We provide the first evidence demonstrating that single-nucleotide polymorphisms in macrophage function-related genes may predict prognosis in locoregional gastric cancer patients. Our results also suggest that the immune-related component of tumor for progression may be dictated not only by the malignant epithelial component but also by the genetic predisposition of host in gastric cancer.
Keywords: NF-κB, CCL2, single-nucleotide polymorphism, gastric cancer, ethnic difference
Abstract
Background
Nuclear factor-kappaB (NF-κB) and CCL2/CCR2 chemokine axis play a central role in tumor progression such as stimulation of angiogenesis, acceleration of tumor invasion and migration, and suppression of innate immunosurveillance in the macrophage-related functions. There have been few reports regarding association of the macrophage function-related genes with the clinical outcome in gastric cancer. We hypothesized that variants in genes encoding for NF-κB and CCL2/CCR2 axis may predict prognosis in gastric cancer and tested whether the functional single-nucleotide polymorphisms (SNPs) will be associated with clinical outcome in patients with gastric cancer across two independent groups.
Patients and methods
This study enrolled two cohorts which consisted of 160 Japanese patients and 104 US patients with locoregional gastric cancer. Genomic DNA was analyzed for association of 11 SNPs in NFKB1, RELA, CCL2, and CCR2 with clinical outcome using PCR-based direct DNA sequencing.
Results
The univariable analysis showed four SNPs had significant association with clinical outcome in the Japanese cohort, NFKB1 rs230510 remained significant upon multivariable analysis. The patients with the A allele of the NFKB1 rs230510 had significantly longer overall survival (OS) compared with those with the T/T genotype in both the Japanese and US cohort in the univariable analysis. In contrast, genotypes with the T allele of CCL2 rs4586 were significantly associated with shorter OS compared with the C/C genotype in the US cohort [hazard ratio (HR) 2.43; P = 0.015] but longer OS in the Japanese cohort (HR 0.58; P = 0.021), resulting in the statistically significant opposite impact on OS (P = 0.001).
Conclusions
Our study provides the first evidence that the NFKB1 rs230510 and CCL2 rs4586 are significantly associated with the clinical outcome in patients with locoregional gastric cancer. These results also suggest that the genetic predisposition of the host may dictate the immune-related component of the tumor for progression in gastric cancer.
introduction
Macrophages promote cancer initiation by creating an inflammatory environment that is suitable for tumor growth and enhance tumor progression by supporting tumor-associated angiogenesis, promoting tumor cell invasion and migration, and suppressing antitumor immunity. Activated macrophages play a critical role in the inflammatory process to create a mutagenic and growth-promoting microenvironment which potentiates the acquisition of oncogenic mutations in the cancer initiation phase [1]. Depending on the surrounding microenvironment, macrophages can be divided into mainly two phenotypes, classically activated phenotype (M1 macrophage) which plays proinflammatory and tumor suppressive roles, or alternatively activated phenotype (M2 macrophage) which plays immunosuppressive and tumor-promoting roles. The majority of tumor-associated macrophages (TAMs) acquire a phenotype similar to the M2 macrophages which can be modified by the tumor microenvironmental triggers such as chemokines and cytokines [2]. Given the critical effects of the M1 and M2 macrophages for tumor progression, there is a significant interest in elucidating the genes that regulate the two macrophage phenotypes in the tumor microenvironment; moreover, the genes may become a target for drug development in addition to be a clinically useful biomarker to help select cancer patients who benefit from the targeted treatment.
Nuclear factor-kappaB (NF-κB) and CCL2/CCR2 chemokine axis play a central role in the macrophage-related functions, by regulating the cancer-related inflammation and the possession of the M1/M2 macrophages, promoting the recruitment of the TAMs, and providing antiapoptotic or angiogenic signals such as vascular endothelial growth factor (VEGF) to tumor cells in the tumor microenvironment [3, 4]. A heterodimer of RelA (p65) and NF-κB1 (p50) which is the most commonly found complex causes the activation of NF-κB pathway by binding to IκBα [5]. There have been some reports investigating association of NF-κB signaling and CCL2 in gastric cancer [6, 7]. However, to the best of our knowledge, it has remained unclear about association of variants in genes encoding for NF-κB and CCL2/CCR2 axis with the clinical outcome in gastric cancer. It should be a study of great interest to focus on a host related factor such as germline variants contributing innate tumor immunity which plays important roles in the tumor microenvironment. We hypothesized that the NF-κB and CCL2/CCR2 axis-related gene variants may serve as a potential biomarker to predict prognosis in gastric cancer and tested whether functional single-nucleotide polymorphisms (SNPs) in NFKB1, RELA, CCL2, and CCR2, will be associated with the clinical outcome in patients with locoregional gastric cancer across two independent groups with different background.
