Abstract
Protein adsorption onto nanoparticles (NPs) in biological fluids has emerged as an important factor when testing biological responses to NPs, as this may influence both uptake and subsequent toxicity. The aim of the present study was to quantify the adsorption of proteins onto TiO2 NPs and to test the influence on cellular uptake. The surface composition of the particles was characterized by thermal analysis and by X-ray photoelectron spectroscopy. The adsorption of three blood proteins, ie, human serum albumin (HSA), γ-globulins (Glbs), and fibrinogen (Fib), onto three types of anatase NPs of different sizes was quantified for each protein. The concentration of the adsorbed protein was measured by ultraviolet-visible spectrophotometry using the Bradford method. The degree of cellular uptake was quantified by inductivity coupled plasma mass spectroscopy, and visualized by an ultra-high resolution imaging system. The proteins were adsorbed onto all of the anatase NPs. The quantity adsorbed increased with time and was higher for the smaller particles. Fib and Glbs showed the highest affinity to TiO2 NPs, while the lowest was seen for HSA. The adsorption of proteins affected the surface charge and the hydrodynamic diameter of the NPs in cell culture medium. The degree of particle uptake was highest in protein-free medium and in the presence HSA, followed by culture medium supplemented with Glbs, and lowest in the presence of Fib. The results indicate that the uptake of anatase NPs by fibroblasts is influenced by the identity of the adsorbed protein.
Keywords: nanoparticles, titanium dioxide, proteins, adsorption, cellular uptake
Introduction
Nano-sized particles are more biologically active than their micron-sized counterparts.1 The toxicity of nanoparticles (NPs) is influenced by physicochemical characteristics such as: the primary particle size, agglomeration state, specific surface area, zeta potential, and surface chemistry.2 Protein adsorption onto NPs in biological fluids has emerged as an important factor to be taken into consideration with regard to the assessment of biological responses to NPs.3–5
Human plasma contains different types and variants of proteins, of which human serum albumin (HSA), fibrinogen (Fib), and γ-globulins (Glbs) are the most abundant.6,7 Cells used in cytotoxicity studies are usually cultured in media supplemented with fetal bovine serum (FBS), which is a mixture of various proteins, similar to what is found in human plasma, except for Fib, which is removed from FBS. NPs introduced to the biological environment will inevitably interact with proteins, leading to their adsorption onto the particles. A “protein corona” may be formed, which is a complex mixture of adsorbed proteins in equilibrium on the surface of NPs.4,8–10 The protein corona may alter the size, agglomeration state, and interfacial properties of the NPs,10 which could potentially affect the cellular uptake of NPs.3,11
Protein adsorption to a given NP and its influence on the cellular uptake and biological responses in general, are not well elucidated. Until recently, only a few studies have investigated the effect of proteins on the uptake of NPs. Some of these studies revealed that the uptake was enhanced in the presence of serum proteins12,13 on polystyrene NPs, whereas others showed higher uptake in the absence of serum proteins on polystyrene,14 silica,15 polymer,16 and carbon nanotubes.17
TiO2 NPs are being incorporated in a variety of pharmaceutical products, food, and cosmetics. Once inside the body, unintentionally or intentionally, such NPs will inevitably interact with blood proteins. The adsorption of proteins onto TiO2 NPs has been a subject of several studies.18–24 In these studies, the connection between protein adsorption and cellular uptake was not investigated. Previously, we have evaluated the effect of physicochemical characteristics of a panel of TiO2 NPs on uptake by fibroblasts.25 The aim of the present study was to quantify the individual adsorption of HSA, Glbs, and Fib onto anatase TiO2 NPs and their effect on uptake by L929 fibroblasts.
