Abstract
Multiplex Automated Genome Engineering (MAGE) allows simultaneous mutagenesis of multiple target sites in bacterial genomes using short oligonucleotides. However, large-scale mutagenesis requires hundreds to thousands of unique oligos, which are costly to synthesize and impossible to scale-up by traditional phosphoramidite column-based approaches. Here, we describe a novel method to amplify oligos from microarray chips for direct use in MAGE to perturb thousands of genomic sites simultaneously. We demonstrated the feasibility of large-scale mutagenesis by inserting T7 promoters upstream of 2585 operons in E. coli using this method, which we call Microarray-Oligonucleotide (MO)-MAGE. The resulting mutant library was characterized by high-throughput sequencing to show that all attempted insertions were estimated to have occurred at an average frequency of 0.02% per locus with 0.4 average insertions per cell. MO-MAGE enables cost-effective large-scale targeted genome engineering that should be useful for a variety of applications in synthetic biology and metabolic engineering.
Keywords: genome engineering, MAGE, metabolic engineering, microarray, library synthesis
A core aim of metabolic engineering and synthetic biology is to redesign and create biological systems with useful purposes, for example cell factories that produce novel medicines1 and chemicals.2 To achieve these goals, the need of a large set of efficient tools have spurred extensive research efforts dedicated to expanding the synthetic biology toolbox.3,4 The alteration of gene expression and rewiring of metabolic networks are important for basic research and metabolic engineering, and methods such as transposon sequencing (Tn-seq)5 and trackable multiplex recombineering (TRMR)6 allow genomic perturbations into a large number of sites. However, these approaches do not allow combinations of several mutations in individual cells and are confined mostly to generating gene knockouts that require integration of a sizable selectable marker, for which there are limited options. The recent development of Multiplex Automated Genome Engineering (MAGE)7 enables rapid and efficient targeted modification of the genome through iterative cycles of λ-Red mediated recombination using multiple oligonucleotides at once. MAGE with multiple degenerate oligos can be applied toward genome mutagenesis through a semirational approach, where specific targets are randomly altered to limit mutagenesis to only selected targets. Such application of MAGE has been shown to be useful for metabolic optimization to increase the biosynthetic production of lycopene and indigo in E. coli.8,9 In theory, MAGE should be amenable to targeted mutagenesis using thousands of oligos all at once to target hundreds to thousands of chromosomal targets. This capability will open new possibilities for many large-scale genome engineering projects.10
Mutagenesis of thousands of genomic targets by MAGE requires large oligo library pools. However, synthesis of thousands of MAGE oligos by traditional column-based phosphoramidite chemistry is impractical in both time and cost. Recent developments in high-fidelity oligonucleotide microarray technologies have enabled the construction of large libraries (>55 000 oligos) of 200 bp oligos11 at a significantly lower cost and turnaround time compared to oligos produced by column-based synthesis.12−14 Here, we describe Microarray Oligonucleotide (MO)-MAGE, a novel method to generate thousands of oligos suitable for large-scale genome engineering by MAGE and demonstrate its application for fast and robust targeted mutagenesis of the E. coli genome.
Results and Discussion
Using a computational framework for MAGE oligo design (MODEST),15 we first identified perturbation targets of the Escherichia coli genome that included most regulatory and protein coding regions (See Table 1 for details about which genes were targeted). Protein coding perturbations were made through the generation of a nonsense and frameshift mutation within the first 5% of the open-reading frame to functionally introduce a reversible gene knockout. Regulatory perturbations included up-regulation (“RBS up”) using consensus (AGGAGG), down-regulation (“RBS down”) using anticonsensus (TCCTCC) ribosomal binding sites, or insertion of a T7 promoter sequence (TAATACGACTCACTATAGGG) upstream of operons. A summary of the genomic perturbation design is given in Table 1. In all, the designed oligo library constituted 13 000 possible targeted perturbations against the E. coli genome (see Supporting Information).