materials and methods
eligible patients
This study enrolled two independent cohorts of patients with histologically confirmed locoregional gastric adenocarcinoma (stage I–IV; AJCC 6th), one from Japan and another from United States. The Japanese and US cohort consisted of 160 patients treated with surgery alone or surgery followed by S-1 or fluoropyrimidine-based adjuvant chemotherapy [Fukushima Red Cross Hospital (Fukushima) and Kitasato University East Hospital (Sagamihara)] and 104 patients treated with surgery alone or surgery followed by fluoropyrimidine-based adjuvant (radio)-chemotherapy [University of Southern California (USC)/Norris Comprehensive Cancer Center (Los Angeles, CA), Los Angeles County Hospital (Los Angeles, CA)] between 1991 and 2011. Patients were followed as clinically routine every 3 months for the first 2 years and then every 6 months. Patient data were collected retrospectively through chart review. Pathologic stage was decided according to tumor–node–metastasis classification, 6th edition. Histological classification of gastric tumors in the Japanese and US cohort was carried out according to Japanese classification [8] and Lauren classification [9], respectively. The tissue analysis presented in this study was conducted at the USC/Norris Comprehensive Cancer Center following approval by the USC Institutional Review Board of Medical Sciences. All patients signed an informed consent for the analysis of molecular correlates.
single-nucleotide polymorphism selection
Common and potential SNPs in the genes encoding for macrophage-related functions, NFKB1, RELA, CCL2, and CCR2, were selected by using stringent and predefined selection criteria: (i) SNPs, which shown to be of biological significance according to literature review [either published data or predicted function using functional SNP (F-SNP) database [10]] or tagging SNPs which are chosen using the HapMap genotype data with r2 threshold = 0.8: http://snpinfo.niehs.nih.gov/snpinfo/snptag.htm; and (ii) 10% or more of a minor allele frequency in both Asians and Caucasians (in the Ensembl Genome Browser: http://uswest.ensembl.org/index.html). Among all SNPs matching these criteria, we focused on 11 promising SNPs (supplementary Table S1, available at Annals of Oncology online).
DNA extraction and genotyping
Genomic DNA was extracted from peripheral blood or formalin-fixed paraffin-embedded tissue derived from tumor samples obtaining germline DNA using the QIAmp Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol (www.qiagen.com). The candidate SNPs were tested using PCR-based direct DNA sequence analysis by ABI 3100A Capillary Genetic Analyzer and Sequencing Scanner v1.0 (Applied Biosystems). For quality control purposes, a random selection of 10% of the samples was examined for each SNP.
statistical analysis
The end points of current study were overall survival (OS) and disease-free survival (DFS) or time-to-tumor recurrence (TTR). The OS, DFS, and TTR were defined as the period from the date of surgery or diagnosis to death in the both cohort, to the first observation of relapse or death in the Japanese cohort, and to the first observation of tumor recurrence in the US cohort, respectively. If events were not observed, the end points were censored at the last time of contact or follow-up.
χ2 Tests were carried out to examine the differences in baseline patient characteristics between the two cohorts. Allelic distribution of all SNPs in each race/ethnic group was examined for deviation from Hardy–Weinberg equilibrium (HWE) using Fisher's exact test. Linkage disequilibrium among selected SNPs was assessed using D′ and r2 values, and the haplotype frequencies were inferred using Haploview version 4.2 (www.broad.mit.edu/mpg/haploview).
Kaplan–Meier curves and log-rank tests were carried out in univariable analysis of the association between the candidate SNPs and OS and DFS or TTR using both dominant and recessive genetic model. Stage, gender, age, and type of adjuvant chemotherapy were adjusted in the Japanese cohort; tumor site, tumor stage, and lymph node stage were adjusted, and type of adjuvant chemotherapy and race were stratified in the US cohort for multivariable Cox regression models to re-evaluate the independent effects of the polymorphisms (supplementary Tables S2 and S3, available at Annals of Oncology online). With 160 patients in the Japanese cohort and 104 patients in the US cohort, we would have 80% power to detect a minimum hazard ratio (HR) of 1.93–2.21 and 2.27–2.66, respectively, in DFS or TTR across the variant allele frequencies of 10%–40% in a dominant model using a 0.05-level two-sided log-rank test. For a recessive model, the minimum HR is about 2.89 and 3.71 in the Japanese and US cohorts, respectively, when the variant allele frequency is 30% and approaches 2.07 and 2.45, respectively, when the allele frequency is 50%.
All tests were carried out using the SAS 9.4 (SAS Institute, Cary, NC). All tests were two-sided at a significance level of 0.05. P values were not adjusted for multiple testing.
results
The baseline characteristics of the two cohorts included in this study were summarized in Table 1. Patients in the US cohort, median follow-up time of 3.3 years, were significantly younger and with higher incidence of gastroesophageal junction (GEJ) cancer and less frequent undifferentiated type adenocarcinoma compared with the Japanese cohort, median follow-up time of 4.1 years. The genotyping quality control by direct DNA sequencing provided a genotype concordance of 99% or more. Genotyping was successful in at least 90% of cases in each polymorphism analyzed. In failed cases, genotyping was not successful because of limited quantity and/or quality of extracted genomic DNA. The allelic frequencies for all SNPs were within the probability limits of HWE (P > 0.05) in each race group.