Materials and methods
TiO2 nanopowders
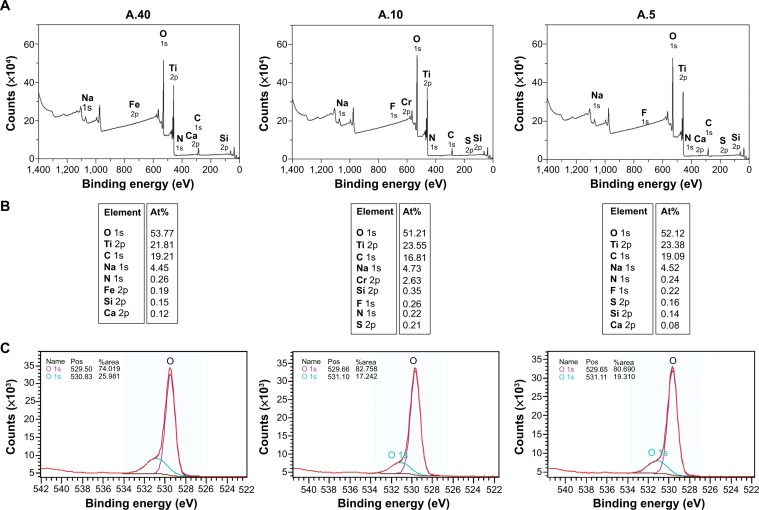
Three types of commercially available anatase TiO2 NPs with different sizes were used in this study, termed A.40, A.10, and A.5. Their physicochemical characteristics and transmission electron micrographs were reported by us in a previous paper.25 The NP samples were 100% anatase, except for A.5, which comprised 14% rutile phase. The smallest particles (A.5) had the highest specific surface area (Braunauer-Emmett-Teller, 131 m2/g), then A.10 (105 m2/g), whereas the largest particles (A.40) had the lowest (49 m2/g). Transmission electron microscopy of the NP samples showed that A.5 had average diameter of 11 nm, A.10 was 24 nm, and A.40 was 43 nm, and they were spherical. A thermogravimetric analysis was carried out in order to reveal the presence of surface contaminants and was performed in a Stanton Redcroft thermobalance (Model STA-780) (Figure 1A). The temperature programmed desorption–mass spectrometry analyses were carried out on an experimental setup coupled to a Pfeiffer quadrupole mass spectrometer (Figure 1B).26 To gain further insight on the particles’ surface chemistry, the NPs’ surface was characterized by X-ray photoelectron spectroscopy (XPS) (Kratos Axix Ultra DLD, Kratos Analytical Ltd., Manchester, UK with CasaXPS software, Casa Software Ltd., UK) (Figure 2).
Figure 1.
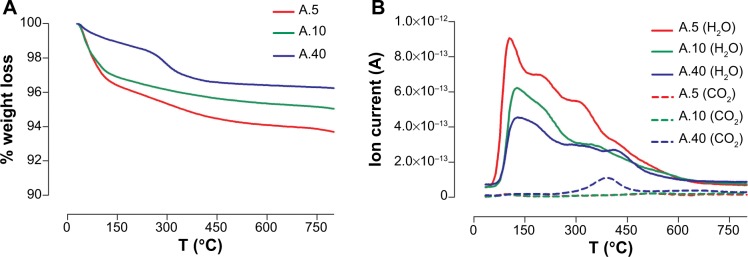
Thermal analysis of TiO2 NP samples A.5, A.10, and A.40.
Notes: (A) TGA; and (B) TPD–MS.
Abbreviations: NP, nanoparticle; TGA, thermogravimetric analysis; TPD–MS, temperature programmed desorption–mass spectrometry; T, temperature.
Figure 2.
XPS of TiO2 NP samples A.5, A.10, and A.40.
Notes: (A) Survey scan; (B) atomic percent of surface elements; (C) high resolution scan of O 1s.
Abbreviations: NP, nanoparticle; XPS, X-ray photoelectron spectroscopy; At%, atomic percent; pos, position.
Suspension of NPs
The suspensions were made following a protocol that was shown to provide a good NP dispersion.25,27 Briefly, 1 g/L TiO2 NPs in deionized H2O stock solution was sonicated (VCX130, Vibra-Cell, 130 watts; Sonics & Materials Inc, Newtown, CT, USA), pulse at 70% duty for 2 minutes. Right after sonication, the volumes needed to prepare the end-point concentrations were transferred to 10 mL test tubes containing Roswell Park Memorial Institute (RPMI) 1640 cell culture medium (BE12-918F; Lonza Group Ltd, Basel, Switzerland), without L-glutamine and phenol red, either alone or with individual proteins.
