of the autoinducer N-(3-oxodecanoyl)-L-homoserine lactone. J Bacteriol. 1997;179:3004–3012. doi: 10.1128/jb.179.9.3004-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed NM, Cicirelli EM, Kan J, Chen F, Fuqua C. Hill RT. Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ Microbiol. 2008;10:75–86. doi: 10.1111/j.1462-2920.2007.01431.x. [DOI] [PubMed] [Google Scholar]
- Muhling M, Woolven-Allen J, Murrell JC. Joint I. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J. 2008;2:379–392. doi: 10.1038/ismej.2007.97. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Nishijima M, Nishimura M, Kuwano K. Saga N. Bacteria that induce morphogenesis in Ulva pertusaChlorophyta) grown under axenic conditions. J Phycol. 1996;32:479–482. [Google Scholar]
- OrtÍz-Castro R, MartÍNez-Trujillo M. LÓPez-Bucio J. N-acyl-L-homoserine lactones: a class of bacterial quorum-sensing signals alter post-embryonic root development in Arabidopsis thaliana. Plant Cell Environ. 2008;31:1497–1509. doi: 10.1111/j.1365-3040.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- Ortori C, Dubern J-F, Chhabra S, Cámara M, Hardie K, Williams P. Barrett D. Simultaneous quantitative profiling of N-acyl-L-homoserine lactone and 2-alkyl-4(1H)-quinolone families of quorum-sensing signalling molecules using LC-MS/MS. Anal Bioanal Chem. 2011;399:839–850. doi: 10.1007/s00216-010-4341-0. [DOI] [PubMed] [Google Scholar]
- Ortori CA, Atkinson S, Chhabra SR, Camara M, Williams P. Barrett DA. Comprehensive profiling of N-acylhomoserine lactones produced by Yersinia pseudotuberculosis using liquid chromatography coupled to hybrid quadrupole-linear ion trap mass spectrometry. Anal Bioanal Chem. 2007;387:497–511. doi: 10.1007/s00216-006-0710-0. [DOI] [PubMed] [Google Scholar]
- Patel P, Callow ME, Joint I. Callow JA. Specificity in the settlement – modifying response of bacterial biofilms towards zoospores of the marine alga Enteromorpha. Environ Microbiol. 2003;5:338–349. doi: 10.1046/j.1462-2920.2003.00407.x. [DOI] [PubMed] [Google Scholar]
- Pedersen MF. Borum J. Nutrient control of algal growth in estuarine waters. Nutrient limitation and the importance of nitrogen requirements and nitrogen storage among phytoplankton and species of macroalgae. Mar Ecol Prog Ser. 1996;142:261–272. [Google Scholar]
- Provasoli L. Pintner IJ. Bacteria induced polymophism in an axenic laboratory strain of Ulva lactucaChlorophyceae. J Phycol. 1980;16:196–201. [Google Scholar]
- von Rad U, Klein I, Dobrev P, Kottova J, Zazimalova E, Fekete A, et al. Response of Arabidopsis thaliana to N-hexanoyl-dl-homoserine-lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta. 2008;229:73–85. doi: 10.1007/s00425-008-0811-4. [DOI] [PubMed] [Google Scholar]
- Romero M, Avendano-Herrera R, Magarinos B, Camara M. Otero A. Acylhomoserine lactone production and degradation by the fish pathogen Tenacibaculum maritimum, a member of the Cytophaga-Flavobacterium-Bacteroides (CFB) group. FEMS Microbiol Lett. 2010;304:131–139. doi: 10.1111/j.1574-6968.2009.01889.x. [DOI] [PubMed] [Google Scholar]
- Sharif DI, Gallon J, Smith CJ. Dudley E. Quorum sensing in CyanobacteriaN-octanoyl-homoserine lactone release and response, by the epilithic colonial cyanobacterium Gloeothece PCC6909. ISME J. 2008;2:1171–1182. doi: 10.1038/ismej.2008.68. [DOI] [PubMed] [Google Scholar]
- Sousa AI, Martins I, Lillebø AI, Flindt MR. Perdall MA. Influence of salinity, nutrients and light on the germination and growth of Enteromorpha sp Spores. J Exp Mar Bio Ecol. 2007;341:142–150. [Google Scholar]
- Swift S, Karlyshev AV, Fish L, Durant EL, Winson MK, Chhabra SR, et al. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait K. Havenhand J. Investigating a possible role for the bacterial signal molicules N-acylhomoserine lactones in Balanus improvisus cyprid settlement. Mol Ecol. doi: 10.1111/mec.12273. doi: 10.1111/mec.12273. [DOI] [PubMed] [Google Scholar]
