Abstract
Deubiquitinases (DUBs) play important roles and therefore are potential drug targets in various diseases including cancer and neurodegeneration. In this review, we recapitulate structure–function studies of the most studied DUBs including USP7, USP22, CYLD, UCHL1, BAP1, A20, as well as ataxin 3 and connect them to regulatory mechanisms and their growing protein interaction networks. We then describe DUBs that have been associated with endocrine carcinogenesis with a focus on prostate, ovarian, and thyroid cancer, pheochromocytoma, and adrenocortical carcinoma. The goal is enhancing our understanding of the connection between dysregulated DUBs and cancer to permit the design of therapeutics and to establish biomarkers that could be used in diagnosis and prognosis.
Keywords: deubiquitinases, USP7, USP22, CYLD, UCHL1, BAP1, A20, ataxin 3
Ubiquitination
The ubiquitin–proteasome system (UPS) has many critical regulatory roles in eukaryotic cellular processes including cell cycle progression, stress response, signal transduction, transcriptional activation, and DNA repair (Ciechanover et al. 2000, Ciechanover 2006). Malfunction, dysregulation, or molecular defects within the UPS lead to several different disease states including various cancers as well as neurodegenerative disorders (Ciechanover 2003, Ciechanover & Schwartz 2004). The UPS is responsible for the regulation, stability, and turnover of many different substrate proteins. However, the post-translational ubiquitination of a target protein has many other fates within eukaryotic cells as well.
Ubiquitin is a 76-residue eukaryotic polypeptide that is covalently attached to target proteins through an isopeptide bond formed between its terminal Gly residue and the ε-amino group of a lysine residue on the target protein. The ubiquitin superfamily of proteins possesses a similar three-dimensional structure, called the ββαββ fold or β-grasp fold, in the absence of sequence similarity. Proteins other than ubiquitin that possess this fold are called ubiquitin-like proteins and ubiquitin-like domains are found in many different proteins involved in the ubiquitination system. Ubiquitin is encoded by four different genes (UBB, UBC, UBA52, and UBA80 (RPS27A)) and is produced as a precursor that must be proteolytically processed to release mature ubiquitin. The ubiquitin amino acid sequence is much conserved between eukaryotic organisms with only two residue changes between the yeast and human proteins. A single ubiquitin moiety can be attached to substrate proteins resulting in mono-ubiquitination. Mono-ubiquitination is involved in a variety of cellular processing including protein translocation, kinase activation, epigenetic regulation, and DNA damage signaling (Miranda & Sorkin 2007, Wang et al. 2012). The N-terminal methionine and the seven lysine residues in ubiquitin (K6, K11, K27, K29, K33 K48, and K63) can form polyubiquitin chains. The attachment of a polyubiquitin chain to substrate proteins results in a variety of cellular effects. The best characterized is Lys48 (K48) polyubiquitination, which targets proteins for degradation by the 26S proteasome complex (Hochstrasser 1996, Hicke 2001). Lys63 (K63) polyubiquitination plays important roles in DNA damage signaling and recruitment of DNA repair proteins to the site of damage (Panier & Durocher 2009). K63 and linear polyubiquitination function in the activation of the nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) signaling pathway (Chen 2005, Terzic et al. 2007, Iwai & Tokunaga 2009). Ubiquitination plays many additional roles depending on the chain configuration and linkage (Komander 2009). There is increasing evidence for the formation of more complex topologies with mixed linkages with unknown functions.
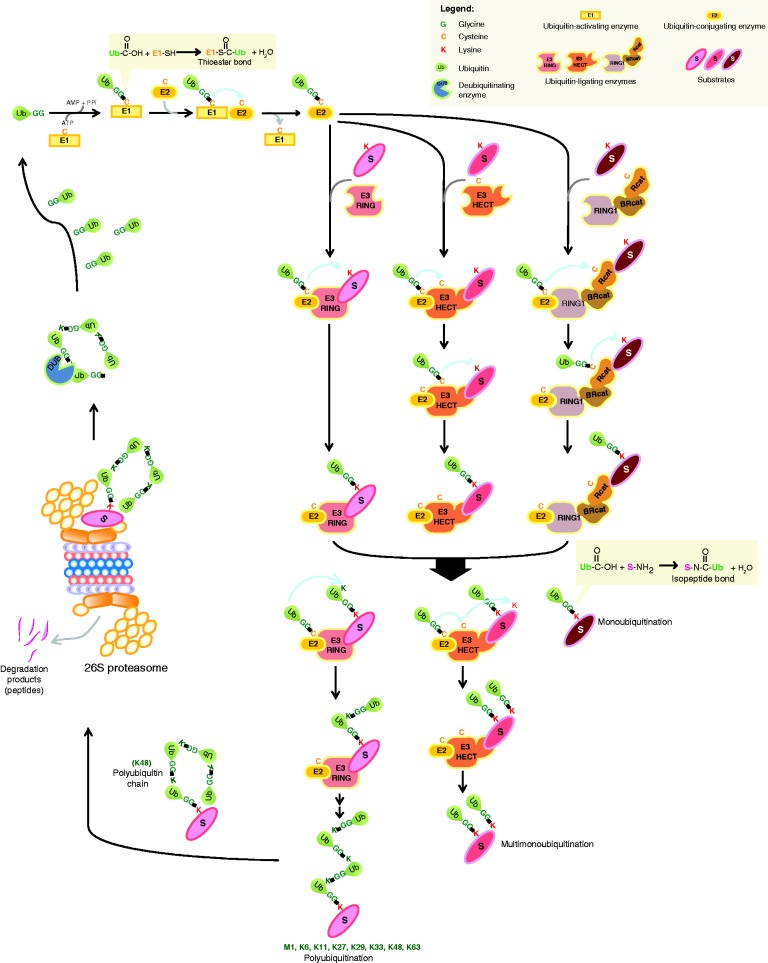
The attachment of ubiquitin to target proteins is mediated by an enzymatic cascade consisting of E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzymes), and E3 (ubiquitin-protein ligases) proteins (Fig. 1; Fang & Weissman 2004). E1 is the ubiquitin-activating enzyme that catalyzes the ATP-dependent activation of ubiquitin resulting in the formation of a thioester bond between the C-terminus of ubiquitin and a cysteine residue in the E1 active site. Activated ubiquitin is subsequently transferred to an E2 ubiquitin-conjugating enzyme and again forms a thioester bond between its C-terminus and a cysteine residue in the E2 active site. There are at least four different classes of E3 ubiquitin protein ligases whose function is to recognize and catalyze the ubiquitination of the substrate protein (Pickart 2001). The first class of E3 ubiquitin protein ligases contains a Really Interesting New Gene (RING) domain and acts as a scaffold between the ubiquitin-conjugated E2 and the target protein to be ubiquitinated. The RING domain-containing protein is essential for target protein recognition and mediates the transfer of the ubiquitin from the E2 onto the target protein. Another class of E3 ligases contains a homologous to the E6-AP carboxyl terminus (HECT) domain. Unlike RING domain proteins, HECT domain proteins actually catalyze the transfer of ubiquitin from the loaded E2 onto a cysteine residue in their active site forming a thioester bond and then finally onto the substrate protein. The third class is called U-box, whose members function like RING E3 ligases and have a RING-like fold; however, they lack the cysteine and histidine Zn-coordination sites (Aravind & Koonin 2000). The latest identified class of E3 ligases is the RING-inbetween-RING (RBR) family, which are hybrid RING/HECT type E3 ligases containing three variant RING domains, known as RING1, IBR/BRcat, and RING2/Rcat (Smit & Sixma 2014, Spratt et al. 2014). Similar to HECT proteins, they catalyze the transfer of ubiquitin to a cysteine in the active site of the RING2/Rcat domain. The RING1 domain mediates interactions with the E2 enzyme. The specificity and type of ubiquitination is generated from substrate recognition by the E3 and the formation of different E2–E3 complex combinations that mediate ubiquitination (Weissman 2001).
Figure 1.
Enzymatic cascades resulting in the ubiquitination, deubiquitination, and 26S proteasome degradation of substrate proteins.
Deubiquitination
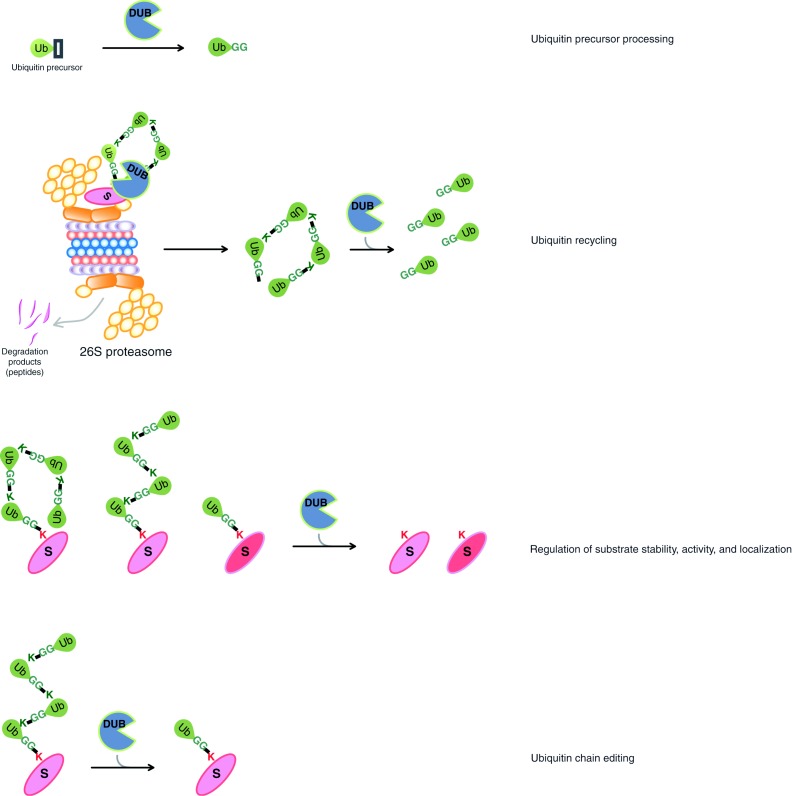
Protein ubiquitination is both dynamic and reversible. Deubiquitinases or deubiquitinating enzymes (DUBs) catalyze the removal of ubiquitin from target proteins and are also involved in ubiquitin maturation, recycling, and editing (Fig. 2; Kim et al. 2003, Amerik & Hochstrasser 2004, Nijman et al. 2005, Reyes-Turcu & Wilkinson 2009, Clague et al. 2013). Processes that can be regulated by deubiquitination include the rescue of proteins that are destined for degradation, the cleavage to release mature ubiquitin, the editing of a polyubiquitin on a substrate protein as well as the removal of ubiquitin to terminate or alter a biological event. DUBs are associated with the 26S proteasome to rescue ubiquitin chains before the degradation of the substrate protein. These unanchored polyubiquitin chains are then processed by other DUBs to replenish the free ubiquitin pool in the cell. DUBs are divided into two main classes according to their enzymatic cleavage mechanism: cysteine proteases and zinc metalloproteases. The larger group of cysteine proteases is further divided into four classes according to sequence conservation in the catalytic domain. Currently, the five known DUB families include ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs), Machado–Joseph disease (MJD) protein domain proteases, ovarian tumor proteases (OTUs), and JAMM motif zinc metalloproteases (Nijman et al. 2005). There are ∼90 human DUBs; however, the biological roles and target proteins are unknown for many of them (Nijman et al. 2005, Clague et al. 2013). The importance of DUBs in cellular functions as well as carcinogenesis has been recently reviewed (Luise et al. 2011, D'Arcy & Linder 2012, Fraile et al. 2012, Bhattacharya & Ghosh 2014a, Pal et al. 2014). By far, USPs are the largest class of DUBs with ∼60 proteases in humans, their sizes ranging from ∼50–300 kDa and most of them containing several domains in addition to the catalytic domain. Sequence conservation among these proteases is limited to the catalytic domain, which is characterized by an active site catalytic triad containing Cys, His, and Asp (or Asn) residues. As the non-catalytic domains are highly diverse at the amino acid sequence level, it is hypothesized that they are important for conferring substrate specificity, regulation of the catalytic activity, and mediating protein interactions to the individual USPs. UCHs were the first identified family of DUBs with four members in humans. The role of UCHs was initially proposed to be in the salvage and maturation of ubiquitin, as they are unable to cleave long polyubiquitin chains. The remaining families of DUBs, OTU, Josephin, and JAMM are described later in this review. DUB specificity and regulation are currently being investigated. With so many DUBs, it is easy to imagine that each DUB will have different modes of regulation and substrates. Most importantly, the majority of DUBs are found in protein complexes, whereby specificity and regulation can be controlled. Other DUBs are activated by post-translational modifications including phosphorylation, sumoylation, and ubiquitination. Some DUBs are specific for their target substrates and some DUBs are also specific for the ubiquitin chain type (K48 vs K63 linked). Most DUBs specifically recognize ubiquitin and cannot catalyze the proteolysis of ubiquitin-like proteins. DUBs also have specificity that determines where proteolysis occurs with some DUBs cleaving from the end of a polyubiquitin chain, whereas others cleave internally.