Quantification of the adsorbed proteins
One g/L TiO2 NPs prepared as described above were added to the test tubes containing 1 g/L proteins in the cell culture medium. The proteins used were HSA (A1653-5G; Sigma-Aldrich Co, St Louis, MO, USA), Fib from human plasma (F3879-1G; Sigma-Aldrich Co) and Glbs from human blood (G4386-1G; Sigma-Aldrich Co). The test tubes were rotated for 2 hours or 24 hours at 37°C. The particles were then separated by 2× centrifugation (5810R, Eppendorf AG centrifuge, rotor: A-4-62; Hamburg, Germany) at 3,220× g, for 15 minutes. The concentration of proteins in the supernatant was determined by adding 1.5 mL of the Bradford reagent (B6916-500ML; Sigma-Aldrich Co) to 50 μL taken from the supernatant.28 The mixture was vortexed and allowed to react for 5 minutes, and then the optical absorption was measured in a ultraviolet-visible spectrophotometer (BioMate 3S; Thermo Fisher Scientific, Waltham, MA, USA) at the wavelength of 595 nm. The protein concentration was obtained from a reference standard curve, made for each protein. The amount of protein adsorbed on the particle surface was obtained by subtracting the protein concentration in the supernatant from the initial protein concentration.29
Diameter and zeta potentials of particles in suspension
Dynamic light scattering (DLS) was used in order to reveal the hydrodynamic diameter of the agglomerates (Table 1) using a Zetasizer NS (Malvern Instruments, Malvern, UK). Dilute samples based on the suspensions described above (maximum 100 mg/L) were used in order to avoid multiple scattering. The electrophoretic mobilities (Uef =150 V) of the particles were converted to apparent zeta-potentials (ζ-potentials) using the Helmholtz–Smoluchowski relationship (Table 2).30
Table 1.
Agglomerate sizes expressed as hydrodynamic diameter of TiO2 NPs (100 mg/L) in RPMI 1640 cell culture medium with (100 mg/L) and without proteins, after 24 h and 3 h (in brackets) rotation at 37°C
| Sample | Hydrodynamic diameter ± SD (nm)
|
|||
|---|---|---|---|---|
| Without proteins | HSA | Glbs | Fib | |
| A.5 | 2,779±130 | 573±6 | * | * |
| (2,874±62) | (547±6) | (*) | (660±24) | |
| A.10 | 3,242±244 | 572±14 | * | * |
| (3,211±39) | (580±10) | (*) | (573±17) | |
| A.40 | 1,439±41 | 1,275±30 | * | * |
| (1,800±70) | (1,500±64) | (*) | (962±121) | |
Note:
It was impossible to measure the hydrodynamic diameter because the samples were too polydispersed (polydispersity index >0.6).
Abbreviations: NPs, nanoparticles; RPMI, Roswell Park Memorial Institute; h, hours; SD, standard deviation; HSA, human serum albumin; Glbs, γ-globulins; Fib, fibrinogen.
Table 2.
Zeta potentials of TiO2 NPs in RPMI medium with and without proteins, after 24 h rotation at 37°C
| Sample | Zeta potential ± SD (mV)
|
|||
|---|---|---|---|---|
| Without proteins | HSA | Glbs | Fib | |
| A.5 | −18.2±1.6 | −12.0±1.0 | −5.4±0.5 | −9.0±0.5 |
| A.10 | −18.0±1.3 | −13.4±0.6 | −5.9±0.4 | −7.6±0.5 |
| A.40 | −20.6±1.7 | −17.6±1.1 | −16.4±1.1 | −18.4±1.4 |
Abbreviations: NPs, nanoparticles; RPMI, Roswell Park Memorial Institute; h, hours; SD, standard deviation; HSA, human serum albumin; Glbs, γ-globulins; Fib, fibrinogen.
Cell culture
The National Collection of Type Cultures (NCTC) clone 929 (L929 mouse fibroblasts) from the American Type Culture Collection, Manassas, VA, USA were employed because fibroblasts constitute the major cellular component of fibrous connective tissue surrounding the implants. L929 cells were maintained in culture at 20,000 cells/cm2, in 25 cm2 polystyrene flasks in RPMI 1640 with 10% FBS, 2% penicillin/streptomycin/fungisone, and 1% L-glutamine (all from MedProbe AS, Lysaker, Norway), at 37°C, 5% CO2. The cells were trypsinized every 3–4 days and then transferred to new flasks. Only cells cultures with a viability >90% (tested by exclusion of 0.2% trypan blue) and below 30 passages were used in the experiments.