Table 1.
Baseline clinical characteristics of Japanese and US patient cohort
| Japanese (N = 160) |
US (N = 104) |
P valuea | |||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Gender | |||||
| Male | 102 | 64 | 64 | 62 | |
| Female | 58 | 36 | 40 | 38 | 0.79 |
| Age (years) | |||||
| Median (range) | 68 (31–88) | 57 (26–85) | |||
| <65 | 60 | 38 | 84 | 81 | |
| 65–74 | 56 | 35 | 12 | 12 | <0.001 |
| ≥75 | 44 | 27 | 8 | 8 | |
| Stage | |||||
| I–II | 76 | 48 | 38 | 37 | |
| III–IV | 84 | 52 | 66 | 63 | 0.098 |
| Tumor stage | |||||
| T1 | 12 | 8 | 2 | 2 | |
| T2 | 61 | 38 | 36 | 34 | |
| T3 | 84 | 52 | 56 | 54 | 0.003 |
| T4 | 3 | 2 | 10 | 10 | |
| N stage | |||||
| N0 | 36 | 23 | 24 | 23 | |
| N1 | 79 | 49 | 49 | 47 | 0.89 |
| N2 | 31 | 19 | 19 | 18 | |
| N3 | 14 | 9 | 12 | 12 | |
| Tumor site | |||||
| Stomach | 158 | 99 | 62 | 60 | <0.001 |
| GEJ | 2 | 1 | 31 | 30 | |
| Unknown | 11 | 11 | |||
| Histological classification (Japanese/Lauren) | |||||
| Differentiated/intestinal | 64 | 40 | 37 | 36 | |
| Undifferentiated/diffuse | 96 | 60 | 30 | 29 | |
| Mixed | 18 | 17 | <0.001 | ||
| Unknown | 19 | 18 | |||
| Adjuvant chemotherapy | |||||
| Yes | 103 | 64 | 79 | 76 | |
| No | 57 | 36 | 25 | 24 | 0.057 |
| Ethnicity | |||||
| Asian | 160 | 100 | 24 | 23 | |
| Caucasian | 0 | 36 | 35 | N/A | |
| Hispanic | 0 | 43 | 41 | ||
| African American | 0 | 1 | 1 | ||
Based on χ2 test or Fisher's exact test.
GEJ, gastroesophageal junction.
In the Japanese cohort, high linkage disequilibrium was found between NFKB1 rs230510 and rs3821958 (D′ = 0.98, r2 = 0.66), RELA rs11820062 and rs7119750 (D′ = 0.92, r2 = 0.52), and CCL2 rs4586 and rs1024611 (D′ = 0.92, r2 = 0.76). In the US cohort, NFKB1 rs230510 and rs3821958 also showed linkage disequilibrium (D′ = 0.97, r2 = 0.67). Haplotypes were constructed from these SNPs separately. However, there were no significant relations between these variants and clinical outcomes.
univariable and multivariable analyses in the Japanese cohort
The univariable analysis showed that CCL2 rs4586 and RELA rs11820062 had a statistical significance in the dominant genetic model, while NFKB1 rs230510 and NFKB1 rs3821958 had a statistical significance in the recessive genetic model. The NFKB1 rs230510 remained significantly associated with both DFS and OS upon the multivariable analysis (Table 2 and supplementary Table S4, available at Annals of Oncology online).
Table 2.