Figure 2.
Reactions catalyzed by deubiquitinases (DUBs). (A) Mature ubiquitin is generated following DUB cleavage. (B) DUBs recycle ubiquitin on proteins destined for degradation by the 26S proteasome. (C) DUBs deubiquitinate substrate proteins. (D) DUBs regulate ubiquitin chain editing.
Deubiquitinases
All DUBs that play important roles in pathways that are dysregulated in cancer including DNA repair, cell growth, and apoptosis are potential drug targets. However, the difficulty in development of effective drugs may be the challenge in designing specific inhibitors for a single DUB. This challenge is similar to that encountered in the design of inhibitors against protein kinases that have hampered drug development for many years. However, there are currently several potent kinase inhibitor drugs in the market, indicating that these concerns are indeed valid and challenging but certainly not insurmountable. The three-dimensional structures of the catalytic domains of USP2, USP7, USP8, and USP14 have revealed striking structural conservation of their active site, thus substantiating the concerns that drug development may be difficult (Hu et al. 2002, 2005, Avvakumov et al. 2006, Renatus et al. 2006). Interestingly, these crystal structures also revealed that these catalytic domains were in inactive conformations before ubiquitin binding, suggesting alternative targets for drug design. As the binding of ubiquitin activates USPs, it is desirable to identify compounds that bind to the catalytic domains and prevent the binding of ubiquitin. However, in parallel to targeting the catalytic domains of USPs for drug design, it would be advantageous to target individual substrate recognition, regulatory or protein interaction domains, as this would avoid concerns about cross-reactivity. Each USP has at least one or more of these domains and there is no sequence or structure conservation between most USP domains making these attractive drug-interaction sites. The ability to prevent complex formation between USPs and their ubiquitinated substrates is desirable for the design of drug compounds that interfere with specific deubiquitination processes. Therefore, structural analysis to reveal the molecular mechanism of interaction between USPs and their substrates will greatly enhance the development of these inhibitors. However, directly targeting large protein–protein interaction surfaces is quite challenging and requires precise structural knowledge of the interaction sites. The ability to understand the pathways and proteins that are involved in key regulatory events leading to disease will allow the development of therapeutics that targets only these proteins.
The structure–function relationships of selected DUBs will be discussed in the following section. We chose at least one example from each of the five DUB classes (USP-class: USP7, CYLD, and USP22; OTU-class: A20; UCH-class: UCHL1 and BRCA1-associated protein 1 (BAP1); MJD-class: ataxin 3 (ATXN3); and JAMM-class: Rpn11). Within their classes, the DUBS were selected based on the number of publications, disease relevance, as well as the available data on structure–function relationships. Overall, USP7, CYLD, UCHL1, BAP1, A20, and ATXN3 are the most intensively studied DUBS based on the number of citations in PubMed. All of the selected DUBS are related to cancer.
Ubiquitin-specific protease 7
USP7 was initially identified through its interaction with the ICP0 protein of herpes simplex virus (Meredith et al. 1994). Subsequently, USP7 was also shown to interact with the EBNA1 protein from Epstein–Barr virus, vIRF4 and LANA proteins from Kaposi's sarcoma herpesvirus, UL35 from cytomegalovirus, and E1B-55K from adenovirus (Holowaty et al. 2003, Saridakis et al. 2005, Lee et al. 2011, Jager et al. 2012, Salsman et al. 2012, Ching et al. 2013). Therefore, USP7 has emerged as a common target of herpesviruses (and perhaps other DNA viruses) living up to its original name of herpesvirus-associated USP (HAUSP).
USP7 regulates the turnover of many critical proteins involved in tumor suppression and DNA repair. p53, a tumor suppressor that induces cell cycle arrest and apoptosis, was the first protein to be identified as a USP7 substrate (Li et al. 2002a). Apart from p53, USP7 also deubiquitinates and stabilizes Hdm2 and HdmX, which are negative regulators of p53, indicating a complex regulatory mechanism (Meulmeester et al. 2005). Hdm2 is preferentially targeted by USP7 over p53 under normal unstressed conditions; however, USP7 targets p53 in response to DNA damage (Meulmeester et al. 2005). It has recently been shown that both GMPS and Abro1 modulate USP7-mediated deubiquitination of p53 after DNA damage (Reddy et al. 2014, Zhang et al. 2014). The deletion of the USP7 gene resulted in early embryonic lethality partly due to increased p53 levels (Kon et al. 2010). USP7 regulates the localization, rather than the stability, of tumor suppressors phosphatase and tensin homolog (PTEN) and FOXO4 by deubiquitinating their monoubiquitinated, transcriptionally active, nuclear forms (van der Horst et al. 2006, Song et al. 2008). USP7 also plays important roles in genomic stability by deubiquitinating and stabilizing many proteins involved in these pathways including BUB3 (Giovinazzi et al. 2014), claspin (Faustrup et al. 2009), ARF-BP1 (Khoronenkova & Dianov 2013), and others. These USP7 substrate proteins play critical roles in pathways that are often dysregulated in cancer highlighting the importance of investigating USP7 as a therapeutic target in cancers involving these proteins.
GMPS is not only involved in nucleotide biosynthesis but also modulates the function of USP7 deubiquitination of both H2B and p53, resulting in epigenetic silencing and DNA repair respectively (van der Knaap et al. 2005, 2009, Sarkari et al. 2009, Reddy et al. 2014). The role of USP7 in epigenetic silencing is not limited to H2B deubiquitination, as it also deubiquitinates and stabilizes RING1B, BMI1, UHRF1, and DNMT1, proteins that are involved in chromatin remodeling and epigenetic regulation (de Bie et al. 2010, Du et al. 2010, Maertens et al. 2010).
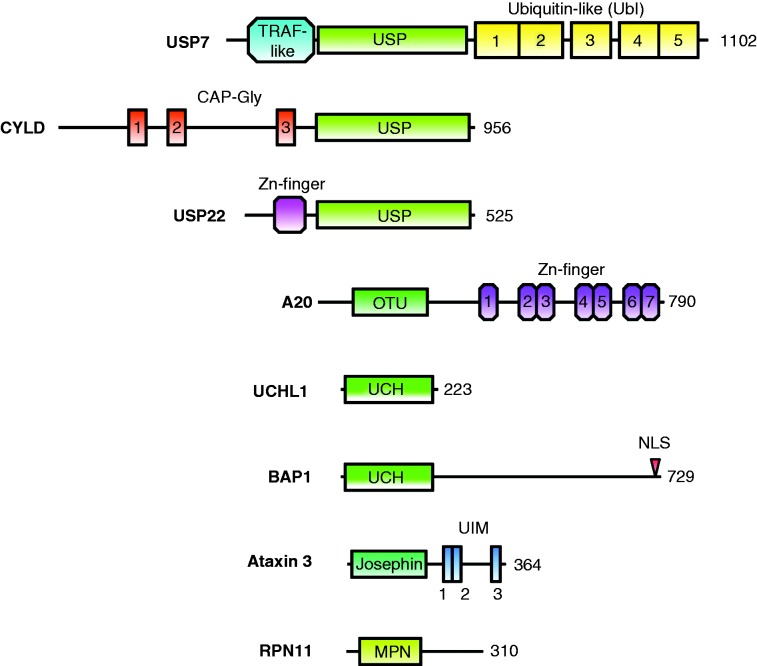
Individual USP7 domains have been structurally characterized, including the catalytic domain with bound ubiquitin-aldehyde (Hu et al. 2002), the TNF receptor-associated factor (TRAF)-like N-terminal domain (USP7-NTD) with various peptides from different substrates (p53, Hdm2, HdmX, and UbE2E1) (Sheng et al. 2006, Sarkari et al. 2010, 2013) as well as the five ubiquitin-like domains of the C-terminus (Faesen et al. 2011) (Fig. 3). USP7-NTD is a protein–protein interaction domain involved primarily in substrate recognition, whereas USP7-CTD is involved in protein–protein interactions and regulation of catalytic activity.
Figure 3.
Domain architecture of selected DUBs: USP7, CYLD, USP22, A20, UCHL1, BAP1, ataxin 3, and RPN11.
The ability to understand how and when USP7 deubiquitinates each substrate is essential to our overall understanding of its function. Owing to its regulation of Hdm2, HdmX, and p53, USP7 is an exemplary USP drug target. In cancers that are caused by a dysregulated Hdm2 protein but retain WT p53, the ability to inhibit USP7 deubiquitination of Hdm2 is considered to be of therapeutic importance because Hdm2 degradation would lead to p53 stabilization and activation. Therefore, a drug that prevents the formation of the Hdm2/USP7 complex or inhibits its catalytic activity will benefit patients with cancer associated with a dysregulated Hdm2 protein. Elevated levels of USP7 have been recently discovered in prostate cancer and glioma (Song et al. 2008, Bhattacharya & Ghosh 2014b). USP7 was also implicated to modulate tumor growth and apoptosis in a colon carcinoma xenograft model (Becker et al. 2008). As well, reduced expression of USP7 was observed in non-small cell lung carcinomas (Masuya et al. 2006). The recent discoveries of new USP7 substrates (such as Rb and BMI) implicate its possible involvement in other cancers indicating a need for USP7 therapeutics (Maertens et al. 2010, Bhattacharya & Ghosh 2014b).
Several USP7 inhibitors have been tested in a variety of in vitro and in vivo assays (Colland et al. 2009, Chauhan et al. 2012, Reverdy et al. 2012, Weinstock et al. 2012, Fan et al. 2013, Yamaguchi et al. 2013). HBX 41,108 and P5091 are USP7 inhibitors with IC50 values of submicromolar concentrations (Colland et al. 2009, Chauhan et al. 2012). The potency of HBX 41,108 has been studied in HCT116, human colon carcinoma cells, where it was shown to inhibit cell growth and induce apoptosis (Colland et al. 2009). The potency of P5091 was shown to induce apoptosis in multiple myeloma cells resistant to bortezomib (Chauhan et al. 2012). However, to date, there have not been any studies evaluating USP7 inhibitors in endocrine-related cancers.
CYLD
Similar to USP7, CYLD belongs to the USP class of cysteine proteases (Fig. 3). Together with A20 (see below), CYLD regulates the pathway of NFκB (Brummelkamp et al. 2003, Trompouki et al. 2003), which is a key regulatory protein in the response of the immune system to infections. Dysregulation of the NFκB pathway is involved not only in cancer (Massoumi 2011), but also in a number of other diseases such as viral infection (Chan & Greene 2011) and inflammatory diseases (Harhaj & Dixit 2012). It has been recently suggested that CYLD down-regulation could increase breast cancer metastasis through NFκB activation (Hayashi et al. 2014). The USP domain of CYLD is located at the C-terminus and contains a zinc-binding B-box domain. Though similar to the RING finger, the zinc-binding domain does not show E3 ligase activity and does not alter enzymatic activity or substrate specificity, but is relevant for the localization of CYLD to the cytoplasm (Komander et al. 2008). Compared with other USP domains (e.g. USP7), the USP finger region involved in ubiquitin binding is shorter, which could explain the preference of CYLD for K63-linked ubiquitin chains (Komander et al. 2008). In addition to the USP domain, CYLD contains three cytoskeleton-associated protein-glycine-conserved (CAP-Gly) domains. Its three-dimensional fold is similar to the SH domain, which is a binding motif for proline-rich sequences (Saito et al. 2004). CYLD negatively regulates the NFκB pathway by deubiquitinating critical factors such as NFκB essential modulator (NEMO, also called IKKγ) and TRAF2 (Kovalenko et al. 2003).
Ubiquitin-specific protease 22
USP22 consists of 525 amino acids and contains an N-terminal UBP-type zinc finger and a C-terminal USP domain. It is involved in the deubiquitination of NAD-dependent protein deacetylase sirtuin 1 (SIRT1; Lin et al. 2012), as well as histones H2A and H2B, as part of the transcriptional regulatory histone acetylation and deubiquitination complex known as Spt-Ada-Gcn5 acetyltransferase (SAGA). Structural data on USP22 are not available, but its yeast ortholog Ubp8 (ubiquitin processing protease 8) has been crystallized within the deubiquitination module of the SAGA complex (Köhler et al. 2010, Samara et al. 2010). The SAGA DUB module is a tight complex between Ubp8, Sgf11, Sgf73, and Sus1, and Ubp8 is allosterically regulated by its binding partners (Samara et al. 2012). USP22 is involved in oncogenesis due to its role in a large variety of cancers (Schrecengost et al. 2014). USP22 expression is increased in laryngeal squamous cell carcinoma (Li et al. 2014), salivary adenoid cystic carcinoma (Dai et al. 2014), cervical cancer (Yang et al. 2014), salivary duct carcinoma (Piao et al. 2013), colorectal carcinoma (Liu et al. 2010), breast cancer (Zhang et al. 2011a), oral squamous cell carcinoma (Piao et al. 2012), and papillary carcinoma (Wang et al. 2013). The overexpression of USP22 is related to poor prognosis and may be used as a biomarker during diagnosis. USP22 has been identified as a cancer stem cell marker in the ‘death-from-cancer’ signature genes study (Glinsky 2005, 2006, Glinsky et al. 2005). Genome-wide expression-profiling analysis identified 11 genes that could be used as markers to predict both metastases and failure of cancer treatment.