Quantification of TiO2 NP cellular uptake
The cells were seeded in 12-well plates (Thermo Fisher Scientific; Nunc™ Nunclon™ Delta, category number 150628) in the same medium as explained above, and then incubated for 48 hours until they reached 70%–80% confluence. The supernatant was removed, washed twice with phosphate-buffered saline (PBS), and exposed for 24 hours to 5 mg/L nano-TiO2 NPs suspended in RPMI 1640 cell culture medium either without proteins or with individual proteins; ie, HSA, Fib, or Glbs, at a concentration of 100 mg/L. The prepared exposure solutions were rotated 1 hour before exposure. After exposure, the cells were washed again three times with PBS to remove unattached particles. The cells were then trypsinized, transferred into new tubes, and sonicated in an ultrasound bath for 30 minutes, at 45°C. The solutions were then digested in a microwave digestion unit (MLS 1200 Mega; Gemini BV, Apeldoorn, the Netherlands) by adding 2 mL nitric acid (60%) (Ultrapure; EMD Millipore, Billerica, MA, USA) and 50 μL hydrofluoric acid (40%) (Suprapur®; EMD Millipore).
The total concentration of Ti, representing the TiO2 uptake, was determined by inductively coupled plasma–mass spectrometry (ICP–MS) (Element 2; Thermo Fisher Scientific). An internal standard of indium (1 μg/L) was added to all the samples to monitor and correct for any instrumental fluctuations. Calibration was performed by standard addition using calibrating solutions (0.2, 0.5, 2, and 10 μg/L) (EMD Millipore).
Visualization of uptake
Prior to exposure, the fibroblasts were seeded in two-well glass chambers (Thermo Fisher Scientific; Nunc™ Lab-Tek™) and kept for 48 hours at 37°C till they became 70%–80% confluent. They were then exposed for 24 hours to 0.5 mg/L of TiO2 NPs by removing the supernatant, washing with PBS, and replacing it with 1 mL of TiO2 NP solutions prepared as described above. At the end of the exposure, cells were washed three times with PBS in order to remove unattached particles, followed by fixation in 4% formaldehyde for 15 minutes at room temperature. The fibroblasts were then washed twice with PBS and once with sterile water. After removing the chambers from the slides, a mounting medium (Eukitt™; Electron Microscopy Sciences, Hatfield, PA, USA) was used to mount a cover-slip. Each experiment was repeated at least three times and run in duplicate. Imaging and image analysis was performed by using an ultra-high resolution dark field condenser (URI [ultra-high resolution imaging] CytoViva™ 130; Warner Instruments, Hamden, CT, USA), on an optical microscope (Olympus BX41 microscope; Olympus Corporation, Tokyo, Japan), with a light source (X-Cite™ 120; EXFO, VANIER, QC, Canada).31 The fixed cells were examined with a 100× oil immersion objective.
Statistical analysis
Data for adsorption and uptake were not normally distributed and thus, non-parametric tests were employed. Results in Figures 3 and 4 were expressed as medians and quartiles. The Kruskal–Wallis H test and Dunn’s test were used for multiple comparisons. P<0.05 was considered statistically significant. Measurements of agglomerates’ sizes (Table 1) and zeta potentials (Table 2) were presented as means ± standard deviations. Data were analyzed by a statistical analysis software package (SPSS for Windows, version 18).
Figure 3.
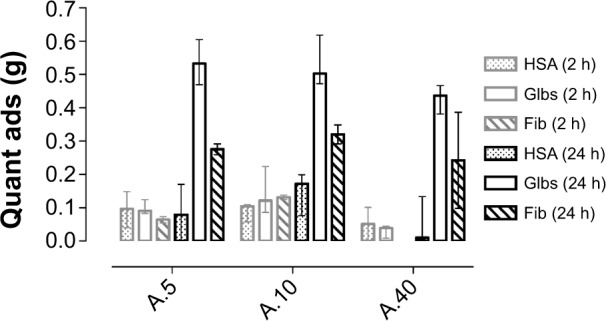
Quantity adsorbed of proteins onto TiO2 NPs after 2-h and 24-h rotations at 37°C. The results for samples A.5, A.10, and A.40 are expressed as medians with interquartile range.
Abbreviations: HSA, human serum albumin; Glbs, γ-globulins; Fib, fibrinogen; h, hours; quant, quantity; ads, adsorbed; NPs, nanoparticles.
Figure 4.