Table 1. Overview of Oligos Designed and Synthesized on the Micro-Array Chipa.
| targets | general | TFs | genes targeted | |
|---|---|---|---|---|
| CDS knock outs | 3798 | 3633 | 167 | all nonessential, non-pseudo-, non-ncRNA |
| T7 promoters | 2723 | 2585 | 138 | all non-pseudo-, non-ncRNA,b |
| RBS up | 3323 | 3172 | 151 | all non-pseudo-, non-ncRNA |
| RBS down | 3099 | 2948 | 151 | all nonessential, non-pseudo-, non-ncRNA |
| total | 12 943 | 12 338 | 607 |
Four oligo subpools were made, intended for knocking out genes by introduction of a nonsense and frameshift mutation within the first 5% of the CDS (“CDS Knock Outs”), upregulation by insertion of T7 promoters upstream of genes (“T7 promoters”) and insertion of a consensus RBS sequence (“RBS up”), and down-regulation by insertion of an anti-consensus RBS sequence (“RBS down”). TFs = Transcription Factors.
Only genes with sufficient spacing to the next gene upstream were targeted, to ensure that the insertion of T7 promoters did not disturb upstream genes.
Traditional column-based synthesis of 90mer MAGE oligos of this library size would take months to years to generate at a cost of aproximately $500,000 USD. Thus, we turned to new approaches in long oligonucleotide synthesis using DNA microarrays.11 A 130mer single-stranded DNA library was generated using the Oligo Library Synthesis (OLS) platform from Agilent Technologies (Santa Clara, CA, U.S.A.). Because the amount of oligos needed for MAGE is much higher than the total yield from microarray synthesis (μM versus pM), we devised a PCR-based amplification protocol to generate a renewable supply of MAGE-compatible oligo pool from our initial microarray-derived oligo library, which we describe in greater detail later.
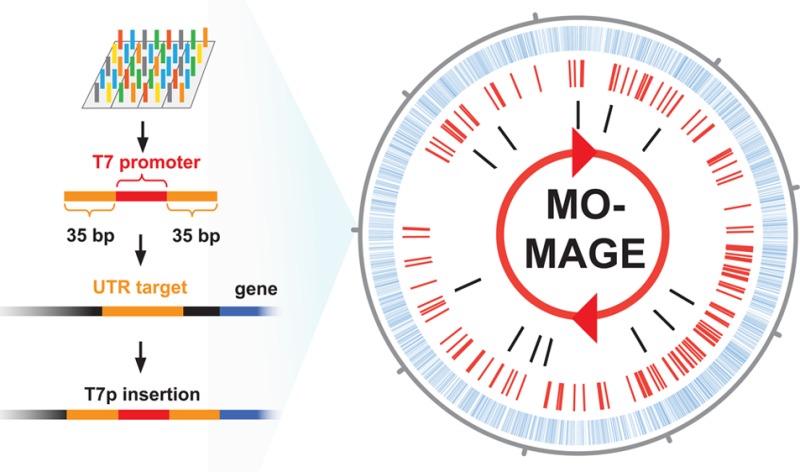
Each oligo library subpool (e.g., CDS Knockout, T7 insertion, RBS up, RBS down) was designed with a unique set of barcodes, which allowed for selective amplification of only the subpool library by using specific primers (see Figure 1). A total volume of 38.4 mL of PCR was first performed to ensure that we generated enough oligos for >10 MAGE cycles. One of the two primers used for the PCR contained a 5′-phosphothioester bond and a 3′-uracil, while the other contained a 5′ phosphorylated group. The resulting amplicons contain a 5′-phosphothioester bond on the MAGE-compatible sense-strand and a 5′-phosphate on the reverse complement strand. This 130 bp double-stranded (ds) DNA amplicon was digested with λ-exonuclease. Since λ-exonuclease has much higher activity for unphosphorylated substrates16 and its activity is blocked by phosphothioester bonds, the digest results in a 130mer single-stranded (ss) DNA library of the MAGE-compatible sense-strand that contained the phosphothioester bond at the 5′ end. Subsequently, the 20 bp flanking barcodes used for PCR amplification of the subpools were removed from each end of the ssDNA to yield unique 90mer oligos needed for high efficiency MAGE. This was accomplished by first digesting the 130mer ssDNA library, which contained an internal uracil at the barcode junction from the previous PCR (see Figure 1), with an uracil DNA glycosylase, endonuclease VIII,17 which removes the uracil from the ssDNA strand. The uracil excision effectively removes the 20mer barcode at the 5′ end of the ssDNA library, yielding a 110mer library. The 3′ barcode was designed with a DpnII restriction site placed immediately after the target oligo sequence. To remove the 3′ barcode, a guide primer complementary to the 3′ barcode including the DpnII site was used to hybridize to the 110mer oligo and was digested with DpnII to yield the designed 90mer oligo library. The use of a guide primer ensured that the remaining part of the single-stranded oligo is not digested by DpnII, which only cuts dsDNA. The resulting oligo library contained unique 90 base single-stranded oligos.