A20
By far, the most intensely studied member of the OTU-class of cysteine proteases is A20 (alternative name TNFAIP3: tumor necrosis factor alpha-induced protein); together with CYLD, it negatively regulates the NFκB pathway (Fig. 3). A20 is regulated by estrogen and A20 levels are highly increased in aggressive forms of breast cancer and involved in tamoxifen resistance (Vendrell et al. 2007), suggesting a potential role both as a marker and as a therapeutic target. However, A20 levels are decreased in B-cell lymphomas, leading to NFκB activation and contributing to their resistance to apoptosis (Hymowitz & Wertz 2010). Unlike most other DUBs, A20 also possesses an E3 ubiquitin ligase function. This dual E3 ligase–DUB function allows A20 to elegantly inhibit NFκB-related signaling by removing K63-linked polyubiquitin chains from substrate molecules such as TRAF6 and receptor-interacting serine/threonine protein kinase 1 (RIP1) and label them for 26S proteasomal degradation by attaching a K48-linked polyubiquitin chain (Wertz et al. 2004). A20 is generated in response to NFκB-induced gene activation (Krikos et al. 1992) and is thus part of the negative feedback mechanism in the NFκB pathway. For distinct functions in the NFκB pathway, A20 requires other proteins such as tax1-binding protein 1 (TAX1BP1), E3 ubiquitin-protein ligase Itchy homolog (ITCH), and RING finger protein 11 (RNF11) to form an A20 ubiquitin-editing complex (Shembade et al. 2011). The C-terminus of A20 contains seven zinc finger (ZnF) domains, of which ZnF4 is essential for E3 ligase activity (Wertz et al. 2004). Furthermore, this domain binds ubiquitin (Bosanac et al. 2010, Tokunaga et al. 2012) and TAX1BP1 (Verstrepen et al. 2011). The ZnF1 domain is essential for recognition of RIP1 (Bosanac et al. 2010). ZnF4 has been crystallized with mono-ubiquitin and shows a high affinity for K63-linked polyubiquitin chains (Bosanac et al. 2010), whereas ZnF7 crystallized with linear di-ubiquitin (Tokunaga et al. 2012). A comparison between the crystal structures of the N-terminal OTU-domain and otubain 2 (OTUB2) revealed different structural features around the catalytic triad, suggesting differences in substrate specificity as well as catalytic activity of the individual DUBs (Nanao et al. 2004, Komander & Barford 2008, Lin et al. 2008). Although the core architecture of the catalytic triad was found to be very similar in the USP- and OTU-domains, A20 showed variations in the third catalytic residue. A more recent crystallographic study has reported reversible oxidation of the catalytic cysteine residue in A20 and other OTU–DUBs (Kulathu et al. 2013), which is stabilized by the specific nature of the catalytic center and has been suggested as a regulatory mechanism.
UCHL1 (PGP 9.5)
UCHL1 (also known as PGP 9.5) is the most studied member of the UCH-class of cysteine protease DUBs and is associated with multiple types of cancers including lung (Hibi et al. 1998), colorectal (Yamazaki et al. 2002), pancreatic (Leiblich et al. 2007), and breast (Xiang et al. 2012) cancers as well as neurodegenerative diseases such as Parkinson's (Leroy et al. 1998) and Alzheimer's (Xue & Jia 2006) (Fig. 3). It is a marker for neuroendocrine cells and tumors (Thompson et al. 1983, Rode et al. 1985) and has been suggested as a detection tool for pancreatic (Kato et al. 2013, Tomita 2013), parathyroid (Howell et al. 2009), ovarian (Brait et al. 2013), and breast cancers (Liu et al. 2014). UCHL1 cleaves single amino acids or small peptides from the C-terminus of ubiquitin precursors to create mono-ubiquitin, but does not cleave ubiquitin from proteins (Larsen et al. 1998). Dimeric UCHL1 displays E3 ubiquitin ligase activity in vitro (Liu et al. 2002). With only 223 amino acids, it is among the shortest DUBs, containing only a UCH domain. UCHL1 adopts a papain-like α–β–α fold (Das et al. 2006) similar to UCHL3 and UCHL5. The crystal structure of apo-UCHL1 showed a distorted catalytic triad (Das et al. 2006), whereas with bound ubiquitin vinyl methyl ester the catalytic triad realigns into an active form (Boudreaux et al. 2010). A similar scenario is present in USP7, where the catalytic triad was only properly aligned with bound ubiquitin-aldehyde (Hu et al. 2002). A tripeptide fluoromethylketone inhibitor has been shown to bind at the active site to the misaligned UCHL1 form (Davies et al. 2012). The precise physiological role of UCHL1 is somewhat elusive. In healthy subjects, it is exclusively expressed in the neurons and testis, but it is found in cancerous tissues. Mutations in UCHL1 have been linked to Parkinson's disease (Leroy et al. 1998). Both protein and mRNA levels of UCHL1 are down-regulated in prostate cancer due to hypo-methylation of the UCHL1 promoter (Ummanni et al. 2011).
BRCA1-associated protein 1
Research into BAP1 has gained attention due to its role in lung and breast cancers (Jensen et al. 1998) and tumor suppression (Ventii et al. 2008) (Fig. 3). An extensive study found mutations and deletions to BAP1 to be particularly associated with the detrimental outcome of various common cancers, especially in renal clear cell carcinoma (Kandoth et al. 2013). BAP1 is named according to its interaction with the RING finger domain of BRCA1, which is an ubiquitin E3 ligase involved in DNA repair. Although BRCA1 itself is not a substrate of BAP1, its function is affected by BAP1. A recent study has linked the deubiquitination of γ-tubulin by BAP1 to breast cancer (Zarrizi et al. 2014). Drosophila BAP1 has been found to form the so-called polycomb repressive DUB (PR-DUB) complex with the polycomb group (PcG) protein ASX (additional sex combs protein), which deubiquitinates mono-ubiquitinated histone H2A (Scheuermann et al. 2010). The corresponding PR-DUB complex in humans involves BAP1 and host cell factor 1 (HCFC1). It had been observed earlier that HCFC1 is a substrate of BAP1, suggesting that BAP1 regulates cell proliferation (Machida et al. 2009, Misaghi et al. 2009). A recent study has found that the transcription factor forkhead box protein K2 (FOXK2) guides the PR-DUB complex to its targets on the genome via BAP1 binding (Ji et al. 2014). There is no structural information for BAP1. In addition to its UCH domain, BAP1 contains a long C-terminal extension. The C-terminal extension contains a functional nuclear core localization (NLS) domain and several binding sites for interaction partners such as BRCA1 and HCFC1. For tumor suppression, both the enzymatic activity and the NLS domain are essential (Ventii et al. 2008).
Ataxin 3
ATXN3 belongs to the MJD class of cysteine proteases and is named after spinocerebellar ataxia type 3, a neurodegenerative disease, which is also called MJD (Fig. 3). A recent study has reported that ATXN3 represses the transcription of PTEN (Sacco et al. 2014), an immensely important tumor suppressor (Alimonti et al. 2010) critically linked to endocrine-related cancers (Fu et al. 2014). The regulation of PTEN by ATXN3 highlights an earlier study that suggested involvement of ATXN3 in gene expression by binding to specific DNA sequences as well as histone deacetylase 3 (HDAC3) and nuclear receptor corepressor (NCoR) (Evert et al. 2006). In addition to the N-terminal catalytic Josephin domain, ATXN3 contains two C-terminal ubiquitin-interaction motifs (UIMs), which are also found in the 26S proteasome (Hofmann & Falquet 2001). ATXN3 shows preferred binding and cleavage of longer poly-ubiquitin, with a minimal chain length of four (Burnett et al. 2003). Neighboring the two UIMs is a poly-glutamine (polyQ) stretch. ATXN3 mutations with extended polyQ stretches cause MJD (Gusella & MacDonald 2000). Ironically, normal ATXN3 is involved in the regulation of protein aggregates and supposedly plays a role in the suppression of polyQ-related neurodegenerative diseases such as Huntington's disease (Warrick et al. 2005). Critical for this suppressive function are the UIM domains, the catalytic activity as well as proteasome activity. It has been suggested that the deubiquitinating function of ATXN3 protects against its own intrinsic polyQ toxicity (Warrick et al. 2005). Binding of ATXN3 to valosin-containing protein (VCP, also called p97) suggested a regulatory involvement of ATXN3 in endoplasmatic reticulum-associated degradation (Zhong & Pittman 2006), which is a quality control system targeting misfolded proteins for proteasomal digestion. ATXN3 regulates C-terminus of Hsc70-interacting protein (Scaglione et al. 2011), which is an E3 ligase that collaborates with chaperones in protein quality control. The catalytic Josephin domain of ATXN3 investigated by solution NMR (Mao et al. 2005, Nicastro et al. 2005) revealed structural similarity to the catalytic domains of other cysteine proteases and ubiquitin binding.
Rpn11
Rpn11 (also known as PSMD14 or POH1) is an essential enzyme of the 26S proteasome and belongs to the 19S regulatory particle non-ATPase (Rpn) subunits (Fig. 3). Rpn11 is located in the lid of the proteasome in a hetero-dimeric complex with Rpn8 (Lander et al. 2012). It cleaves K48-linked ubiquitin chains from proteasome substrates, which is necessary for proteasomal processing and degradation (Yao & Cohen 2002). Only recently, the involvement of the proteasome in DNA repair has been reported (Ben-Aroya et al. 2010). In this respect, Rpn11 has been found to promote the double-strand DNA break response by restricting K63-linked ubiquitin chains attached to histones near damaged DNA sites (Butler et al. 2012). It had been observed earlier that Rpn11 can specifically cleave K63-linked poly-ubiquitin chains (Cooper et al. 2009), which is contrary to its cleavage of K48-linked chains targeted for proteasomal digestion, but fits to the K63 specificity of other zinc metalloproteases such as AMSH (associated molecule with the SH3 domain of signal transducting adapter molecule 1) and AMSH-LP (AMSH-like protein) (Sato et al. 2008). Rpn11 has also been connected to drug resistance in cancer (Spataro et al. 2002) as well as idiopathic nephrotic syndrome (Ma et al. 2009), which is a common kidney disease in children. Rpn11 alone is not active (Yao & Cohen 2002), and in complex with Rpn8 it displays low activity (Pathare et al. 2014), suggesting that it is only fully functional when embedded into the 26S proteasome.
Endocrine cancers
The remainder of this review focuses on DUBs that have already been implicated in endocrine cancers.
Prostate cancer
Prostate cancer is the second most common cancer diagnosed and the second most common cause of cancer death in men worldwide. In the USA, an estimated 233 000 new cases of prostate cancer were predicted to be diagnosed in 2014 and ∼30 000 associated deaths (Siegel et al. 2014). Prostate cancer is more frequently diagnosed in men over the age of 65. Screening for prostate cancer is based on the prostate-specific antigen blood test; however, abnormal levels require prostate biopsies for definitive diagnosis as well as digital rectal examination. Combinations of tumor surveillance, surgery, radiation, hormonal, and chemotherapies are used in the management and treatment of prostate cancer. The molecular basis of disease is quite complex and mutations in many different oncogenes and tumor suppressor genes have been implicated including TP53, RB1, EZH2, BMI, and PTEN (Wang & Wong 1997, Dean & Knudsen 2013, Yang & Yu 2013). All of these genes have also been implicated in most other cancers; therefore, none are directly linked to prostate cancer alone. In light of all of the identified mutations, prostate cancer appears to be caused by dysregulation of many different cellular signaling pathways especially those involved in cell survival and apoptosis.