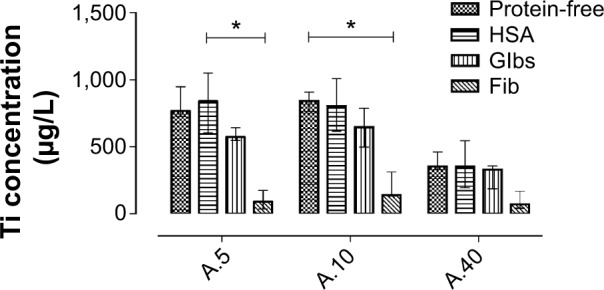
Degree of cellular uptake, showing the concentration (in μg/L) of titanium element associated with L929 fibroblasts, after 24 h exposure to TiO2 NPs suspended in different media. The results are expressed as medians with interquartile range.
Note: *Statistical significance (P<0.05, Dunn’s multiple comparison test).
Abbreviations: HSA, human serum albumin; Glbs, γ-globulins; Fib, fibrinogen; h, hours; NPs, nanoparticles.
Results and discussion
Surface characterization of TiO2 NPs
The thermogravimetric analysis of the nano-TiO2 showed a total weight loss in the whole temperature range of 6.3%, 5.0%, and 3.8% for A.5, A.10, and A.40, respectively (Figure 1A). Two main regions of weight loss could be distinguished, below 150°C and between 150°C and 500°C. This could be due to the removal of H2O and hydroxyl, respectively.32 Thermal desorption studies (Figure 1B) confirmed the existence of H2O onto the particles and showed the dehydroxylation at higher temperatures.32 In addition, carbon traces were observed for A.40 following desorption of CO2 from its surface, at ~400°C.
The XPS survey scan of the NPs further revealed that a high percent of the carbon element was in fact present in all three samples and not just A.40 (Figure 2A, B). Sodium was also present in a relatively high percentage (~4.5%) on all particles. Moreover, a significant level of chromium (2.63%) was found at the surface of A.10 as well as trace elements such as sulfur (0.21% for A.10) and iron (0.19% for A.40). These trace elements are likely impurities from the synthesis process. Storage and transport may also be a source of surface contamination. The concentration ratios of oxygen and titanium were 2.46, 2.17, and 2.23 for A.40, A.10, and A.5, respectively. This means that there are other O atoms than Ti–O in TiO2. A high resolution XPS spectrum of O(1s) binding energy region revealed two peaks, one ~529 eV attributed to Ti–O and one ~231 eV attributed to Ti–OH (Figure 2C), which are hydroxyl groups chemisorbed on the surface of the sample.33 This is in accordance with thermogravimetric analysis. The hydroxyl groups readily adsorb water under ambient conditions, and give an indication on the wettability.
Protein adsorption onto TiO2 NPs
The proteins used in the present study (HSA, Fib, and Glbs) were all able to adsorb onto TiO2 NPs when incubated individually with the particles for a 2–24-hour period at 37°C. The adsorption of Glbs and Fib was time-dependent (Figure 3). After 2 hours, the quantity adsorbed was low and relatively similar for all the proteins used. However, after 24 hours, the adsorbed quantity of Glbs and Fib increased significantly and was much higher than that of HSA. Protein adsorption is a dynamic process in which longer exposure time leads to a higher probability of reaching the surface of the particles and thus, to a higher adsorption over time. The highest adsorption was observed for Glbs, followed by Fib, whereas the lowest was found for HSA (Figure 3).
The highest quantity adsorbed was observed for the smallest NPs (A.5) (Figure 3), partly due to the larger specific surface area of the small particles (Figure 3), which provides increased available adsorption sites.4,17,34,35 However, the increase in protein adsorption with the increase in surface area was not substantial and not statistically significant in the present study. This may be due to agglomeration, which decreases the exposed effective surface for adsorption. DLS experiments revealed that in the absence of proteins the particles were highly agglomerated and reached micron-sized agglomerates (Table 1). This may be caused by the increase in the ionic strength resulting in a decrease of the electrical double layer.27,36 In contrast, in the presence of HSA, the agglomerates’ sizes were much smaller and their hydrodynamic diameters did not increase from 3 to 24 hours (Table 1). The low zeta potentials (absolute values) following protein adsorption (Table 2) support the hypothesis that the stabilizing effect of proteins is not related to electrostatic repulsion, but rather to steric repulsion.37–39 Some agglomeration over time is however inevitable, even in the presence of proteins. In the presence of Glbs and Fib, the polydispersity index was high, which means a broad size distribution. In that case, the accuracy of the DLS method decreases substantially and therefore, results with a polydispersity index >0.6 were not reported.