Figure 1.
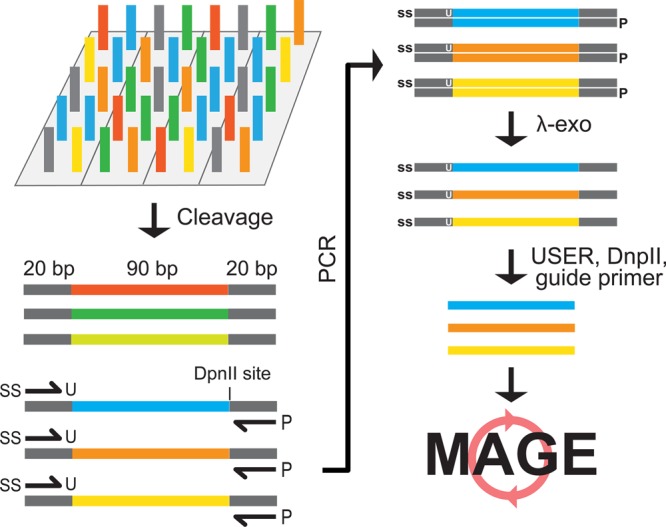
MO-MAGE method for targeted whole genome mutagenesis. 130 base oligonucleotides were designed and synthesized on a DNA microarray, which can be ordered from several commercial vendors. The oligos can be designed with different barcodes, which allow selective PCR amplification of a desired subpool. One of the primers are 5′ phosphorylated, which allow the degradation of only one of the strands by λ-exonuclease, resulting in single stranded oligos. The barcodes are removed by enzymatic treatment with endonuclease VIII (cutting the barcode by removing a uracil from the modified primer), DpnII and a guide primer (to make a double stranded cut site for DpnII). The final 90 bp single stranded oligos are directly applicable for MAGE.
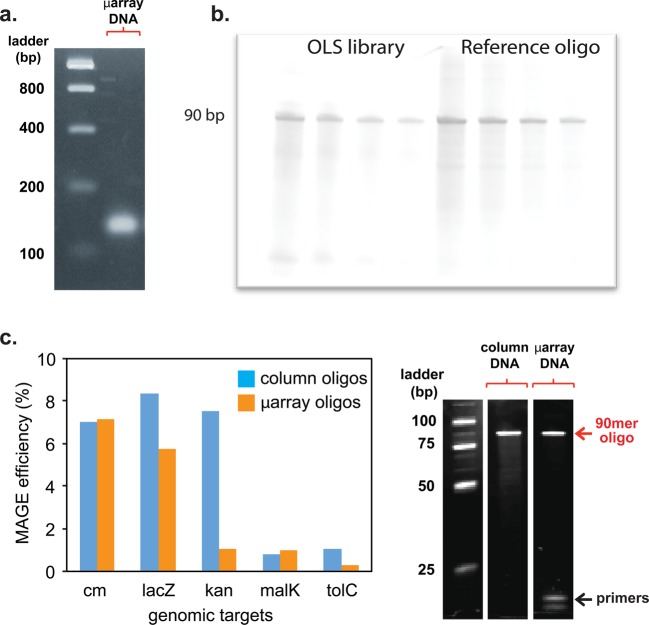
Gel electrophoresis of the oligos was performed after each step to ensure that the processing resulted in the expected product (see Figure 2a and b). Serial dilutions of the library were further visualized on a TBE-UREA gel to estimate library concentration using ImageJ.18 We estimate that a typical amplification generates 1.4 nmols of oligos (∼12 μM in 115 μL), which is sufficient for 14 MAGE cycles at 2 μM per 50 μL reaction per cycle.