Interestingly, the DUB USP7 has been implicated in the regulation of several of these genes including TP53, RB1, BMI, and PTEN (Li et al. 2002a, Song et al. 2008, Maertens et al. 2010, Bhattacharya & Ghosh 2014b). The involvement of overexpressed USP7 has already been described in prostate cancer via dysregulation of PTEN (Song et al. 2008). PTEN is a lipid and protein phosphatase that catalyzes the dephosphorylation of phosphatidylinositol (3,4,5)-triphosphate inhibiting the Akt/PKB signaling pathway. However, PTEN also has nuclear functions, whereby it regulates chromatin and DNA repair processes. It has recently become clear that in many cancers, PTEN is excluded from the nucleus (Planchon et al. 2008). The ubiquitination of PTEN is one mechanism that leads to its nuclear translocation. In the nucleus, USP7 catalyzes the deubiquitination of PTEN leading to its export from the nucleus (Song et al. 2008). This USP7-dependent deubiquitination and removal of PTEN from the nucleus is associated with many cancers (Song et al. 2008). More recently, the ability of USP7 to deubiquitinate PTEN was found to be under the control of BCR-ABL (Morotti et al. 2014). BCR-ABL phosphorylates USP7 and activates the deubiquitination of PTEN. A USP7 inhibitor could target prostate cancers; however, the effectiveness of such an inhibitor needs further study. Single nucleotide polymorphisms in Hdm2, HdmX, and USP7 have also been associated with aggressive prostate cancer (Sun et al. 2010). The involvement of USP7 in other mechanisms of prostate cancer awaits further investigations.
USP2a (also known as USP2) has been implicated in prostate cancer through its deubiquitination and stabilization of fatty acid synthase (Graner et al. 2004) and more recently Hdm2 (Stevenson et al. 2007) as well as its closely related functional homolog HdmX (Allende-Vega et al. 2010). Furthermore, overexpressed USP2a can transform prostate epithelial cells in vitro and NIH3T3 cells overexpressing USP2a caused the growth of tumors in all 12 injected nude mice in vivo (Priolo et al. 2006). More importantly, USP2a was found to be overexpressed in prostate cancer and is regulated by androgens in a concentration-dependent manner (Graner et al. 2004). The inactivation of USP2a via knockdown, mutations, or low expression has a negative effect on fatty acid synthase levels and p53-regulated genes, which enhances apoptosis (Graner et al. 2004, Priolo et al. 2006). USP2a levels were found to be up-regulated along with fatty acid synthase-related genes in almost half of all prostate cancers. These studies suggested an important role for USP2a in prostate cancer. It is therefore expected that an USP2a inhibitor could have important therapeutic applications. The c-myc oncogene is overexpressed in a subset of USP2a overexpressing prostate cancer cells (Benassi et al. 2012). The stabilization of Hdm2 by USP2a results in enhanced degradation of p53, thus preventing apoptosis. This enhanced p53 degradation has a direct consequence in MYC up-regulation by repressing the expression of regulatory microRNAs that target the MYC mRNA. The derepression of microRNAs miR-34b/c, miR-98, and let-7c resulting in increased levels of MYC is attributed to increased levels of USP2a (Benassi et al. 2012). Inhibitors targeting the catalytic activity of USP2a could attack prostate cancer cells via several different mechanisms, such as i) reducing MYC levels, thus reducing proliferation and ii) increasing p53 levels leading to increased apoptosis.
USP19 silencing directly affected the growth of several prostate cancer cell lines, suggesting a putative role in carcinogenesis (Lu et al. 2011). USP19 deubiquitinates and stabilizes KPC1, an E3 ligase for p27. Interestingly, the effects of decreased nuclear levels of p27, resulting in poor prognosis, have already been described in prostate cancer (Chu et al. 2008). USP19 regulates the levels of p27, although p27 is not a USP19 substrate.
One of the functions of USP26 is to regulate the activity and stability of the androgen receptor in the nucleus (Dirac & Bernards 2010). The androgen receptor is a steroid hormone receptor that is responsible for male development and maturation. It is activated by binding androgens such as testosterone or dihydrotestosterone and is transported to the nucleus where it functions as a transcription factor. The androgen receptor has been implicated in several cancers including testicular (Garolla et al. 2005), prostate (Heinlein & Chang 2004), breast (Cochrane et al. 2014), and thyroid cancers (Stanley et al. 2012). Investigation of the effects of USP26 in prostate cancer is necessary given its role in activity and stability of the androgen receptor and the role of the androgen receptor in prostate cancer (Wosnitzer et al. 2014). Similar to USP26, USP12 catalyzes the deubiquitination and stabilization of the androgen receptor (Burska et al. 2013). Furthermore, the functional role of USP12 in the increased transcriptional activation of the androgen receptor has been extended to the Akt pathway through the pro-apoptotic phosphatases (PHLPP and PHLPPL). The USP12/Uaf-1/WDR20 deubiquitinating complex also stabilizes both PHLPP and PHLPPL eventually resulting in increased transcriptional activation by the androgen receptor (Burska et al. 2013). Even though the role of USP10 in prostate cancer has not been investigated, it is interesting to note that USP10 is also involved in the regulation of the androgen receptor (Draker et al. 2011).
Betulinic acid, derived from the bark of white birch trees, induces apoptosis quite selectively in a variety of cancer cells (Gheorgheosu et al. 2014). Betulinic acid had already been described to inhibit prostate cancer growth though a mechanism of action involving the decreased levels of VEGF and survivin due to the increased ubiquitin-mediated degradation of specificity protein transcription factors (Sp1, Sp3, and Sp4; Chintharlapalli et al. 2007). Another study determined that betulinic acid increases the levels of poly-ubiquitinated proteins and their degradation and concluded that betulinic acid most probably inhibits the actions of several as yet unidentified DUBs (Reiner et al. 2013).
Ovarian cancer
In the USA, an estimated 22 000 new cases of ovarian cancer were predicted to be diagnosed in 2014 resulting in ∼14 000 deaths associated with this disease (Siegel et al. 2014). Ovarian cancer is asymptomatic until later stages making it quite difficult to diagnose. The number of deaths is quite high because of the late stages of disease and metastases. The greatest risk factor for ovarian cancer is genetic with several common mutations. The current understanding of the involvement of DUBs in ovarian cancer is rather limited. Many tumor suppressor genes including TP53, PTEN, RB1, and BRCA1 are either mutated or dysregulated in ovarian cancer. Therefore, as USP7 regulates all of these proteins, the role of USP7 in ovarian cancer needs to be investigated. The ubiquitin carboxyl terminal hydrolases UCH37 (also known as UCHL5) and UCHL1 have both been implicated in ovarian cancer. As in other cancers, UCH37 has been found to be up-regulated and linked to poor prognosis (Wang et al. 2014). UCHL1 has been identified as being both up-regulated (Jin et al. 2013) and down-regulated in ovarian cancer (Okochi-Takada et al. 2006, Jin et al. 2013). UCHL1 knockdown in ovarian cancer cell lines where it was overexpressed caused increased proliferation. Another study that set out to identify both up- and down-regulated genes in ovarian cancer for use in diagnosis determined that USP36 was overexpressed (Li et al. 2008). More importantly, USP36 was identified in sera of 36% of cases from ovarian cancer patients compared with none from healthy subjects. Further investigations are needed to define the role of USP36 in ovarian and other cancers and confirm its ability to be useful in diagnosis. USP15 is amplified in ovarian and breast cancers as well as glioblastoma (Eichhorn et al. 2012). USP15 regulates the TGFβ and NFκB signaling pathways by deubiquitinating the type 1 TGFβ receptor as well as IκBα (Schweitzer et al. 2007, Eichhorn et al. 2012). Proteins from both of these pathways have already been implicated as possible cancer therapeutic targets. Down-regulation of USP15 has been observed in paclitaxel-resistant ovarian cancer (Xu et al. 2009). Furthermore, knockdown of USP15 in HeLa cells also lead to paclitaxel resistance corroborating its important role in this resistance. Krüppel-like factor 8 (KLF8) is a transcription factor that is highly overexpressed in ovarian and other cancers. KLF8 can transform ovarian epithelial cells alone and in combination with other oncogenes by down-regulating USP44 (Lu et al. 2014). USP44 plays a role in cancer through its dysregulation of the DNA damage response mediated by the E3 ligase RNF168 on histone H2A (Mosbech et al. 2013). USP44 is a tumor suppressor protein that is silenced in a variety of other cancers (Holland & Cleveland 2012, Sloane et al. 2014).
Pheochromocytoma
Pheochromocytoma is a usually benign cancer originating in the adrenal glands that is associated with Von Hippel–Lindau disease, neurofibromatosis, and multiple endocrine neoplasia type 2 and characterized by excessive secretion of catecholamines (Tsirlin et al. 2014). Von Hippel–Landau disease is associated with various other malignancies as well (Tootee & Hasani-Ranjbar 2012). The following discussion continues only with Von Hippel–Landau disease, as it is related to the ubiquitin system. Von Hippel–Lindau disease is caused by mutations in the gene encoding the VHL protein (pVHL). The VHL protein associates with elongin B, elongin C, and cullin 2 to form an E3 ligase complex that is responsible for the regulation of hypoxia inducible factor 1 alpha (HIF1α). Under normoxia, pVHL catalyzes the poly-ubiquitination and degradation of HIF1α by interacting with a 2-hydroxylated proline on HIF1α. However, under conditions of hypoxia, HIF1α is not hydroxylated, therefore pVHL is unable to interact and catalyze its poly-ubiquitination. As a transcription factor, HIF1α induces expression of genes containing hypoxia response elements that are involved in angiogenesis, glucose metabolism, and invasion/metastasis (Li et al. 2005). Mutations in pVHL disrupt the ubiquitination of HIF1α. Dysregulated degradation of HIF1α results in enhanced expression of its target genes. Two DUBs, VDU1 and VDU2 (pVHL-interacting deubiquitinating enzymes 1 and 2), are also known substrates of the pVHL E3 ligase complex (Li et al. 2002b). VDU1 is also known as USP33 and VDU2 as USP20. Both of these DUBs share sequence and functional similarities and are ubiquitinated by the pVHL–E3 ligase complex and targeted for degradation (Li et al. 2002b). In addition, VDU2, but not VDU1, is able to interact with and deubiquitinate HIF1α, and rescue it from proteasome-mediated degradation. Interestingly, pVHL regulates the levels of both HIF1α and VDU2 leading to a vital equilibrium between the degradation and rescue of HIF1α. The inability of the pVHL–E3 ligase complex to catalyze the ubiquitination of not only HIF1α but also VDU1 and VDU2 may lead to disease. As DUBs are substrates of the pVHL–E3 ligase complex, it is important to note that both of these DUBs are emerging targets in the development of therapeutics against Von Hippel–Landau disease. The design of inhibitors against VDU1 and VDU2 could prevent non-regulated deubiquitination of substrate proteins to compensate for the inability of the pVHL–E3 ligase complex to degrade it. USP20 is involved in the deubiquitination and rescue of RAD17, a protein involved in the DNA damage response pathway (Shanmugam et al. 2014). The knockdown of USP20 caused the sensitization of two different cancer cell lines to cisplatin (Shanmugam et al. 2014), strongly suggesting that an inhibitor against USP20 may function as a cancer therapeutic drug.
Thyroid cancer
In the USA, an estimated 63 000 new cases of thyroid cancer were predicted to be diagnosed in 2014 resulting in ∼1800 deaths (Siegel et al. 2014). USP22 is overexpressed in papillary carcinoma, a type of thyroid cancer (Wang et al. 2013). Increased USP22 protein levels were shown for papillary carcinoma tissue when compared with noncancerous tissue using immunohistochemistry (Wang et al. 2013). Kaplan–Meier analysis indicated that the survival of patients with papillary carcinoma was significantly lower with high USP22 expression levels. Taken together, the authors concluded that USP22 could be functioning as an oncogene and used in the prognosis of papillary carcinoma. There is one case of MALT lymphoma of the thyroid that has been associated with the deletion of A20 (Chanudet et al. 2009). The expression of OTUD1 is highly up-regulated in various carcinomas of the thyroid and could potentially be used as a biomarker to distinguish between cancerous samples (Carneiro et al. 2014).
UCHL1 belongs to the ubiquitin-carboxyl terminal hydrolases family of DUBs. This family of proteases catalyzes the hydrolysis of ubiquitin from small adducts to maintain cellular ubiquitin homeostasis. It is the main DUB responsible for maintaining cellular levels of monomeric ubiquitin primarily by hydrolyzing ester and amide forms of ubiquitin. It is highly expressed in neuronal and endocrine cells; however, its expression has been detected in many non-neuronal cancers. The role of UCHL1 in endocrine cancers is well established. The up-regulation of UCHL1 has been observed in one of the six cases of granular cell tumors located in the thyroid gland (Espinosa-de-Los-Monteros-Franco et al. 2009). UCHL1 is also known as PGP9.5, a well-established marker for diagnosis and prognosis of a variety of cancers. PGP9.5 was evaluated as a prognostic marker in invasive colorectal, pancreatic, parathyroid, and lung cancers (Hibi et al. 1999, Tezel et al. 2000, Yamazaki et al. 2002, Howell et al. 2009). Patients with pancreatic cancer that were positive for PGP9.5 had lower survival rates than those who were negative for PGP9.5 (Tezel et al. 2000). There were significant differences in maximal tumor size and the extent of tumor in patients with invasive colorectal cancer expressing PGP9.5 (Yamazaki et al. 2002). PGP9.5 was evaluated as a diagnostic marker in medullary thyroid carcinoma (Takano et al. 2004). Overexpression of PGP9.5 mRNA was identified in all eleven medullary thyroid carcinoma samples examined. This study showed that levels of PGP9.5 mRNA were similar to normal thyroid tissues in other thyroid cancers including anaplastic, papillary, and follicular carcinomas as well as follicular adenoma, suggesting that overexpression of PGP9.5 could not be used as a biomarker for these cancers.