All the particles used in this study had a negative net charge in all the media, and their surface potentials were relatively similar in protein-free medium (Table 2). If we assume electrostatic interactions as the main mechanism governing the adsorption of proteins onto NPs,40 proteins will then adsorb favorably onto positively charged NPs.41 However, proteins can also adsorb to negatively charged particles, like in the present study. The suggested mechanism in this case is bridging by divalent cations (eg, Ca2+).18,19
A distinct shift in the zeta potential of the particles was observed following adsorption of proteins (Table 2), eg, for A.5, from −18.2 (protein-free medium) to −12 (HSA), -9.0 (Fib), and −5.4 (Glbs). The shift in the zeta potential appears to correlate with the amount of the protein adsorbed for all the particles used, ie, the highest adsorption was observed for Glbs, followed by Fib, and the lowest for HSA. The zeta potential is a measure of net surface charge density. When the proteins adsorb onto NPs, the measured zeta potential may reflect the net surface charge density of the adsorbed protein. There is a prevailing opinion that NPs tend to adopt the physicochemical properties of the adsorbed protein.3,4,10,42 Thus, protein adsorption affects the surface charge of the particles, which may influence their uptake.41,43,44 The zeta potential for A.40 was less affected by protein adsorption than for the other anatase NPs. This may be due to the low adsorption of proteins.
Cellular uptake of TiO2 NPs
Due to their small size, NPs can enter the cells and eventually induce harmful effects. The main reported effects of TiO2 NPs are inflammation, cell death, and DNA damage.45–49 The three anatase samples were all taken up, to different degrees, by the fibroblasts after 24 hours’ incubation in all media used (Figure 4). The uptake of TiO2 NPs was higher for the smallest (A.5 and A.10) particles, both in the presence and absence of proteins. Several other studies clearly indicate that size is an important parameter for uptake.25,50–52 It is difficult to say if the difference in the degree of uptake was size-related in our study, since the primary particle size range was small.25,50–52 However, small primary NPs are taken up by other mechanisms than larger particles or agglomerates of the same compound.53,54
The highest uptake, measured by ICP–MS, was observed when the cells were exposed to NPs in protein-free medium and in the presence of HSA (Figure 4). In presence of Fib, and to some extent Glbs, the uptake was lower.
These findings indicate that the protein adsorption influences cellular uptake. In an earlier study, the uptake of polystyrene NPs by endothelial cells did not depend on the identity of protein adsorbed, but rather on the quantity adsorbed.12 In the present study, this was particularly apparent in the case of HSA vs Glbs, and HSA vs Fib. Increased protein adsorption retains the particles longer in suspension, and consequently, they are in less contact with the cell in an in vitro system where the cells are attached to the bottom of the well. Fast sedimentation will increase particle–cell contact, which probably is the case for particles in protein-free medium.
The finding that the uptake was low in the presence of Fib should be interpreted with caution, as this might be due to bridge flocculation (interfacial agglomeration), derived from the adsorption of Fib.55,56 This could explain why it was not possible to measure the hydrodynamic diameter of TiO2 NPs suspended in medium containing Fib (Table 1), as the resulting interfacial agglomerates were probably responsible for the high polydispersity index of the Fib–TiO2 suspension. Thus, the lower uptake of Fib-bound particles may be due to the interfacial agglomeration, which is relevant for in vivo studies as the interaction of NPs with the blood coagulant may reduce the bioavailable surface area of the particles and the concentration presented to the cells.55
URI imaging supports the results from ICP–MS and showed that the cells were associated with TiO2 NPs’ agglomerates in all the media (Figure 5B–E), but to a lesser extent in media supplemented with Fib (Figure 5E) compared with HSA (Figure 5C) and Glbs (Figure 5D). The agglomerates were observed by URI as bright spots due to high light scattering. No such optical traits were observed with cells in media without NPs (Figure 5A). It was difficult to judge if the NPs were placed inside the cells and/or attached to their cellular membrane. Both is the probably the case, as demonstrated by scanning electron microscopy and transmission electron microscopy imaging in our previous study.25
Figure 5.