Association between macrophage-related gene polymorphisms and the clinical outcome in the Japanese and US cohorts
| Japanese cohort | N | Disease-free survival |
Overall survival |
||||
|---|---|---|---|---|---|---|---|
| Gene rs number | 3-year recurrence rate ± SE | HR (95% CI) | HR (95% CI)a | 5-year survival rate ± SE | HR (95% CI) | HR (95% CI)a | |
| NFKB1 rs230510 | |||||||
| A/A | 49 | 0.45 ± 0.07 | 1 (Reference) | 1 (Reference) | 0.55 ± 0.07 | 1 (Reference) | 1 (Reference) |
| A/T | 69 | 0.34 ± 0.06 | 0.75 (0.42–1.32) | 0.78 (0.43–1.42) | 0.64 ± 0.07 | 0.66 (0.36–1.21) | 0.64 (0.34–1.21) |
| T/T | 38 | 0.68 ± 0.08 | 1.71 (0.96–3.04) | 1.51 (0.82–2.77) | 0.28 ± 0.08 | 1.64 (0.91–2.95) | 1.35 (0.72–2.54) |
| P value | 0.010 | 0.080 | 0.007 | 0.055 | |||
| A/T, T/Tb | 107 | 0.46 ± 0.05 | 1.04 (0.63–1.72) | 1.03 (0.61–1.76) | 0.51 ± 0.06 | 0.97 (0.58–1.62) | 0.89 (0.51–1.56) |
| P valueb | 0.88 | 0.90 | 0.90 | 0.70 | |||
| A/T, A/Ac | 118 | 0.39 ± 0.05 | 0.50 (0.30–0.81) | 0.58 (0.35–0.96) | 0.60 ± 0.05 | 0.48 (0.29–0.80) | 0.57 (0.34–0.98) |
| P valuec | 0.004 | 0.034 | 0.004 | 0.040 | |||
| NFKB1 rs3821958 | |||||||
| A/A | 62 | 0.46 ± 0.06 | 1 (Reference) | 1 (Reference) | 0.56 ± 0.07 | 1 (Reference) | 1 (Reference) |
| A/G | 72 | 0.38 ± 0.06 | 0.81 (0.48–1.37) | 0.85 (0.50–1.47) | 0.58 ± 0.07 | 0.75 (0.43–1.29) | 0.73 (0.41–1.30) |
| G/G | 22 | 0.71 ± 0.10 | 1.76 (0.94–3.30) | 1.30 (0.67–2.52) | 0.26 ± 0.10 | 1.75 (0.93–3.31) | 1.21 (0.62–2.39) |
| P value | 0.045 | 0.43 | 0.028 | 0.27 | |||
| A/G, G/Gb | 94 | 0.46 ± 0.05 | 1.00 (0.62–1.60) | 0.96 (0.58–1.59) | 0.50 ± 0.06 | 0.95 (0.58–1.56) | 0.85 (0.50–1.44) |
| P valueb | 0.99 | 0.87 | 0.84 | 0.55 | |||
| A/G, A/Ac | 134 | 0.42 ± 0.04 | 0.51 (0.29–0.90) | 0.70 (0.39–1.26) | 0.57 ± 0.05 | 0.49 (0.27–0.87) | 0.68 (0.38–1.24) |
| P valuec | 0.018 | 0.24 | 0.013 | 0.21 | |||
| RELA rs11820062 | |||||||
| C/C | 48 | 0.54 ± 0.07 | 1 (Reference) | 1 (Reference) | 0.44 ± 0.09 | 1 (Reference) | 1 (Reference) |
| C/T | 71 | 0.33 ± 0.06 | 0.45 (0.26–0.80) | 0.65 (0.36–1.19) | 0.66 ± 0.06 | 0.48 (0.27–0.86) | 0.60 (0.32–1.11) |
| T/T | 31 | 0.53 ± 0.09 | 0.92 (0.50–1.70) | 1.38 (0.72–2.61) | 0.46 ± 0.10 | 0.81 (0.43–1.55) | 1.09 (0.56–2.12) |
| P value | 0.011 | 0.069 | 0.037 | 0.14 | |||
| C/T, T/Tb | 102 | 0.39 ± 0.05 | 0.58 (0.36–0.95) | 0.85 (0.50–1.44) | 0.59 ± 0.05 | 0.58 (0.34–0.97) | 0.74 (0.43–1.29) |
| P valueb | 0.028 | 0.54 | 0.034 | 0.29 | |||
| C/T, C/Cc | 119 | 0.41 ± 0.05 | 0.70 (0.40–1.22) | 0.58 (0.33–1.01) | 0.57 ± 0.05 | 0.81 (0.45–1.46) | 0.69 (0.38–1.25) |
| P valuec | 0.20 | 0.055 | 0.49 | 0.23 | |||
| CCL2 rs4586 | |||||||
| C/C | 68 | 0.54 ± 0.06 | 1 (Reference) | 1 (Reference) | 0.44 ± 0.07 | 1 (Reference) | 1 (Reference) |
| C/T | 72 | 0.36 ± 0.06 | 0.51 (0.31–0.86) | 0.61 (0.35–1.06) | 0.64 ± 0.06 | 0.47 (0.27–0.81) | 0.67 (0.37–1.19) |
| T/T | 19 | 0.60 ± 0.12 | 0.99 (0.52–1.90) | 0.77 (0.38–1.59) | 0.38 ± 0.12 | 0.99 (0.51–1.92) | 0.83 (0.40–1.74) |
| P value | 0.022 | 0.21 | 0.012 | 0.39 | |||
| C/T, T/Tb | 91 | 0.41 ± 0.05 | 0.61 (0.39–0.96) | 0.65 (0.40–1.08) | 0.58 ± 0.06 | 0.58 (0.36–0.93) | 0.71 (0.42–1.21) |