Figure 2.
(a) PCR amplicons of the T7 promoter oligo pool can be seen as the strong bands around 130 bp (4% Agarose E-Gel EX with Low Range Quantitative DNA Ladder). (b) Serial dilutions of the processed single stranded T7 promoter oligo library (left 4 lanes) compared to a reference oligo of 90 bp (right 4 lanes), which indicates correct processing of the oligos from 130 bp to 90 bp oligos ready for MAGE (TBE-Urea gel 4% from Invitrogen). (c) Comparison of MAGE efficiency using column-synthesized oligos and microarray-processed oligos by MO-MAGE. Gel shows size distribution of the two processed oligo pools (TBE-Urea gel 4% from Invitrogen).
We performed a side-by-side comparison of the microarray-derived oligos with those obtained through standard column-synthesis from a commercial vendor (Integrated DNA Technologies, Iowa, U.S.A.). A test group of 5 oligos generated from the microarray pool and the commercial vendor was used for MAGE and the efficiency of oligo incorporation was determined (see Figure 2c). We find that 3 out of the 5 tested oligos showed slightly lower incorporation efficiencies in the microarray library compared to the column library. We attribute the decreased efficiency to differences in individual oligo concentrations that may result during amplification from the microarray library. Nonetheless, these results offered a convincing proof-of-concept that microarray-derived oligos are compatible with MAGE mutagenesis. Since the introduction of synthetic regulation to native genomic loci has been an outstanding challenge in synthetic biology, we sought to further explore our sublibrary that generated T7 promoter insertions in the untranslated regions (UTR) upstream of each of 2585 operons in the E. coli genome. This multiplexed promoter insertion perturbation enables the generation of a mutant library that contains new transcriptional regulation in the presence of an inducible T7 polymerase system.9,20 Thus, in this current work, we focused on characterizing our methods in greater detail using this T7 insertion library.
The T7 promoter insertion library was designed using MODEST15 to target the UTR region 40 bp upstream of all nonessential and nontranscription-factor operons in E. coli. Downstream polycistronic open reading frames without adequate intergenic space were not targeted. In all, 2585 unique insertion target sites were identified. We designed 90 bp oligos by flanking a 20 bp T7 promoter (TAATACGACTCACTATAGGG) sequence with 35 bp of homologous sequence at each end to the target genomic integration site. In general, introduction of a 20 bp insertion is expected to have lower incorporation efficiency than smaller point mutations or deletions.7 Nonetheless, these larger insertions represent a unique opportunity to introduce synthetic regulation and to challenge the limits of our MAGE capabilities using large oligo library pools derived from microarrays. Furthermore, effective selection of desirable mutants from a smaller genomic library with T7 promoter insertions have previously been performed successfully.9
Following the generation of the 2585-oligo library, we applied this T7 promoter pool to mutagenize the E. coli genome by MAGE for 12 cycles. The entire oligo processing pipeline and MAGE mutagenesis was performed twice in parallel to generate two separate cell libraries to test the reproducibility of our protocol. To assess the success of T7 insertion in these combinatorial libraries, we performed deep sequencing of the final cell populations. We first extracted genomic DNA of the cell populations and hybridized the genomic DNA with a biotinylated oligo containing the T7 promoter sequence. Genomic regions that contained the T7 promoter insertions can thus be enriched from total genomic DNA when it is applied to streptavidin beads that bind to the biotinylated oligo-genomic hybrid. We then sequenced the enriched genomic library by deep-sequenced to identify T7 insertion sites.
Next-generation sequencing analysis of the data showed that the two separate libraries contained 87 reads and 208 reads with the T7 promoter sequence, with the reads mapping to 56 and 98 targeted operons respectively in the two libraries. Only 4 of the total 154 targets were redundant, resulting in identification of 150 unique targets out of 2585 possible. The sequencing coverage (the number of times the genome has been sequenced) of the libraries was 2064× and 1364× respectively. We used a Monte Carlo model to simulate the insertion frequency of the total library, and found that the total cell library is expected to contain between 2250 and 3500 modified genes (95% confidence interval, see Supporting Information Figure 1). These results show that all or most of the targeted sites are predicted to have T7 promoter insertions within the cell library.