- Tait K, Joint I, Daykin M, Milton DL, Williams P. Camara M. Disruption of quorum sensing in seawater abolishes attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ Microbiol. 2005;7:229–240. doi: 10.1111/j.1462-2920.2004.00706.x. [DOI] [PubMed] [Google Scholar]
- Tait K, Williamson H, Atkinson S, Williams P, Camara M. Joint I. Turnover of quorum sensing signal molecules modulates cross-kingdom signalling. Environ Microbiol. 2009;11:1792–1802. doi: 10.1111/j.1462-2920.2009.01904.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M. Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatewaki M, Provasoli L. Pintner IJ. Morphogenesis of Monostroma oxyspermum (Kütz.) Doty (Chlorophyceae) in axenic culture, especially in bialgal culture. J Phycol. 1983;19:409–416. [Google Scholar]
- Taylor MW, Schupp PJ, Baillie HJ, Charlton TS, de Nys R, Kjelleberg S. Steinberg PD. Evidence for acyl homoserine lactone signal production in bacteria associated with marine sponges. Appl Environ Microbiol. 2004;70:4387–4389. doi: 10.1128/AEM.70.7.4387-4389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Döbler I, Thiel V, Eberl L, Allgaier M, Bodor A, Meyer S, et al. Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine Alphaproteobacteria. Chembiochem. 2005;6:2195–2206. doi: 10.1002/cbic.200500189. [DOI] [PubMed] [Google Scholar]
- Wheeler GL, Tait K, Taylor A, Brownlee C. Joint I. Acyl-homoserine lactones modulate the settlement rate of zoospores of the marine alga Ulva intestinalis via a novel chemokinetic mechanism. Plant Cell Environ. 2006;29:608–618. doi: 10.1111/j.1365-3040.2005.01440.x. [DOI] [PubMed] [Google Scholar]
- Williams P, Winzer K, Chan WC. Camara M. Look who's talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond B Biol Sci. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winson MK, Swift S, Fish L, Throup JP, Jørgensen F, Chhabra SR, et al. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, Sockett RE, et al. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect Immun. 2002;70:5635–5646. doi: 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Phylogenetic tree of the Ulva thallus bacterial population. The tree resulted from a sequence alignment of 16S rDNA from bacterial clones and isolates obtained from the Ulva thallus at 97% sequence similarity. The reference strains were taken from GenBank. In brackets is the number of clones in each OTU. The tree topology is based on neighbour- joining and bootstrap analysis was performed with 1000 replications.
Fig. S2. AHL bioreporter activation assay.
A. Cell-free supernatant extracts from Shewanella spp. 79 pBBRIMCS and pMT01 were assayed with lux-based AHL bioreporters E. coli pSB536 (clear bars) and pSB1075 (cross-hatched bars).
B. Cell-free supernatant extracts from Sulfitobacter spp. 376 pBBRIMCS and pMT01 were assayed with lux-based AHL bioreporter E. coli pSB401. All strains harbouring pMT01 and therefore expressing the aiiA lactonase failed to activate the bioreporters showing that their cognate AHLs were being deactivated.
Fig. S3. AHL germination assays slides. Example images of Ulva zoospores germinated on (A) Sulfitobacter sp. pMT01 biofilms (AHL-deficient) and (B) Sulfitobacter sp. pBBRIMCS biofilms (AHL-producing). Bar = 50 μm.
Table S1. Phylogenetic identification and AHL signal molecule production in Ulva-associated marine bacteria. The ability of each strain to produce AHLs was assayed using lux-based AHL bioreporters, a positive result is indicated by + and a negative result by –. AHL production and identity was confirmed by LC-MS/MS.
Table S2. Non-marine bacterial strains used in this study.
Table S3. Plasmids used in this study.