| P valueb | 0.031 | 0.095 | 0.021 | 0.21 | |||
| C/T, C/Cc | 140 | 0.45 ± 0.04 | 0.73 (0.39–1.36) | 1.03 (0.52–2.02) | 0.54 ± 0.05 | 0.70 (0.37–1.31) | 0.99 (0.50–1.97) |
| P valuec | 0.32 | 0.94 | 0.26 | 0.98 | |||
| US cohort | |||||||
| Gene rs number | N | Time-to-tumor recurrence |
Overall survival |
||||
| 3-year recurrence rate ± SE | HR (95% CI) | HR (95% CI)a | 5-year survival rate ± SE | HR (95% CI) | HR (95% CI)a | ||
| NFKB1 rs230510 | |||||||
| T/T | 31 | 0.72 ± 0.11 | 1 (Reference) | 1 (Reference) | 0.21 ± 0.12 | 1 (Reference) | 1 (Reference) |
| T/A | 39 | 0.57 ± 0.10 | 0.71 (0.36–1.40) | 1.51 (0.67–3.42) | 0.81 ± 0.09 | 0.52 (0.24–1.11) | 0.81 (0.31–2.06) |
| A/A | 26 | 0.38 ± 0.10 | 0.52 (0.23–1.17) | 0.83 (0.31–2.23) | 0.78 ± 0.10 | 0.33 (0.13–0.88) | 0.37 (0.12–1.19) |
| P value | 0.23 | 0.38 | 0.040 | 0.24 | |||
| T/A, A/Ab | 65 | 0.50 ± 0.08 | 0.62 (0.33–1.17) | 1.04 (0.48–2.28) | 0.53 ± 0.09 | 0.44 (0.22–0.89) | 0.58 (0.24–1.38) |
| P valueb | 0.13 | 0.92 | 0.016 | 0.22 | |||
| T/A, T/Tc | 70 | 0.63 ± 0.07 | 1.59 (0.77–3.29) | 1.32 (0.53–3.25) | 0.38 ± 0.09 | 2.06 (0.85–4.99) | 2.40 (0.82–7.06) |
| P valuec | 0.18 | 0.55 | 0.10 | 0.11 | |||
| NFKB1 rs3821958 | |||||||
| A/A | 32 | 0.53 ± 0.11 | 1 (Reference) | 1 (Reference) | 0.54 ± 0.12 | 1 (Reference) | 1 (Reference) |
| A/G | 41 | 0.67 ± 0.09 | 1.80 (0.92–3.53) | 1.33 (0.62–2.85) | 0.59 ± 0.10 | 2.47 (1.13–5.40) | 2.09 (0.82–5.34) |
| G/G | 20 | 0.45 ± 0.13 | 1.15 (0.48–2.76) | 0.79 (0.30–2.12) | 0.70 ± 0.13 | 1.65 (0.61–4.42) | 1.68 (0.54–5.21) |
| P value | 0.17 | 0.53 | 0.060 | 0.31 | |||
| A/G, G/Gb | 61 | 0.60 ± 0.08 | 1.57 (0.83–2.99) | 1.01 (0.49–2.12) | 0.36 ± 0.08 | 2.20 (1.03–4.66) | 1.85 (0.75–4.55) |
| P valueb | 0.15 | 0.97 | 0.034 | 0.18 | |||
| A/G, A/Ac | 73 | 0.60 ± 0.07 | 1.21 (0.56–2.61) | 1.18 (0.47–2.92) | 0.43 ± 0.08 | 1.04 (0.46–2.37) | 0.85 (0.34–2.17) |
| P valuec | 0.62 | 0.73 | 0.92 | 0.74 | |||
| RELA rs11820062 | |||||||
| C/C | 28 | 0.59 ± 0.15 | 1 (Reference) | 1 (Reference) | 0.50 ± 0.18 | 1 (Reference) | 1 (Reference) |
| C/T | 45 | 0.54 ± 0.09 | 1.68 (0.75–3.76) | 1.15 (0.43–3.11) | 0.71 ± 0.08 | 1.16 (0.43–3.17) | 0.76 (0.22–2.58) |
| T/T | 25 | 0.64 ± 0.12 | 1.67 (0.70–4.00) | 0.64 (0.19–2.18) | 0.67 ± 0.11 | 1.17 (0.40–3.41) | 0.42 (0.10–1.72) |
| P value | 0.40 | 0.42 | 0.95 | 0.41 | |||
| C/T, T/Tb | 70 | 0.57 ± 0.07 | 1.67 (0.78–3.61) | 0.73 (0.25–2.18) | 0.45 ± 0.08 | 1.17 (0.44–3.05) | 0.52 (0.14–1.91) |
| P valueb | 0.17 | 0.58 | 0.75 | 0.32 | |||
| C/T, C/Cc | 73 | 0.53 ± 0.07 | 0.86 (0.45–1.62) | 2.26 (0.87–5.87) | 0.41 ± 0.09 | 0.96 (0.47–1.96) | 1.95 (0.71–5.37) |
| P valuec | 0.63 | 0.095 | 0.91 | 0.19 | |||
| CCL2 rs4586 | |||||||
| C/C | 33 | 0.45 ± 0.11 | 1 (Reference) | 1 (Reference) | 0.62 ± 0.12 | 1 (Reference) | 1 (Reference) |
| C/T | 50 | 0.69 ± 0.08 | 2.07 (1.06–4.04) | 1.76 (0.78–4.01) | 0.37 ± 0.10 | 2.53 (1.15–5.58) | 1.67 (0.59–4.76) |
| T/T | 21 | 0.32 ± 0.12 | 0.88 (0.32–2.45) | 1.00 (0.31–3.18) | 0.36 ± 0.18 | 2.18 (0.81–5.87) | 1.89 (0.57–6.27) |
| P value | 0.024 | 0.27 | 0.046 | 0.54 | |||