Both VDU1 (USP33) and VDU2 (USP20) also play important biological roles related to the thyroid. VDU1 and VDU2 deubiquitinate and thus reactivate the hormone-activating type 2 deiodinase (D2), which is an endoplasmic reticulum integral membrane protein (Curcio-Morelli et al. 2003, Gereben et al. 2008). D2 functions in the conversion of the inactive precursor thyroxine, into triiodothyroxine (T3), the active hormone responsible for cellular energy and metabolism homeostasis. Therefore, very tight control of D2 levels is critical. The mechanism by which levels of T3 are controlled involves the ubiquitination leading to inactivation and the subsequent degradation of D2. D2 is ubiquitinated by the WSB-1 and TEB4 E3 ligases in response to D2 activation and increased levels of T3 (Dentice et al. 2005, Zavacki et al. 2009). However, the process of D2 degradation can be reversed by VDU1- and VDU2-catalyzed deubiquitination, resulting in D2 rescue and reactivation. It is unknown whether the deubiquitination of D2 has any roles in VHL disease or cancer (Curcio-Morelli et al. 2003).
Adrenocortical carcinoma
Adrenocortical adenoma and carcinoma are tumors of the adrenal cortex. Adrenocortical carcinoma is a rare but very aggressive cancer with a 5-year survival rate of 30%. Adenomas on the other hand are benign tumors. The up-regulation of USP4 and USP38 was identified in adrenocortical carcinoma using microarray gene expression analysis (Laurell et al. 2009). USP4 had previously been identified as being up-regulated in adrenocortical carcinoma using transcriptional profiling (Velazquez-Fernandez et al. 2005). Several USP4 deubiquitinating targets have been identified including ARF-BP1, type 1 TGFβ receptor, and PDK1 (Zhang et al. 2011b, 2012, Uras et al. 2012). The roles of ARF-BP1 and PDK1 in adrenocortical carcinomas have not yet been investigated. The TGF signaling pathway has been implicated in the tumorigenicity of adrenocortical carcinomas (Yamamoto et al. 2006, Parviainen et al. 2013).
Therapeutic targeting of DUBs for the treatment of cancer
The studies thus far describing the involvement of DUBs in endocrine cancers have only scratched the surface of the many roles that DUBs may play in these cancers (Table 1). Additional studies are needed to define the actual landscape of DUBs that are mutated, deleted, or differentially regulated in these cancers. These studies, along with the design of targeted, small molecule DUB inhibitors, will most probably lead to new opportunities for endocrine cancer therapeutics and diagnostics.
Table 1.
DUBs in endocrine cancers
| Cancer | DUBs | References |
|---|---|---|
| Prostate | USP2 | Graner et al. (2004), Priolo et al. (2006) and Benassi et al. (2012) |
| USP7 | Song et al. (2008) | |
| USP10 | Draker et al. (2011) | |
| USP12 | Leiblich et al. (2007) and Burska et al. (2013) | |
| USP14 | Du et al. (2015) | |
| USP19 | Lu et al. (2011) | |
| USP22 | Schrecengost et al. (2014) | |
| USP26 | Dirac & Bernards (2010) | |
| USP39 | Wen et al. (2014) | |
| UCHL1 | Leiblich et al. (2007) | |
| Ovarian | USP2a | Yang et al. (2007) |
| USP36 | Li et al. (2008) | |
| USP44 | Lu et al. (2014) | |
| OTUD1 | Carneiro et al. (2014) | |
| Thyroid | USP20 | Curcio-Morelli et al. (2003) and Li et al. (2005) |
| USP22 | Wang et al. (2013) | |
| USP33 | Li et al. (2002c) and Curcio-Morelli et al. (2003) | |
| A20 | Chanudet et al. (2009) | |
| UCHL1 | Takano et al. (2004) | |
| Adrenal cortex | USP4 | Laurell et al. (2009) |
| USP38 | Laurell et al. (2009) | |
| Parathyroid | UCHL1 | Howell et al. (2009) |
DUBs have been implicated in several types of cancers and neurodegenerative diseases. As such, targeting DUBs can lead to effective therapies in the treatment, diagnosis, and prevention of these diseases. The UPS regulates many important proteins; therefore, it is not surprising that, when UPS components are acting aberrantly, accelerated degradation or mislocalization of substrate proteins results in disease. Various studies have suggested the dual roles of DUBs, both as oncogenes and as tumor suppressors. With DUBs having multiple substrates, the nature of their substrates may distinguish their oncogenic vs suppressive roles.
Velcade (bortezomib) was the first clinically approved UPS inhibitor and is used as a therapeutic agent for B cell lymphoma and multiple myeloma, exemplifying the importance of the UPS as an effective therapeutic target (Bold 2004). Kyprolis (carfilzomib), another proteasome inhibitor, was FDA approved in 2012 for multiple myeloma (Steele 2013). The biggest challenge in therapeutics is that a proteasome inhibitor is non-specific and indeed bortezomib and carfilzomib treatments are quite toxic. A potentially more attractive therapeutic approach would be inhibitors designed with increased specificity against DUBs.
Concluding remarks
A more comprehensive understanding of the roles, substrates, and regulation of DUBs will lead to a better understanding of their emerging roles in carcinogenesis and clinical applications of their inhibitors. The continued and accelerated development of small molecule inhibitors against DUBs will lead to increased success in the treatment of various cancers and other diseases.
Acknowledgements
The authors tried to be as thorough as possible but regret if some publications were not included in this review. They thank Dinesh Christendat, Lori Frappier, and Feroz Sarkari for critically reading the manuscript.
Footnotes
This paper forms part of a thematic review section on Ubiquitination and Cancer. The guest editor for this section was Deborah Marsh, The University of Sydney, Sydney, Australia.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this review.
Funding
This research was supported by an operating grant to V Saridakis from the Canadian Institutes of Health Research (CIHR; grant number 106583).
References
- Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, Salmena L, Sampieri K, Haveman WJ, Brogi E, et al. Subtle variations in Pten dose determine cancer susceptibility. Nature Genetics. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende-Vega N, Sparks A, Lane DP, Saville MK. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene. 2010;29:432–441. doi: 10.1038/onc.2009.330. [DOI] [PubMed] [Google Scholar]
- Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochimica et Biophysica Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The U box is a modified RING finger – a common domain in ubiquitination. Current Biology. 2000;10:R132–R134. doi: 10.1016/S0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Finerty PJ, Jr, Mackenzie F, Newman EM, Dhe-Paganon S. Amino-terminal dimerization, NRDP1–rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) Journal of Biological Chemistry. 2006;281:38061–38070. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- Becker K, Marchenko ND, Palacios G, Moll UM. A role of HAUSP in tumor suppression in a human colon carcinoma xenograft model. Cell Cycle. 2008;7:1205–1213. doi: 10.4161/cc.7.9.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S, Agmon N, Yuen K, Kwok T, McManus K, Kupiec M, Hieter P. Proteasome nuclear activity affects chromosome stability by controlling the turnover of Mms22, a protein important for DNA repair. PLoS Genetics. 2010;6:e1000852. doi: 10.1371/journal.pgen.1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benassi B, Flavin R, Marchionni L, Zanata S, Pan Y, Chowdhury D, Marani M, Strano S, Muti P, Blandino G, et al. MYC is activated by USP2a-mediated modulation of microRNAs in prostate cancer. Cancer Discovery. 2012;2:236–247. doi: 10.1158/2159-8290.CD-11-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Ghosh MK. Cell death and deubiquitinases: perspectives in cancer. BioMed Research International. 2014a;2014:435197. doi: 10.1155/2014/435197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Ghosh MK. HAUSP, a novel deubiquitinase for Rb – MDM2 the critical regulator. FEBS Journal. 2014b;281:3061–3078. doi: 10.1111/febs.12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bie P, Zaaroor-Regev D, Ciechanover A. Regulation of the polycomb protein RING1B ubiquitination by USP7. Biochemical and Biophysical Research Communications. 2010;400:389–395. doi: 10.1016/j.bbrc.2010.08.082. [DOI] [PubMed] [Google Scholar]
- Bold R. “Development of the proteasome inhibitor Velcade (Bortezomib)” by Julian Adams, Ph.D., and Michael Kauffman, M.D., Ph.D. Cancer Investigation. 2004;22:328–329. doi: 10.1081/CNV-120030223. [DOI] [PubMed] [Google Scholar]
- Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, Phu L, Phung Q, Maurer B, Arnott D, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Molecular Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Boudreaux DA, Maiti TK, Davies CW, Das C. Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation. PNAS. 2010;107:9117–9122. doi: 10.1073/pnas.0910870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brait M, Maldonado L, Noordhuis MG, Begum S, Loyo M, Poeta ML, Barbosa A, Fazio VM, Angioli R, Rabitti C, et al. Association of promoter methylation of VGF and PGP9.5 with ovarian cancer progression. PLoS ONE. 2013;8:e70878. doi: 10.1371/journal.pone.0070878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Burnett B, Li F, Pittman RN. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Human Molecular Genetics. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- Burska UL, Harle VJ, Coffey K, Darby S, Ramsey H, O'Neill D, Logan IR, Gaughan L, Robson CN. Deubiquitinating enzyme Usp12 is a novel co-activator of the androgen receptor. Journal of Biological Chemistry. 2013;288:32641–32650. doi: 10.1074/jbc.M113.485912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler LR, Densham RM, Jia J, Garvin AJ, Stone HR, Shah V, Weekes D, Festy F, Beesley J, Morris JR. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO Journal. 2012;31:3918–3934. doi: 10.1038/emboj.2012.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro AP, Reis CF, Morari EC, Maia YC, Nascimento R, Bonatto JM, de Souza MA, Goulart LR, Ward LS. A putative OTU domain-containing protein 1 deubiquitinating enzyme is differentially expressed in thyroid cancer and identifies less-aggressive tumours. British Journal of Cancer. 2014;111:551–558. doi: 10.1038/bjc.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JK, Greene WC. NF-κB/Rel: agonist and antagonist roles in HIV-1 latency. Current Opinion in HIV and AIDS. 2011;6:12–18. doi: 10.1097/COH.0b013e32834124fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanudet E, Ye H, Ferry J, Bacon CM, Adam P, Muller-Hermelink HK, Radford J, Pileri SA, Ichimura K, Collins VP, et al. A20 deletion is associated with copy number gain at the TNFA/B/C locus and occurs preferentially in translocation-negative MALT lymphoma of the ocular adnexa and salivary glands. Journal of Pathology. 2009;217:420–430. doi: 10.1002/path.2466. [DOI] [PubMed] [Google Scholar]
- Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, McDermott JL, Leach CA, Fulcinniti M, Kodrasov MP, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. Ubiquitin signalling in the NF-κB pathway. Nature Cell Biology. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching W, Koyuncu E, Singh S, Arbelo-Roman C, Hartl B, Kremmer E, Speiseder T, Meier C, Dobner T. A ubiquitin-specific protease possesses a decisive role for adenovirus replication and oncogene-mediated transformation. PLoS Pathogens. 2013;9:e1003273. doi: 10.1371/journal.ppat.1003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintharlapalli S, Papineni S, Ramaiah SK, Safe S. Betulinic acid inhibits prostate cancer growth through inhibition of specificity protein transcription factors. Cancer Research. 2007;67:2816–2823. doi: 10.1158/0008-5472.CAN-06-3735. [DOI] [PubMed] [Google Scholar]
- Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nature Reviews. Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin proteolytic system and pathogenesis of human diseases: a novel platform for mechanism-based drug targeting. Biochemical Society Transactions. 2003;31:474–481. doi: 10.1042/BST0310474. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin proteolytic system: from an idea to the patient bed. Proceedings of the American Thoracic Society. 2006;3:21–31. doi: 10.1513/pats.200510-106JH. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Schwartz AL. The ubiquitin system: pathogenesis of human diseases and drug targeting. Biochimica et Biophysica Acta. 2004;1695:3–17. doi: 10.1016/j.bbamcr.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5%3c442::AID-BIES6%3e3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbe S. Deubiquitylases from genes to organism. Physiological Reviews. 2013;93:1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- Cochrane DR, Bernales S, Jacobsen BM, Cittelly DM, Howe EN, D'Amato NC, Spoelstra NS, Edgerton SM, Jean A, Guerrero J, et al. Role of the androgen receptor in breast cancer and preclinical analysis of enzalutamide. Breast Cancer Research. 2014;16:R7. doi: 10.1186/bcr3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, Trouplin V, Bianchi J, Aushev VN, Camonis J, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Molecular Cancer Therapeutics. 2009;8:2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, Cohen RE. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO Journal. 2009;28:621–631. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio-Morelli C, Zavacki AM, Christofollete M, Gereben B, de Freitas BC, Harney JW, Li Z, Wu G, Bianco AC. Deubiquitination of type 2 iodothyronine deiodinase by von Hippel–Lindau protein-interacting deubiquitinating enzymes regulates thyroid hormone activation. Journal of Clinical Investigation. 2003;112:189–196. doi: 10.1172/JCI18348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Yao Y, Zhou Q, Sun CF. Ubiquitin-specific peptidase 22, a histone deubiquitinating enzyme, is a novel poor prognostic factor for salivary adenoid cystic carcinoma. PLoS ONE. 2014;9:e87148. doi: 10.1371/journal.pone.0087148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcy P, Linder S. Proteasome deubiquitinases as novel targets for cancer therapy. International Journal of Biochemistry & Cell Biology. 2012;44:1729–1738. doi: 10.1016/j.biocel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- Das C, Hoang QQ, Kreinbring CA, Luchansky SJ, Meray RK, Ray SS, Lansbury PT, Ringe D, Petsko GA. Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1. PNAS. 2006;103:4675–4680. doi: 10.1073/pnas.0510403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies CW, Chaney J, Korbel G, Ringe D, Petsko GA, Ploegh H, Das C. The co-crystal structure of ubiquitin carboxy-terminal hydrolase L1 (UCHL1) with a tripeptide fluoromethyl ketone (Z-VAE(OMe)-FMK) Bioorganic & Medicinal Chemistry Letters. 2012;22:3900–3904. doi: 10.1016/j.bmcl.2012.04.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JL, Knudsen KE. The role of tumor suppressor dysregulation in prostate cancer progression. Current Drug Targets. 2013;14:460–471. doi: 10.2174/1389450111314040007. [DOI] [PubMed] [Google Scholar]
- Dentice M, Bandyopadhyay A, Gereben B, Callebaut I, Christoffolete MA, Kim BW, Nissim S, Mornon JP, Zavacki AM, Zeold A, et al. The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate. Nature Cell Biology. 2005;7:698–705. doi: 10.1038/ncb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirac AM, Bernards R. The deubiquitinating enzyme USP26 is a regulator of androgen receptor signaling. Molecular Cancer Research. 2010;8:844–854. doi: 10.1158/1541-7786.MCR-09-0424. [DOI] [PubMed] [Google Scholar]
- Draker R, Sarcinella E, Cheung P. USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Research. 2011;39:3529–3542. doi: 10.1093/nar/gkq1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Science Signaling. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Yuan T, Schilter KF, Dittmar RL, Mackinnon A, Huang X, Tschannen M, Worthey E, Jacob H, Xia S, et al. Prostate cancer risk locus at 8q24 as a regulatory hub by physical interactions with multiple genomic loci across the genome. Human Molecular Genetics. 2015;24:154–166. doi: 10.1093/hmg/ddu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn PJ, Rodon L, Gonzalez-Junca A, Dirac A, Gili M, Martinez-Saez E, Aura C, Barba I, Peg V, Prat A, et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nature Medicine. 2012;18:429–435. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- Espinosa-de-Los-Monteros-Franco VA, Martinez-Madrigal F, Ortiz-Hidalgo C. Granular cell tumor (Abrikossoff tumor) of the thyroid gland. Annals of Diagnostic Pathology. 2009;13:269–271. doi: 10.1016/j.anndiagpath.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Evert BO, Araujo J, Vieira-Saecker AM, de Vos RA, Harendza S, Klockgether T, Wullner U. Ataxin-3 represses transcription via chromatin binding, interaction with histone deacetylase 3, and histone deacetylation. Journal of Neuroscience. 2006;26:11474–11486. doi: 10.1523/JNEUROSCI.2053-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Molecular Cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- Fan YH, Cheng J, Vasudevan SA, Dou J, Zhang H, Patel RH, Ma IT, Rojas Y, Zhao Y, Yu Y, et al. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death & Disease. 2013;4:e867. doi: 10.1038/cddis.2013.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Weissman AM. A field guide to ubiquitylation. Cellular and Molecular Life Sciences. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustrup H, Bekker-Jensen S, Bartek J, Lukas J, Mailand N. USP7 counteracts SCFβTrCP – but not APCCdh1-mediated proteolysis of Claspin. Journal of Cell Biology. 2009;184:13–19. doi: 10.1083/jcb.200807137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- Fu X, Creighton CJ, Biswal NC, Kumar V, Shea M, Herrera S, Contreras A, Gutierrez C, Wang T, Nanda S, et al. Overcoming endocrine resistance due to reduced PTEN levels in estrogen receptor-positive breast cancer by co-targeting mammalian target of rapamycin, protein kinase B, or mitogen-activated protein kinase kinase. Breast Cancer Research. 2014;16:430. doi: 10.1186/s13058-014-0430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garolla A, Ferlin A, Vinanzi C, Roverato A, Sotti G, Artibani W, Foresta C. Molecular analysis of the androgen receptor gene in testicular cancer. Endocrine-Related Cancer. 2005;12:645–655. doi: 10.1677/erc.1.00954. [DOI] [PubMed] [Google Scholar]
- Gereben B, Zavacki AM, Ribich S, Kim BW, Huang SA, Simonides WS, Zeold A, Bianco AC. Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocrine Reviews. 2008;29:898–938. doi: 10.1210/er.2008-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheorgheosu D, Duicu O, Dehelean C, Soica C, Muntean D. Betulinic acid as a potent and complex antitumor phytochemical: a minireview. Anti-Cancer Agents in Medicinal Chemistry. 2014;14:936–945. doi: 10.2174/1871520614666140223192148. [DOI] [PubMed] [Google Scholar]
- Giovinazzi S, Sirleto P, Aksenova V, Morozov VM, Zori R, Reinhold WC, Ishov AM. Usp7 protects genomic stability by regulating Bub3. Oncotarget. 2014;5:3728–3742. doi: 10.18632/oncotarget.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky GV. Death-from-cancer signatures and stem cell contribution to metastatic cancer. Cell Cycle. 2005;4:1171–1175. doi: 10.4161/cc.4.9.2001. [DOI] [PubMed] [Google Scholar]
- Glinsky GV. Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated polycomb group (PcG) protein chromatin silencing pathway. Cell Cycle. 2006;5:1208–1216. doi: 10.4161/cc.5.11.2796. [DOI] [PubMed] [Google Scholar]
- Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. Journal of Clinical Investigation. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graner E, Tang D, Rossi S, Baron A, Migita T, Weinstein LJ, Lechpammer M, Huesken D, Zimmermann J, Signoretti S, et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5:253–261. doi: 10.1016/S1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME. Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nature Reviews. Neuroscience. 2000;1:109–115. doi: 10.1038/35039051. [DOI] [PubMed] [Google Scholar]
- Harhaj EW, Dixit VM. Regulation of NF-κB by deubiquitinases. Immunological Reviews. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Jono H, Shinriki S, Nakamura T, Guo J, Sueta A, Tomiguchi M, Fujiwara S, Yamamoto-Ibusuki M, Murakami K, et al. Clinical significance of CYLD downregulation in breast cancer. Breast Cancer Research and Treatment. 2014;143:447–457. doi: 10.1007/s10549-013-2824-3. [DOI] [PubMed] [Google Scholar]
- Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocrine Reviews. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- Hibi K, Liu Q, Beaudry GA, Madden SL, Westra WH, Wehage SL, Yang SC, Heitmiller RF, Bertelsen AH, Sidransky D, et al. Serial analysis of gene expression in non-small cell lung cancer. Cancer Research. 1998;58:5690–5694. [PubMed] [Google Scholar]
- Hibi K, Westra WH, Borges M, Goodman S, Sidransky D, Jen J. PGP9.5 as a candidate tumor marker for non-small-cell lung cancer. American Journal of Pathology. 1999;155:711–715. doi: 10.1016/S0002-9440(10)65169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L. Protein regulation by monoubiquitin. Nature Reviews. Molecular Cell Biology. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annual Review of Genetics. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends in Biochemical Sciences. 2001;26:347–350. doi: 10.1016/S0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. The deubiquitinase USP44 is a tumor suppressor that protects against chromosome missegregation. Journal of Clinical Investigation. 2012;122:4325–4328. doi: 10.1172/JCI66420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowaty MN, Zeghouf M, Wu H, Tellam J, Athanasopoulos V, Greenblatt J, Frappier L. Protein profiling with Epstein–Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. Journal of Biological Chemistry. 2003;278:29987–29994. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nature Cell Biology. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- Howell VM, Gill A, Clarkson A, Nelson AE, Dunne R, Delbridge LW, Robinson BG, Teh BT, Gimm O, Marsh DJ. Accuracy of combined protein gene product 9.5 and parafibromin markers for immunohistochemical diagnosis of parathyroid carcinoma. Journal of Clinical Endocrinology and Metabolism. 2009;94:434–441. doi: 10.1210/jc.2008-1740. [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/S0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO Journal. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nature Reviews. Cancer. 2010;10:332–341. doi: 10.1038/nrc2775. [DOI] [PubMed] [Google Scholar]
- Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-κB activation. EMBO Reports. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager W, Santag S, Weidner-Glunde M, Gellermann E, Kati S, Pietrek M, Viejo-Borbolla A, Schulz TF. The ubiquitin-specific protease USP7 modulates the replication of Kaposi's sarcoma-associated herpesvirus latent episomal DNA. Journal of Virology. 2012;86:6745–6757. doi: 10.1128/JVI.06840-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- Ji Z, Mohammed H, Webber A, Ridsdale J, Han N, Carroll JS, Sharrocks AD. The forkhead transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex. Nucleic Acids Research. 2014;42:6232–6242. doi: 10.1093/nar/gku274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Yu W, Lou X, Zhou F, Han X, Zhao N, Lin B. UCHL1 is a putative tumor suppressor in ovarian cancer cells and contributes to cisplatin resistance. Journal of Cancer. 2013;4:662–670. doi: 10.7150/jca.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N, Yamamoto H, Adachi Y, Ohashi H, Taniguchi H, Suzuki H, Nakazawa M, Kaneto H, Sasaki S, Imai K, et al. Cancer detection by ubiquitin carboxyl-terminal esterase L1 methylation in pancreatobiliary fluids. World Journal of Gastroenterology. 2013;19:1718–1727. doi: 10.3748/wjg.v19.i11.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoronenkova SV, Dianov GL. USP7S-dependent inactivation of Mule regulates DNA damage signalling and repair. Nucleic Acids Research. 2013;41:1750–1756. doi: 10.1093/nar/gks1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Park KC, Chung SS, Bang O, Chung CH. Deubiquitinating enzymes as cellular regulators. Journal of Biochemistry. 2003;134:9–18. doi: 10.1093/jb/mvg107. [DOI] [PubMed] [Google Scholar]
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Molecular Cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- van der Knaap JA, Kozhevnikova E, Langenberg K, Moshkin YM, Verrijzer CP. Biosynthetic enzyme GMP synthetase cooperates with ubiquitin-specific protease 7 in transcriptional regulation of ecdysteroid target genes. Molecular and Cellular Biology. 2009;30:736–744. doi: 10.1128/MCB.01121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell. 2010;141:606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. The emerging complexity of protein ubiquitination. Biochemical Society Transactions. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- Komander D, Barford D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochemical Journal. 2008;409:77–85. doi: 10.1042/BJ20071399. [DOI] [PubMed] [Google Scholar]
- Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Molecular Cell. 2008;29:451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, Gu W. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29:1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Krikos A, Laherty CD, Dixit VM. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by κB elements. Journal of Biological Chemistry. 1992;267:17971–17976. [PubMed] [Google Scholar]
- Kulathu Y, Garcia FJ, Mevissen TE, Busch M, Arnaudo N, Carroll KS, Barford D, Komander D. Regulation of A20 and other OTU deubiquitinases by reversible oxidation. Nature Communications. 2013;4:1569. doi: 10.1038/ncomms2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- Laurell C, Velazquez-Fernandez D, Lindsten K, Juhlin C, Enberg U, Geli J, Hoog A, Kjellman M, Lundeberg J, Hamberger B, et al. Transcriptional profiling enables molecular classification of adrenocortical tumours. European Journal of Endocrinology. 2009;161:141–152. doi: 10.1530/EJE-09-0068. [DOI] [PubMed] [Google Scholar]