We proceeded to further validate our mutagenesis results and to estimate the insertion efficiency. We randomly selected 12 targets, including 8 that had not been detected by population sequencing. We amplified ∼200 bp PCR fragments spanning the T7 insertion sites of each of the 12 genes from the cell library and sequenced the PCR products by Next Generation Sequencing. For all 12 loci, we found reads containing the T7 promoter insertion sequence. This provides further support that the cell library contains a majority of the 2585 T7 promoter insertions (see Table 2 and Figure 3).
Table 2. Twelve Genes Randomly Selected for Deep Amplicon Sequencing and Analysisa.
| reads with T7 insertion | reads total | insertion frequency | |
|---|---|---|---|
| acrD | 549 | 452814 | 0.1212% |
| edd | 59 | 157788 | 0.0374% |
| osmC | 13 | 85100 | 0.0153% |
| fryB | 26 | 299019 | 0.0087% |
| SodB | 25 | 450003 | 0.0056% |
| pssA | 6 | 151518 | 0.0040% |
| secE | 15 | 561640 | 0.0027% |
| thrL | 4 | 197901 | 0.0020% |
| GlnD | 4 | 263023 | 0.0015% |
| acrA | 7 | 481259 | 0.0015% |
| mdaB | 2 | 237727 | 0.0008% |
| hemC | 1 | 129353 | 0.0008% |
The number of reads with a T7 promoter insertion was compared to reads without an insertion to calculate the insertion frequency.
Figure 3.
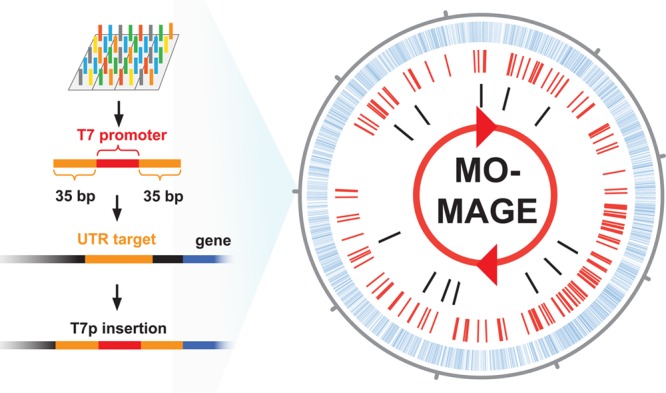
MO-MAGE of 2585 genomic targets corresponding to untranslated regions (UTR) upstream of genes for insertion of 20-bp T7 synthetic promoter. Designed targets are shown in blue. Mutated targets verified by whole-genome sequencing are shown in red (see Supporting Information Table 3 for complete list). Mutated targets verified by amplicon sequencing are shown in black.
The insertion frequencies for the 12 loci were estimated by comparing the number of reads with T7 promoter insertions to the number of reads without the insertion. The average frequency was 0.017%, and thus, the average number of insertions per cell can be estimated as μ = k × p = 0.434 where p = 0.00017 and k = 2585 targets. This means that 43% of the cell library is expected to have at least one insertion on average. The top 1% of the population is expected to have at least four (4.3) insertions based on the calculations m = μ + 2.326(k × p(1 – p))1/2 = 4.3 (for details about the MAGE efficiency calculations see the Supporting Information as well as Wang and Church, 20117). The MAGE efficiency for insertions has previously been predicted based on fitting of empirically determined efficiencies.7,8 Using such approach, the insertion efficiency (IE) is IE = 0.15 × e–0.075 × (b–1) = 0.0361, where b is the size of the insertion in number of bases (b = 20). Based on this insertion efficiency, the predicted average frequency of each insertion can be calculated by pN–predicted = 1 – (1 – IE/k)N = 1 – (1 – 0.0361/2585)12 = 0.000167, where N is the number of MAGE cycles. The predicted average insertion frequency of 0.0167% matches very well with the measured frequency of 0.017%, providing confidence to our predicted population mutagenesis profile and its applications to analyze MAGE mutagenesis of complex oligo pools. These results further highlight that that cell libraries with combinations of multiple insertions per genome can be generated using this massively multiplexed approach.