| C/T, T/Tb | 71 | 0.61 ± 0.07 | 1.73 (0.90–3.29) | 1.63 (0.68–3.89) | 0.36 ± 0.09 | 2.43 (1.14–5.18) | 1.52 (0.54–4.26) |
| P valueb | 0.071 | 0.27 | 0.015 | 0.42 | |||
| C/T, C/Cc | 83 | 0.59 ± 0.07 | 1.75 (0.69–4.41) | 1.29 (0.47–3.53) | 0.47 ± 0.08 | 0.80 (0.35–1.83) | 0.70 (0.28–1.79) |
| P valuec | 0.23 | 0.62 | 0.58 | 0.45 | |||
Based on the log-rank test in the univariable analysis and Wald test in the multivariable analysis within Cox regression model.
Stage (I, II, III, and IV), gender, age (<65, 65–74, ≥75 years as continuous), and type of adjuvant therapy (no versus yes) were adjusted in Japanese cohort; tumor site, tumor stage, and lymph node stage were adjusted; type of adjuvant chemotherapy and race were stratified in US cohort.
Combined in the analysis in the dominant genetic model.
Recessive genetic model when considering a genotype with two minor alleles as a reference.
evaluation of impact of the macrophage function-related gene SNPs on clinical outcome between the Japanese and US cohort
We sequentially carried out analyses whether four SNPs which were significant in the Japanese cohort will be associated with clinical outcome in the US cohort. The univariable analysis showed that three SNPs, NFKB1 rs230510, NFKB1 rs3821958, and CCL2 rs4586 were significantly associated with OS in the dominant genetic model (Table 2).
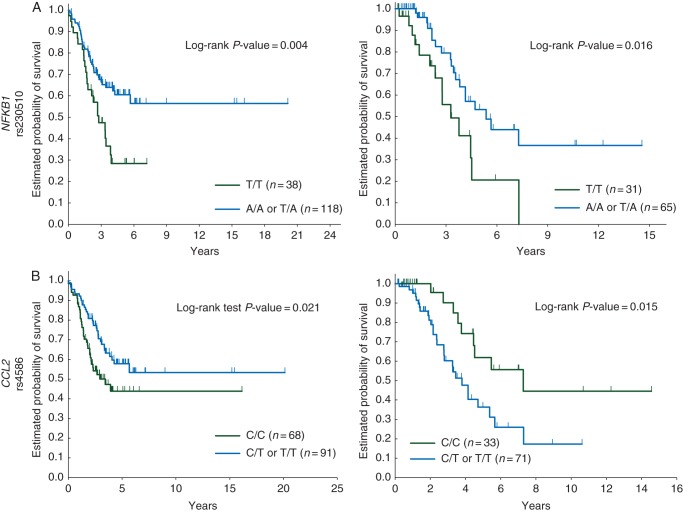
In univariable analysis, genotypes with the A allele of the NFKB1 rs230510 correlated with longer OS in both the Japanese and US cohorts. Interestingly, genotypes with the T allele of the CCL2 rs4586 showed association with longer OS in the Japanese cohort, whereas those showed association with shorter OS in the US cohort (Figure 1). The impact of the T allele of the CCL2 rs4586 on OS in the US cohort was opposite to that in the Japanese cohort and reached statistical significance by the likelihood ratio test of the Cox regression model including the interaction term of cohort group and SNP (P = 0.001).
Figure 1.
Comparison of clinical outcome by macrophage function-related gene variants in two cohorts. Overall survival probability by (A) NFKB1 rs230510, left; Japanese cohort, right; US cohort and (B) CCL2 rs4586, left; Japanese cohort, right; US cohort.
All significant SNPs were included in the multivariable models in two cohorts separately, and backward elimination method was used to find the best predictive models. In the Japanese cohort, the NFKB1 rs230510T/T versus A/A or A/T showed significant association with DFS (P = 0.042) and marginally significant association with OS (P = 0.060). However, no good predictive model including SNPs was found in the US cohort.
discussion
Our study provides the first evidence suggesting that variants in genes encoding for macrophage-related functions may predict prognosis in patients with locoregional gastric cancer. Our results also suggest that the immune-related component of the tumor for progression may be dictated not only by the malignant epithelial component, but also by the genetic predisposition of the host in gastric cancer.