- Lee HR, Choi WC, Lee S, Hwang J, Hwang E, Guchhait K, Haas J, Toth Z, Jeon YH, Oh TK, et al. Bilateral inhibition of HAUSP deubiquitinase by a viral interferon regulatory factor protein. Nature Structural & Molecular Biology. 2011;18:1336–1344. doi: 10.1038/nsmb.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiblich A, Cross SS, Catto JW, Pesce G, Hamdy FC, Rehman I. Human prostate cancer cells express neuroendocrine cell markers PGP 9.5 and chromogranin A. Prostate. 2007;67:1761–1769. doi: 10.1002/pros.20654. [DOI] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, et al. The ubiquitin pathway in Parkinson's disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002a;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang D, Na X, Schoen SR, Messing EM, Wu G. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel–Lindau tumor suppressor. Biochemical and Biophysical Research Communications. 2002b;294:700–709. doi: 10.1016/S0006-291X(02)00534-X. [DOI] [PubMed] [Google Scholar]
- Li Z, Na X, Wang D, Schoen SR, Messing EM, Wu G. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel–Lindau tumor suppressor protein. Journal of Biological Chemistry. 2002c;277:4656–4662. doi: 10.1074/jbc.M108269200. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1α. EMBO Reports. 2005;6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Olson LM, Zhang Z, Li L, Bidder M, Nguyen L, Pfeifer J, Rader JS. Differential display identifies overexpression of the USP36 gene, encoding a deubiquitinating enzyme, in ovarian cancer. International Journal of Medical Sciences. 2008;5:133–142. doi: 10.7150/ijms.5.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wen S, Wang B, Gao W, Zhang W, Meng X, Yang L, Kong L. [Expression of cancer stem cell marker USP22 in laryngeal squamous cell carcinoma] Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2014;49:479–482. [PubMed] [Google Scholar]
- Lin SC, Chung JY, Lamothe B, Rajashankar K, Lu M, Lo YC, Lam AY, Darnay BG, Wu H. Molecular basis for the unique deubiquitinating activity of the NF-κB inhibitor A20. Journal of Molecular Biology. 2008;376:526–540. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Yang H, Kong Q, Li J, Lee SM, Gao B, Dong H, Wei J, Song J, Zhang DD, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Molecular Cell. 2012;46:484–494. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect α-synuclein degradation and Parkinson's disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/S0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yang Y, Xu H, Dong X. Implication of USP22 in the regulation of BMI-1, c-Myc, p16INK4a, p14ARF, and cyclin D2 expression in primary colorectal carcinomas. Diagnostic Molecular Pathology. 2010;19:194–200. doi: 10.1097/PDM.0b013e3181e202f2. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhao Q, Liang X, Du G, Lu L, Dong J, Han H. [Expression of neuronal marker protein gene product 9.5 and its clinicopathologic significance in breast cancer] Zhonghua Bing Li Xue Za Zhi. 2014;43:318–320. [PubMed] [Google Scholar]
- Lu Y, Bedard N, Chevalier S, Wing SS. Identification of distinctive patterns of USP19-mediated growth regulation in normal and malignant cells. PLoS ONE. 2011;6:e15936. doi: 10.1371/journal.pone.0015936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Wang X, Urvalek AM, Li T, Xie H, Yu L, Zhao J. Transformation of human ovarian surface epithelial cells by Kruppel-like factor 8. Oncogene. 2014;33:10–18. doi: 10.1038/onc.2012.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luise C, Capra M, Donzelli M, Mazzarol G, Jodice MG, Nuciforo P, Viale G, Di Fiore PP, Confalonieri S. An atlas of altered expression of deubiquitinating enzymes in human cancer. PLoS ONE. 2011;6:e15891. doi: 10.1371/journal.pone.0015891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Gao X, Zhao W, Li Y, Li C. Relationship between expression of Pad1 homologue and multidrug resistance of idiopathic nephrotic syndrome. Pediatrics International. 2009;51:732–735. doi: 10.1111/j.1442-200X.2009.02845.x. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. Journal of Biological Chemistry. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. Ubiquitin-specific proteases 7 and 11 modulate polycomb regulation of the INK4a tumour suppressor. EMBO Journal. 2010;29:2553–2565. doi: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Senic-Matuglia F, Di Fiore PP, Polo S, Hodsdon ME, De Camilli P. Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. PNAS. 2005;102:12700–12705. doi: 10.1073/pnas.0506344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi R. CYLD: a deubiquitination enzyme with multiple roles in cancer. Future Oncology. 2011;7:285–297. doi: 10.2217/fon.10.187. [DOI] [PubMed] [Google Scholar]
- Masuya D, Huang C, Liu D, Nakashima T, Yokomise H, Ueno M, Nakashima N, Sumitomo S. The HAUSP gene plays an important role in non-small cell lung carcinogenesis through p53-dependent pathways. Journal of Pathology. 2006;208:724–732. doi: 10.1002/path.1931. [DOI] [PubMed] [Google Scholar]
- Meredith M, Orr A, Everett R. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Molecular Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Miranda M, Sorkin A. Regulation of receptors and transporters by ubiquitination: new insights into surprisingly similar mechanisms. Molecular Interventions. 2007;7:157–167. doi: 10.1124/mi.7.3.7. [DOI] [PubMed] [Google Scholar]
- Misaghi S, Ottosen S, Izrael-Tomasevic A, Arnott D, Lamkanfi M, Lee J, Liu J, O'Rourke K, Dixit VM, Wilson AC. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Molecular and Cellular Biology. 2009;29:2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morotti A, Panuzzo C, Crivellaro S, Pergolizzi B, Familiari U, Berger AH, Saglio G, Pandolfi PP. BCR-ABL disrupts PTEN nuclear-cytoplasmic shuttling through phosphorylation-dependent activation of HAUSP. Leukemia. 2014;28:1326–1333. doi: 10.1038/leu.2013.370. [DOI] [PubMed] [Google Scholar]
- Mosbech A, Lukas C, Bekker-Jensen S, Mailand N. The deubiquitylating enzyme USP44 counteracts the DNA double-strand break response mediated by the RNF8 and RNF168 ubiquitin ligases. Journal of Biological Chemistry. 2013;288:16579–16587. doi: 10.1074/jbc.M113.459917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanao MH, Tcherniuk SO, Chroboczek J, Dideberg O, Dessen A, Balakirev MY. Crystal structure of human otubain 2. EMBO Reports. 2004;5:783–788. doi: 10.1038/sj.embor.7400201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro G, Menon RP, Masino L, Knowles PP, McDonald NQ, Pastore A. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. PNAS. 2005;102:10493–10498. doi: 10.1073/pnas.0501732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Okochi-Takada E, Nakazawa K, Wakabayashi M, Mori A, Ichimura S, Yasugi T, Ushijima T. Silencing of the UCHL1 gene in human colorectal and ovarian cancers. International Journal of Cancer. Journal International du Cancer. 2006;119:1338–1344. doi: 10.1002/ijc.22025. [DOI] [PubMed] [Google Scholar]
- Pal A, Young MA, Donato NJ. Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Research. 2014;74:4955–4966. doi: 10.1158/0008-5472.CAN-14-1211. [DOI] [PubMed] [Google Scholar]
- Panier S, Durocher D. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair. 2009;8:436–443. doi: 10.1016/j.dnarep.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Parviainen H, Schrade A, Kiiveri S, Prunskaite-Hyyrylainen R, Haglund C, Vainio S, Wilson DB, Arola J, Heikinheimo M. Expression of Wnt and TGF-β pathway components and key adrenal transcription factors in adrenocortical tumors: association to carcinoma aggressiveness. Pathology, Research and Practice. 2013;209:503–509. doi: 10.1016/j.prp.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathare GR, Nagy I, Sledz P, Anderson DJ, Zhou HJ, Pardon E, Steyaert J, Forster F, Bracher A, Baumeister W. Crystal structure of the proteasomal deubiquitylation module Rpn8–Rpn11. PNAS. 2014;111:2984–2989. doi: 10.1073/pnas.1400546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao S, Liu Y, Hu J, Guo F, Ma J, Sun Y, Zhang B. USP22 is useful as a novel molecular marker for predicting disease progression and patient prognosis of oral squamous cell carcinoma. PLoS ONE. 2012;7:e42540. doi: 10.1371/journal.pone.0042540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao S, Ma J, Wang W, Liu Y, Zhang M, Chen H, Guo F, Zhang B. Increased expression of USP22 is associated with disease progression and patient prognosis of salivary duct carcinoma. Oral Oncology. 2013;49:796–801. doi: 10.1016/j.oraloncology.2013.03.454. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annual Review of Biochemistry. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Planchon SM, Waite KA, Eng C. The nuclear affairs of PTEN. Journal of Cell Science. 2008;121:249–253. doi: 10.1242/jcs.022459. [DOI] [PubMed] [Google Scholar]
- Priolo C, Tang D, Brahamandan M, Benassi B, Sicinska E, Ogino S, Farsetti A, Porrello A, Finn S, Zimmermann J, et al. The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Research. 2006;66:8625–8632. doi: 10.1158/0008-5472.CAN-06-1374. [DOI] [PubMed] [Google Scholar]
- Reddy BA, van der Knaap JA, Bot AG, Mohd-Sarip A, Dekkers DH, Timmermans MA, Martens JW, Demmers JA, Verrijzer CP. Nucleotide biosynthetic enzyme GMP synthase is a TRIM21-controlled relay of p53 stabilization. Molecular Cell. 2014;53:458–470. doi: 10.1016/j.molcel.2013.12.017. [DOI] [PubMed] [Google Scholar]
- Reiner T, Parrondo R, de Las Pozas A, Palenzuela D, Perez-Stable C. Betulinic acid selectively increases protein degradation and enhances prostate cancer-specific apoptosis: possible role for inhibition of deubiquitinase activity. PLoS ONE. 2013;8:e56234. doi: 10.1371/journal.pone.0056234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renatus M, Parrado SG, D'Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, Pierrat B, Riedl R, Vinzenz D, Worpenberg S, et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, Harpon J, Battaglia V, Vivat V, Sippl W, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chemistry & Biology. 2012;19:467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Wilkinson KD. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chemical Reviews. 2009;109:1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode J, Dhillon AP, Doran JF, Jackson P, Thompson RJ. PGP 9.5, a new marker for human neuroendocrine tumours. Histopathology. 1985;9:147–158. doi: 10.1111/j.1365-2559.1985.tb02431.x. [DOI] [PubMed] [Google Scholar]
- Sacco JJ, Yau TY, Darling S, Patel V, Liu H, Urbe S, Clague MJ, Coulson JM. The deubiquitylase ataxin-3 restricts PTEN transcription in lung cancer cells. Oncogene. 2014;33:4265–4272. doi: 10.1038/onc.2013.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Kigawa T, Koshiba S, Sato K, Matsuo Y, Sakamoto A, Takagi T, Shirouzu M, Yabuki T, Nunokawa E, et al. The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKγ. Structure. 2004;12:1719–1728. doi: 10.1016/j.str.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Salsman J, Jagannathan M, Paladino P, Chan PK, Dellaire G, Raught B, Frappier L. Proteomic profiling of the human cytomegalovirus UL35 gene products reveals a role for UL35 in the DNA repair response. Journal of Virology. 2012;86:806–820. doi: 10.1128/JVI.05442-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010;328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara NL, Ringel AE, Wolberger C. A role for intersubunit interactions in maintaining SAGA deubiquitinating module structure and activity. Structure. 2012;20:1414–1424. doi: 10.1016/j.str.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, Zhang RG, Liao J, Lee W, Edwards AM, et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein–Barr nuclear antigen 1 implications for EBV-mediated immortalization. Molecular Cell. 2005;18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Sarkari F, Sanchez-Alcaraz T, Wang S, Holowaty MN, Sheng Y, Frappier L. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein–Barr virus latent origin of DNA replication. PLoS Pathogens. 2009;5:e1000624. doi: 10.1371/journal.ppat.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkari F, La Delfa A, Arrowsmith CH, Frappier L, Sheng Y, Saridakis V. Further insight into substrate recognition by USP7: structural and biochemical analysis of the HdmX and Hdm2 interactions with USP7. Journal of Molecular Biology. 2010;402:825–837. doi: 10.1016/j.jmb.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Sarkari F, Wheaton K, La Delfa A, Mohamed M, Shaikh F, Khatun R, Arrowsmith CH, Frappier L, Saridakis V, Sheng Y. Ubiquitin-specific protease 7 is a regulator of ubiquitin-conjugating enzyme UbE2E1. Journal of Biological Chemistry. 2013;288:16975–16985. doi: 10.1074/jbc.M113.469262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- Scaglione KM, Zavodszky E, Todi SV, Patury S, Xu P, Rodriguez-Lebron E, Fischer S, Konen J, Djarmati A, Peng J, et al. Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Molecular Cell. 2011;43:599–612. doi: 10.1016/j.molcel.2011.