Here, we have presented a proof-of-concept demonstration for the generation and application of oligo pools that specifically target thousands of unique chromosomal loci across a cell population to introduce promoters amenable for synthetic regulation. The Microarray Oligonucleotide MAGE enables rapid mutagenesis of bacterial genomes using oligos derived directly from microarrays without intermediate cloning or cassette selection steps, which can significantly expand the combinatorial genomic diversity of the resulting cell population. A resulting cell library will be useful for various screens for metabolic engineering purposes such as increased production of biochemicals or tolerance toward biomass inhibitors. The MO-MAGE process described here allow researchers to limit the combinatorial space to only specific mutations that are expected to have a much higher chance of leading to a desired phenotype than random mutations. Thus, the effect of rationally designed targets can be assessed very effectively because the mutagenesis quality (i.e., the proportion of interesting mutants to other cells) is much higher. Whereas current approaches are limited to only creating few genomic changes at a time, our method could be used to target all predicted beneficial mutations predicted from metabolic models, and screen for optimal phenotypes.
As the complexity of engineering tasks of synthetic biology and metabolic engineering increases, the need to reduce cost of genome engineering becomes more important. MO-MAGE allows the synthesis of oligos at a fraction of the cost (>1000×) compared to traditional column based oligo synthesis to make large-scale genome engineering accessible to most laboratories. At a price of 36 USD per column-based oligo, MO-MAGE (2800 USD) is currently cost competitive when using more than 78 oligos. This method could provide a paradigm shift by making large-scale genome engineering of many thousands of targets available as a standard tool for strain optimization and other projects where large-scale targeted mutagenesis searches are needed. New advances in DNA and gene synthesis will further foster growth in genome engineering of microbial and eukaryotic systems,23 and in theory, oligo pools containing millions of oligos can be applied for MO-MAGE. However, since increasing oligo diversity leads to lower replacement efficiency per genomic target, there is a practical limitation in the number of oligos that can be meaningfully applied depending on the required replacement efficiency per site for a given experiment.
For some projects, a higher integration frequency might be of interest, to allow more combinations of insertions and a higher quality library. The amount of MAGE cycles performed can be increased to higher levels, and automated solutions could increase the feasible number of MAGE cycles to several hundreds. For creation of complex libraries where many combinations per cell are desired, single base pair substitutions and small insertions and deletions can be applied to increase the replacement efficiency. For instance if 1000 oligos designed for making single base pair substitutions are used for 100 cycles of MAGE, a cell library with 25 average replacements per cell is predicted. This allows creation of unprecedented targeted combinatorial libraries of specific chromosomal modifications. Ultimately, we envision MO-MAGE method will be used to make thousands of specific chromosomal changes predicted to result in a desired phenotype and combined with selection or screening of the cell library for interesting mutants.
Methods
Strains and Culture Conditions
We used a strain based on the EcNR28 strain for all experiments, which is based on E. coli K12 MG1655. The genotype is λ-Red+bla+bioA–/bioB– mutS–zeo+ Lac_T7pol+Spec+ LacIQ+. All strains for MAGE were grown in low salt LB-min medium (10 g tryptone, 5 g yeast extract, 5 g NaCl in 1 L dH2O) for optimal electroporation efficiency with addition of specified antibiotics. All cells for liquid cultures were grown in standard LB-min medium (10 g tryptone, 5 g yeast extract, 10 g NaCl in 1 L dH2O) with addition of specified antibiotics.
Standard Oligonucleotides
All standard oligonucleotides were purchased from Integrated DNA Technologies with standard purification.
Oligo Library Synthesis
OLS pools were synthesized by Agilent Technologies and are available upon signing a Collaborative Technology Development agreement with Agilent. Costs of OLS pools are a function of the number of unique oligos synthesized and of the length of the oligos The OLS pool were synthesized, cleaved, and delivered as lyophilized ∼1–10 picomole pools.
Oligo Library Processing
Please refer to the Supporting Information.