We found that the NFKB1 rs230510 and CCL2 rs4586 were significantly associated with clinical outcome in patients with locoregional gastric cancer in both cohorts. In particular, the A allele of the NFKB1 rs230510 significantly correlated with favorable OS in the Japanese cohort in the univariable and multivariable analyses as well as in the US cohort in the univariable analysis. This finding indicates that the NFKB1 rs230510 may be a promising prognostic marker in gastric cancer. Some investigations have reported conflicting data about the relationship of NF-κB overexpression with the clinical outcome in gastric cancer [6, 11]. These may have resulted from the evidence that the NF-κB has a dual role, proinflammatory and anti-inflammatory role, depending on the stage in cancer development. Therefore, genotypes of NFKB1 may become more clinically useful as a biomarker than immunohistochemistry or overexpression status of the NF-κB since genetic information is independent of the tumor microenvironment. Given a role of the NFKB1 rs230510 as tagging SNP located on intron of the gene, it is biologically plausible that this SNP may affect the transcription of the gene. Further mechanistic studies confirming the functional role of the SNP are warranted.
Our study also indicated that the impact of the T allele of the CCL2 rs4586 on OS was statistically and significantly opposite between two cohorts. The difference in the impact may result from histopathological or etiological differences between the Japanese and US cohort. Gastric cancer has been considered a heterogeneous disease which may be classified into at least three distinct subtypes based on pathology and epidemiology, each with different initiating pathologic processes, and each possibly having different tumor biology [12, 13]. Proximal gastric cancer predominates in Europe and United States, whereas distal gastric cancer is more prevalent in Asia and Eastern Europe [14]. In our study, there appeared to be more frequent diffuse adenocarcinoma and a significantly lower incidence of GEJ cancer in the Japanese than the US cohort, in which the GEJ cancer had significantly shorter prognosis than stomach cancer at the baseline (supplementary Table S3, available at Annals of Oncology online). Each histological type holds different prognostic values [15], and one subtype of gastric cancer enriched by TP53 mutations and receptor tyrosine kinases-RAS activation with intestinal histology are more frequent in the GEJ [16], implying we may have observed the difference caused by one of possible limitations of our study design. On the other hand, in epidemiologic aspect, proximal nondiffuse gastric cancer strongly correlates with obesity and gastroesophageal reflux disease, while the development of distal nondiffuse gastric cancer requires chronic inflammation mainly caused by Helicobacter pylori infection or correlates with dietary factors [14]. There has been shown to be a difference in the prevalence of H. pylori infection between Japan and United States [17, 18], suggesting etiological differences related to inflammation in gastric cancer between Japan and United States may affect the outcome of patients from those regions. Additionally, there have been several reports regarding ethnic differences in CCL2 serum level, suggesting that there may be significantly different profiles of circulating inflammatory mediators among different ethnic groups [19, 20]. The differences that we observed in current study between the two cohorts may contribute that gastric cancer is a complex and enigmatic disease with different etiologies. The histopathologic or epidemiologic distinctions to subdivide gastric cancer should be taken into account in not only future prospective clinical trials but also biomarker studies.
Some macrophage function-related pathways including VEGF and phosphatidylinositol 3-kinase pathway also may cause the differences in the outcome between patients from different regions [3, 21]. Polymorphisms in angiogenic pathway gene had different association with increased cancer risk and different allele frequency between ethnicity in gastric cancer patients [22, 23]. In addition, East Asian and Caucasian gastric cancer patients differed significantly in frequencies of PIK3CA exon 9 and 20 mutations [24]. In our study, the impact of the genetic variant of the RELA rs11820062 on TTR in the US cohort had a strong trend toward opposite to that on DFS in the Japanese cohort (P = 0.07), indicating a result consistent with the difference found in the CCL2 rs4586 (supplementary Figure S1, available at Annals of Oncology online). Additionally, we tested the association of the CCL2 rs4586 and NFKB1 rs230510 with clinical outcome in the US cohort according to ethnicity. The Hispanic patients, but not Caucasian, with the T allele of the CCL2 rs4586 had significantly worse OS (P = 0.04), it was opposite to the result in the Japanese cohort, though small sample size. In contrast, the Hispanic patients had no significant difference in OS by the NFKB1 rs230510 genotype (P = 0.62) (supplementary Figure S2, available at Annals of Oncology online). Taken together, these findings may suggest that some of macrophage-related functions have intrinsic ethnic differences and also have a different impact on the outcome in patients with different background. Our results are hypothesis generating but warrant validation in larger patient cohorts.
Our study demonstrated significant results across the two independent cohorts despite the small sample size. However, there may be some possibility that the patient number of our study has no adequate ability to assess the association between the macrophage function-related gene SNPs and clinical outcome. A selection bias cannot be excluded because of the retrospective study design. Therefore, these results should be confirmed in larger prospective studies. A better understanding of the functional SNPs will be critical for potential new biomarkers.