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J. Histone H2A deubiquitinase activity of the polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrecengost RS, Dean JL, Goodwin JF, Schiewer MJ, Urban MW, Stanek TJ, Sussman RT, Hicks JL, Birbe RC, Draganova-Tacheva RA, et al. USP22 regulates oncogenic signaling pathways to drive lethal cancer progression. Cancer Research. 2014;74:272–286. doi: 10.1158/0008-5472.CAN-13-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer K, Bozko PM, Dubiel W, Naumann M. CSN controls NF-κB by deubiquitinylation of IκBα. EMBO Journal. 2007;26:1532–1541. doi: 10.1038/sj.emboj.7601600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam I, Abbas M, Ayoub F, Mirabal S, Bsaili M, Caulder EK, Weinstock DM, Tomkinson AE, Hromas R, Shaheen M. Ubiquitin-specific peptidase 20 regulates Rad17 stability, checkpoint kinase 1 phosphorylation and DNA repair by homologous recombination. Journal of Biological Chemistry. 2014;289:22739–22748. doi: 10.1074/jbc.M114.550459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N, Pujari R, Harhaj NS, Abbott DW, Harhaj EW. The kinase IKKα inhibits activation of the transcription factor NF-κB by phosphorylating the regulatory molecule TAX1BP1. Nature Immunology. 2011;12:834–843. doi: 10.1038/ni.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, Frappier L. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nature Structural & Molecular Biology. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: A Cancer Journal for Clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- Sloane MA, Wong JW, Perera D, Nunez AC, Pimanda JE, Hawkins NJ, Sieber OM, Bourke MJ, Hesson LB, Ward RL. Epigenetic inactivation of the candidate tumor suppressor USP44 is a frequent and early event in colorectal neoplasia. Epigenetics. 2014;9:1092–1100. doi: 10.4161/epi.29222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JJ, Sixma TK. RBR E3-ligases at work. EMBO Reports. 2014;15:142–154. doi: 10.1002/embr.201338166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP–PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spataro V, Simmen K, Realini CA. The essential 26S proteasome subunit Rpn11 confers multidrug resistance to mammalian cells. Anticancer Research. 2002;22:3905–3909. [PubMed] [Google Scholar]
- Spratt DE, Walden H, Shaw GS. RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochemical Journal. 2014;458:421–437. doi: 10.1042/BJ20140006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley JA, Aruldhas MM, Chandrasekaran M, Neelamohan R, Suthagar E, Annapoorna K, Sharmila S, Jayakumar J, Jayaraman G, Srinivasan N, et al. Androgen receptor expression in human thyroid cancer tissues: a potential mechanism underlying the gender bias in the incidence of thyroid cancers. Journal of Steroid Biochemistry and Molecular Biology. 2012;130:105–124. doi: 10.1016/j.jsbmb.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Steele JM. Carfilzomib: a new proteasome inhibitor for relapsed or refractory multiple myeloma. Journal of Oncology Pharmacy Practice. 2013;19:348–354. doi: 10.1177/1078155212470388. [DOI] [PubMed] [Google Scholar]
- Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO Journal. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Lee GS, Oh WK, Pomerantz M, Yang M, Xie W, Freedman ML, Kantoff PW. Single-nucleotide polymorphisms in p53 pathway and aggressiveness of prostate cancer in a Caucasian population. Clinical Cancer Research. 2010;16:5244–5251. doi: 10.1158/1078-0432.CCR-10-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Miyauchi A, Matsuzuka F, Yoshida H, Nakata Y, Kuma K, Amino N. PGP9.5 mRNA could contribute to the molecular-based diagnosis of medullary thyroid carcinoma. European Journal of Cancer. 2004;40:614–618. doi: 10.1016/j.ejca.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Terzic J, Marinovic-Terzic I, Ikeda F, Dikic I. Ubiquitin signals in the NF-κB pathway. Biochemical Society Transactions. 2007;35:942–945. doi: 10.1042/BST0350942. [DOI] [PubMed] [Google Scholar]
- Tezel E, Hibi K, Nagasaka T, Nakao A. PGP9.5 as a prognostic factor in pancreatic cancer. Clinical Cancer Research. 2000;6:4764–4767. [PubMed] [Google Scholar]
- Thompson RJ, Doran JF, Jackson P, Dhillon AP, Rode J. PGP9.5 – a new marker for vertebrate neurons and neuroendocrine cells. Brain Research. 1983;278:224–228. doi: 10.1016/0006-8993(83)90241-X. [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Nishimasu H, Ishitani R, Goto E, Noguchi T, Mio K, Kamei K, Ma A, Iwai K, Nureki O. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO Journal. 2012;31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T. PGP 9.5 immunocytochemical staining for pancreatic endocrine tumors. Islets. 2013;5:122–128. doi: 10.4161/isl.25351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootee A, Hasani-Ranjbar S. Von Hippel–Lindau disease: a new approach to an old problem. International Journal of Endocrinology and Metabolism. 2012;10:619–624. doi: 10.5812/ijem.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- Tsirlin A, Oo Y, Sharma R, Kansara A, Gliwa A, Banerji MA. Pheochromocytoma: a review. Maturitas. 2014;77:229–238. doi: 10.1016/j.maturitas.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Ummanni R, Jost E, Braig M, Lohmann F, Mundt F, Barett C, Schlomm T, Sauter G, Senff T, Bokemeyer C, et al. Ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) is a potential tumour suppressor in prostate cancer and is frequently silenced by promoter methylation. Molecular Cancer. 2011;10:129. doi: 10.1186/1476-4598-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uras IZ, List T, Nijman SM. Ubiquitin-specific protease 4 inhibits mono-ubiquitination of the master growth factor signaling kinase PDK1. PLoS ONE. 2012;7:e31003. doi: 10.1371/journal.pone.0031003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Fernandez D, Laurell C, Geli J, Hoog A, Odeberg J, Kjellman M, Lundeberg J, Hamberger B, Nilsson P, Backdahl M. Expression profiling of adrenocortical neoplasms suggests a molecular signature of malignancy. Surgery. 2005;138:1087–1094. doi: 10.1016/j.surg.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Vendrell JA, Ghayad S, Ben-Larbi S, Dumontet C, Mechti N, Cohen PA. A20/TNFAIP3, a new estrogen-regulated gene that confers tamoxifen resistance in breast cancer cells. Oncogene. 2007;26:4656–4667. doi: 10.1038/sj.onc.1210269. [DOI] [PubMed] [Google Scholar]
- Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, Wilkinson KD. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Research. 2008;68:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen L, Verhelst K, Carpentier I, Beyaert R. TAX1BP1, a ubiquitin-binding adaptor protein in innate immunity and beyond. Trends in Biochemical Sciences. 2011;36:347–354. doi: 10.1016/j.tibs.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Wang YZ, Wong YC. Oncogenes and tumor suppressor genes in prostate cancer: a review. Urologic Oncology. 1997;3:41–46. doi: 10.1016/S1078-1439(97)00021-5. [DOI] [PubMed] [Google Scholar]
- Wang G, Gao Y, Li L, Jin G, Cai Z, Chao JI, Lin HK. K63-linked ubiquitination in kinase activation and cancer. Frontiers in Oncology. 2012;2:5. doi: 10.3389/fonc.2012.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li YP, Chen JH, Yuan SF, Wang L, Zhang JL, Yao Q, Li NL, Bian JF, Fan J, et al. Prognostic significance of USP22 as an oncogene in papillary thyroid carcinoma. Tumour Biology. 2013;34:1635–1639. doi: 10.1007/s13277-013-0696-0. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen YJ, Xu K, Wang YY, Shen XZ, Tu RQ. High expression of UCH37 is significantly associated with poor prognosis in human epithelial ovarian cancer. Tumour Biology. 2014;35:11427–11433. doi: 10.1007/s13277-014-2446-3. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Morabito LM, Bilen J, Gordesky-Gold B, Faust LZ, Paulson HL, Bonini NM. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Molecular Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Weinstock J, Wu J, Cao P, Kingsbury WD, McDermott JL, Kodrasov MP, McKelvey DM, Suresh Kumar KG, Goldenberg SJ, Mattern MR, et al. Selective dual inhibitors of the cancer-related deubiquitylating proteases USP7 and USP47. ACS Medicinal Chemistry Letters. 2012;3:789–792. doi: 10.1021/ml200276j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nature Reviews. Molecular Cell Biology. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Wen D, Xu Z, Xia L, Liu X, Tu Y, Lei H, Wang W, Wang T, Song L, Ma C, et al. Important role of SUMOylation of Spliceosome factors in prostate cancer cells. Journal of Proteome Research. 2014;13:3571–3582. doi: 10.1021/pr4012848. [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Wosnitzer MS, Mielnik A, Dabaja A, Robinson B, Schlegel PN, Paduch DA. Ubiquitin specific protease 26 (USP26) expression analysis in human testicular and extragonadal tissues indicates diverse action of USP26 in cell differentiation and tumorigenesis. PLoS ONE. 2014;9:e98638. doi: 10.1371/journal.pone.0098638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Li L, Yin X, Yuan C, Tan C, Su X, Xiong L, Putti TC, Oberst M, Kelly K, et al. The ubiquitin peptidase UCHL1 induces G0/G1 cell cycle arrest and apoptosis through stabilizing p53 and is frequently silenced in breast cancer. PLoS ONE. 2012;7:e29783. doi: 10.1371/journal.pone.0029783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Takanashi M, Oikawa K, Tanaka M, Nishi H, Isaka K, Kudo M, Kuroda M. USP15 plays an essential role for caspase-3 activation during paclitaxel-induced apoptosis. Biochemical and Biophysical Research Communications. 2009;388:366–371. doi: 10.1016/j.bbrc.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Xue S, Jia J. Genetic association between ubiquitin carboxy-terminal hydrolase-L1 gene S18Y polymorphism and sporadic Alzheimer's disease in a Chinese Han population. Brain Research. 2006;1087:28–32. doi: 10.1016/j.brainres.2006.02.121. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Miyazaki M, Kodrasov MP, Rotinsulu H, Losung F, Mangindaan RE, de Voogd NJ, Yokosawa H, Nicholson B, Tsukamoto S. Spongiacidin C, a pyrrole alkaloid from the marine sponge Stylissa massa, functions as a USP7 inhibitor. Bioorganic & Medicinal Chemistry Letters. 2013;23:3884–3886. doi: 10.1016/j.bmcl.2013.04.066. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Imai J, Watanabe M, Hiroi N, Sugano S, Yoshino G. Restoration of transforming growth factor-β type II receptor reduces tumorigenicity in the human adrenocortical carcinoma SW-13 cell line. Hormone and Metabolic Research. 2006;38:159–166. doi: 10.1055/s-2006-925185. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Hibi K, Takase T, Tezel E, Nakayama H, Kasai Y, Ito K, Akiyama S, Nagasaka T, Nakao A. PGP9.5 as a marker for invasive colorectal cancer. Clinical Cancer Research. 2002;8:192–195. [PubMed] [Google Scholar]
- Yang YA, Yu J. EZH2, an epigenetic driver of prostate cancer. Protein & Cell. 2013;4:331–341. doi: 10.1007/s13238-013-2093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hou JQ, Qu LY, Wang GQ, Ju HW, Zhao ZW, Yu ZH, Yang HJ. [Differential expression of USP2, USP14 and UBE4A between ovarian serous cystadenocarcinoma and adjacent normal tissues] Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2007;23:504–506. [PubMed] [Google Scholar]
- Yang M, Liu YD, Wang YY, Liu TB, Ge TT, Lou G. Ubiquitin-specific protease 22: a novel molecular biomarker in cervical cancer prognosis and therapeutics. Tumour Biology. 2014;35:929–934. doi: 10.1007/s13277-013-1121-4. [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Zarrizi R, Menard J, Belting M, Massoumi R. Deubiquitination of γ-tubulin by BAP1 prevents chromosome instability in breast cancer cells. Cancer Research. 2014;74:6499–6508. doi: 10.1158/0008-5472.CAN-14-0221. [DOI] [PubMed] [Google Scholar]
- Zavacki AM, Arrojo EDR, Freitas BC, Chung M, Harney JW, Egri P, Wittmann G, Fekete C, Gereben B, Bianco AC. The E3 ubiquitin ligase TEB4 mediates degradation of type 2 iodothyronine deiodinase. Molecular and Cellular Biology. 2009;29:5339–5347. doi: 10.1128/MCB.01498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yao L, Zhang X, Ji H, Wang L, Sun S, Pang D. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. Journal of Cancer Research and Clinical Oncology. 2011a;137:1245–1253. doi: 10.1007/s00432-011-0998-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Berger FG, Yang J, Lu X. USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO Journal. 2011b;30:2177–2189. doi: 10.1038/emboj.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska BE, Mickanin C, Huang H, Sheppard KA, Porter JA, Lu CX, et al. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nature Cell Biology. 2012;14:717–726. doi: 10.1038/ncb2522. [DOI] [PubMed] [Google Scholar]
- Zhang J, Cao M, Dong J, Li C, Xu W, Zhan Y, Wang X, Yu M, Ge C, Ge Z, et al. ABRO1 suppresses tumourigenesis and regulates the DNA damage response by stabilizing p53. Nature Communications. 2014;5:5059. doi: 10.1038/ncomms6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Pittman RN. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Human Molecular Genetics. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a