Oligos for T7 Cell Library Generation
A list of all the 2585 oligos synthesized on the microarray chip designed to insert a T7 promoter upstream of E. coli genes can be found in the Supporting Information.
MAGE
MAGE was performed according to the protocol provided in ref (7). Briefly, the cells were grown to midlog phase, whereafter the β-protein of the λ-red system was induced by growing at 42 °C for 15 min whereafter the cells were chilled to 4 °C. The culture were washed to remove salts and resuspended in cold water (<4 °C). The cells were mixed with the 2 μM oligos in 50 μL and electroporated in a Bio-Rad MicroPulser, BTX ECM-830 with 1 mm gap cuvette, whereafter the cells were incubated for 2–3 h at 30 °C. The process were repeated 12 times (12 MAGE cycles) to allow a higher frequency of insertion. After 5 and 10 MAGE cycles, the cells were grown overnight in 50 mL LB low salt medium and stored at −80 °C in a 15% v/v glycerol solution.
Enrichment for T7 Promoter Containing Sequences
The enrichment protocol was based on Gnirke et al.24 and NimbleGen SeqCap EZ Exome Library SR v2.2 protocol.25 A biotinylated oligo targeting the T7 promoter was incubated 66 h with the prepared sequencing library fragments. The T7 promoter containing fragments that hybridized to the biotinylated oligo was enriched by multiple rounds of binding and washing with Invitrogen Streptavidin M-270 Dynabeads and Invitrogen binding and wash buffer.
Illumina DNA Sequencing and Analysis
Samples for sequencing of the 12 individual strains were processed with Illumina TruSeq v2 sample prep kit and standard Illumina adapters. Samples for sequencing of the cell libraries were prepared with the NEBNext DNA Sample Prep Master Mix Set 1 kit for Illumina sequencing and manually ordered adaptors kindly provided by Luhan Yang (Harvard Medical School, George Church Lab).
| adaptor 1 | PE-A1-F | TACACTCTTTCCCTCACGACGCTCTTCGATCTac*T |
| PE-A1-R | /5Phos/gtAGATCGGAGAGCGGTTCAGCGGAATGCCGAG | |
| adaptor 2 | PE-A2-F | TACACTCTTTCCCTCACGACGCTCTTCGATCTtg*T |
| PE-A2-R | /5Phos/caAGATCGGAGAGCGGTTCAGCGGAATGCCGAG |
The libraries were sequenced in two separate lanes, whereas the 12 isolated strains were sequenced in one lane with multiplexing barcodes. The 12 prepared individual strains were sent to the Harvard Biopolymers facility (genome.med.harvard.edu) for Illumina sequencing and downloaded to the cbs.dtu.dk server and processed here. Sequences containing the T7 promoter sequence were extracted with the “grep” command. BLASTn was performed in CLC Bio main Workbench 6.0 with standard settings: “word size” 11, “match” 1, “mismatch” −3, “gap cost existence” 5, “gap cost extension” 2. Bowtie(26) was applied to perform the alignment of the reads to the reference genomes (see parameters in the script in Supporting Information). Samtools(27) was applied to make a consensus reference genome and indexed BAM-file (for visual inspection of read alignment) from the bowtie output (see parameters in the script in Supporting Information)
PCR and freq-seq of 12 Loci
PCR primers were designed for amplification of 12 genomic regions of around 200 bp with the T7 promoter insertion site in the middle. The amplicons were sequenced on an Illumina MiSeq, and a script created to extract all WT and mutant sequences and report the numbers.
Acknowledgments
H.H.W. acknowledges funding from the National Institutes of Health Director’s Early Independence Award (1DP5OD009172-01). G.M.C acknowledges funding from the Department of Energy (DOE) (as above) and the National Science Foundation (NSF) (SA5283-11210). M.T.B., H.J.G., K.S.L. and M.O.A.S. acknowledge funding from the Novo Nordisk Foundation and the European Union Seventh Framework Programme (FP7-KBBE-2013-7-single-stage) under grant agreement no. 613745, Promys.