In conclusion, our data provide the first evidence that the NFKB1 rs230510 and CCL2 rs4586 are associated with clinical outcome in patients with locoregional gastric cancer. These data also suggest that the genetic predisposition of the host may dictate the immune-related component of the tumor for progression in gastric cancer. Biomarker-embedded translational trials are warranted to validate our findings.
funding
This work was partly supported by the National Institutes of Health (grant number 5 P30CA14089-27S1 to H-JL) and Yvonne Bogdanovich.
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
references
- 1.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 4.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29:313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pomerantz JL, Baltimore D. Two pathways to NF-kappaB. Mol Cell. 2002;10:693–695. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 6.Lee BL, Lee HS, Jung J, et al. Nuclear factor-kappaB activation correlates with better prognosis and Akt activation in human gastric cancer. Clin Cancer Res. 2005;11:2518–2525. doi: 10.1158/1078-0432.CCR-04-1282. [DOI] [PubMed] [Google Scholar]
- 7.Tao LL, Shi SJ, Chen LB, et al. Expression of monocyte chemotactic protein-1/CCL2 in gastric cancer and its relationship with tumor hypoxia. World J Gastroenterol. 2014;20:4421–4427. doi: 10.3748/wjg.v20.i15.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer. 2011;14:101–112. doi: 10.1007/s10120-011-0041-5. [DOI] [PubMed] [Google Scholar]
- 9.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 10.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36:D820–D824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki N, Morisaki T, Hashizume K, et al. Nuclear factor-kappaB p65 (RelA) transcription factor is constitutively activated in human gastric carcinoma tissue. Clin Cancer Res. 2001;7:4136–4142. [PubMed] [Google Scholar]
- 12.Shah MA, Kelsen DP. Gastric cancer: a primer on the epidemiology and biology of the disease and an overview of the medical management of advanced disease. J Natl Compr Canc Netw. 2010;8:437–447. doi: 10.6004/jnccn.2010.0033. [DOI] [PubMed] [Google Scholar]
- 13.Shah MA, Khanin R, Tang L, et al. Molecular classification of gastric cancer: a new paradigm. Clin Cancer Res. 2011;17:2693–2701. doi: 10.1158/1078-0432.CCR-10-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–362. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan IB, Ivanova T, Lim KH, et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology. 2011;141:476–485. doi: 10.1053/j.gastro.2011.04.042. 485.e471–485.e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dooley CP, Cohen H, Fitzgibbons PL, et al. Prevalence of Helicobacter pylori infection and histologic gastritis in asymptomatic persons. N Engl J Med. 1989;321:1562–1566. doi: 10.1056/NEJM198912073212302. [DOI] [PubMed] [Google Scholar]
- 18.Asaka M, Kimura T, Kudo M, et al. Relationship of Helicobacter pylori to serum pepsinogens in an asymptomatic Japanese population. Gastroenterology. 1992;102:760–766. doi: 10.1016/0016-5085(92)90156-s. [DOI] [PubMed] [Google Scholar]
- 19.Jia LQ, Shen YC, Guo SJ, et al. The 2518 A/G polymorphism in the MCP-1 gene and cancer risk: a meta-analysis. Asian Pac J Cancer Prev. 2013;14:3575–3579. doi: 10.7314/apjcp.2013.14.6.3575. [DOI] [PubMed] [Google Scholar]
- 20.Coussens AK, Wilkinson RJ, Nikolayevskyy V, et al. Ethnic variation in inflammatory profile in tuberculosis. PLoS Pathog. 2013;9:e1003468. doi: 10.1371/journal.ppat.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roca H, Varsos Z, Pienta KJ. CCL2 protects prostate cancer PC3 cells from autophagic death via phosphatidylinositol 3-kinase/AKT-dependent survivin up-regulation. J Biol Chem. 2008;283:25057–25073. doi: 10.1074/jbc.M801073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wakatsuki T, Zhang W, Yang DY, et al. Ethnic gene profile of genes involved in angiogenesis to predict regional bevacizumab efficacy difference in gastric cancer. J Clin Oncol. 2012;30(Suppl) (abstr 4026) [Google Scholar]
- 23.Xu B, Feng NH, Tong N, et al. VEGF -460C>T polymorphism and cancer risk: a meta-analysis. Med Oncol. 2010;27:1031–1036. doi: 10.1007/s12032-009-9329-2. [DOI] [PubMed] [Google Scholar]
- 24.Chong ML, Loh M, Thakkar B, et al. Phosphatidylinositol-3-kinase pathway aberrations in gastric and colorectal cancer: meta-analysis, co-occurrence and ethnic variation. Int J Cancer. 2014;134:1232–1238. doi: 10.1002/ijc.28444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.