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
# M.T.B. and S.K. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Weber W.; Fussenegger M. (2012) Emerging biomedical applications of synthetic biology. Nat. Rev. Genet. 13, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L. (2007) Synthetic biology: Promises and challenges. Mol. Syst. Biol. 3, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyo K. E.; Alper H. S.; Stephanopoulos G. N. (2007) Expanding the metabolic engineering toolbox: More options to engineer cells. Trends Biotechnol. 25, 132–7. [DOI] [PubMed] [Google Scholar]
- Khalil A. S.; Collins J. J. (2010) Synthetic biology: Applications come of age. Nat. Rev. Genet. 11, 367–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Opijnen T.; Camilli A. (2013) Transposon insertion sequencing: A new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R.; Reeder P. J.; Karimpour-Fard A.; Woodruff L. B. a; Gill R. T. (2010) Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat. Biotechnol. 28, 856–62. [DOI] [PubMed] [Google Scholar]
- Wang H. H.; Church G. M. (2011) Multiplexed genome engineering and genotyping methods applications for synthetic biology and metabolic engineering. Methods Enzymol. 498, 409–26. [DOI] [PubMed] [Google Scholar]
- Wang H. H.; Isaacs F. J.; Carr P. A.; Sun Z. Z.; Xu G.; Forest C. R.; Church G. M. (2009) Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. H.; Kim H.; Cong L.; Jeong J.; Bang D.; Church G. M. (2012) Genome-scale promoter engineering by coselection MAGE. Nat. Methods 9, 591–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt K. M.; Wang H. H. (2013) Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 9, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S.; Eroshenko N.; Leproust E. M.; Super M.; Way J.; Li J. B.; Church G. M. (2010) Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat. Biotechnol. 28, 1295–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J.; Gong H.; Sheng N.; Zhou X.; Gulari E.; Gao X.; Church G. (2004) Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432, 1050–1054. [DOI] [PubMed] [Google Scholar]
- Tian J.; Ma K.; Saaem I. (2009) Advancing high-throughput gene synthesis technology. Mol. Biosyst. 5, 714–22. [DOI] [PubMed] [Google Scholar]
- Richmond K. E.; Li M.-H.; Rodesch M. J.; Patel M.; Lowe A. M.; Kim C.; Chu L. L.; Venkataramaian N.; Flickinger S. F.; Kaysen J.; Belshaw P. J.; Sussman M. R.; Cerrina F. (2004) Amplification and assembly of chip-eluted DNA (AACED): A method for high-throughput gene synthesis. Nucleic Acids Res. 32, 5011–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonde M. T.; Klausen M. S.; Anderson M. V.; Wallin A. I. N.; Sommer M. O. A. (2014) MODEST : A web-based design tool for oligonucleotide-mediated genome engineering and recombineering. Nucleic Acids Res. 10.1093/nar/gku428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsis P. G.; Kwagh J. G. (1999) Characterization of the interaction of lambda exonuclease with the ends of DNA. Nucleic Acids Res. 27, 3057–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitinaite J., and Nichols N. M. (2009) DNA cloning and engineering by uracil excision. Curr. Protoc. Mol. Biol. Chapter 3, DOI: 10.1002/0471142727.mb0321s86. [DOI] [PubMed] [Google Scholar]
- Abramoff M. D.; Magalhaes P. J.; Ram S. J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42. [Google Scholar]
- Temme K.; Hill R.; Segall-Shapiro T. H.; Moser F.; Voigt C. A. (2012) Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 40, 8773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeProust E. (2008) Agilent’s microarray platform: How high-fidelity DNA synthesis maximizes the dynamic range of gene expression measurements. Agilent Technologies, Santa Clara, CA.
- Gnirke A.; Melnikov A.; Maguire J.; Rogov P.; LeProust E. M.; Brockman W.; Fennell T.; Giannoukos G.; Fisher S.; Russ C.; Gabriel S.; Jaffe D. B.; Lander E. S.; Nusbaum C. (2009) Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27, 182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimblegen (2013) NimbleGen SeqCap EZ Library SR User’s Guide; Roche Nimblegen, Madison, WI. [Google Scholar]
- Langmead B.; Trapnell C.; Pop M.; Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P.; Birney E. (2009) Sense from sequence reads: Methods for alignment and assembly. Nat. Methods 6, S6–S12 10.1038/nmeth.1376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.