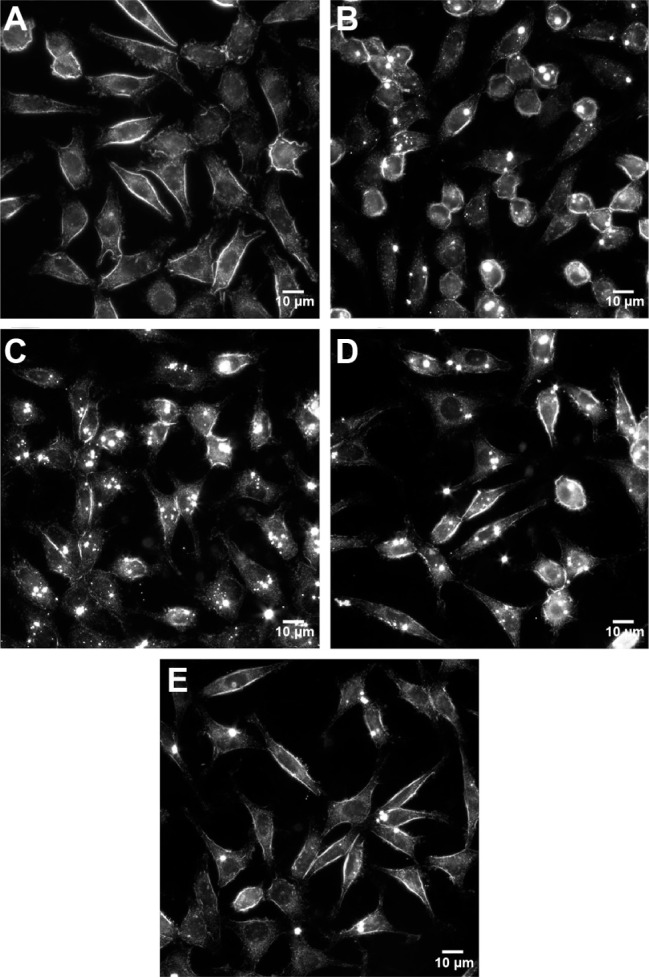
URI images of L929 fibroblasts at 1000× magnification (scale bar: 10 μm), after 24 h exposure to 0.5 mg/L of TiO2 NPs (sample A.5) in different media.
Notes: (A) Control cells not exposed to NPs. (B) Protein-free. (C) HSA. (D) Glb. (E) Fib. Bright spots are NPs/agglomerates either internalized or attached to the cell membrane.
Abbreviations: URI, ultra-high resolution imaging; h, hours; NPs, nanoparticles; HSA, human serum albumin; Glbs, γ-globulins; Fib, fibrinogen.
It seems that both the adsorbed protein and the particles’ native surface may mediate the attachment of NPs to cellular membranes. As mentioned earlier in the discussion, serum proteins adsorb onto the surface of both cationic and anionic NPs. A recent paper by Fleischer and Payne revealed that in the presence of bovine serum albumin (BSA), the cellular binding of BSA−NP complexes containing positively charged NPs was increased, while that of BSA−NP complexes formed from the same NPs, but negatively charged, was inhibited.57 The uptake in absence of serum proteins may be due to direct recognition of the particles at the cell surface.12,58 In the presence of proteins, the uptake proceeds probably by interaction of the adsorbed proteins, specifically with the protein receptors on the cell surface.12 The protein-rich corona may interact with multiple receptors and thus, multiple mechanisms may be involved simultaneously.58,59 In contrast, particles having one type of protein adsorbed at their surface may be restricted to a specific receptor.12
Conclusion
The highest protein adsorption onto the anatase TiO2 NPs was observed for Glbs, followed by Fib, and the lowest for HSA. The adsorption of proteins affected the agglomerate size and surface charge of the NPs, which alters the electrostatic binding affinity with cells.
The presence of Fib, and to some extent Glbs, significantly lowered the uptake in fibroblasts. The lower uptake might be related to the high protein adsorption, which increases steric stabilization of the agglomerates, leading to less sedimentation and cell–particle contact.
Acknowledgments
The authors would like to thank the University of Bergen, the Grieg Foundation, and the Meltzer Research Fund for financial support. A special thanks to Tore Skodvin for DLS and electrophoretic mobility measurements and to Miguel A. Cauqui for the surface characterization of the particles.
Footnotes
Disclosure
The authors declare that they alone are responsible for the content and writing of this paper. The authors report no conflicts of interest in this work.
References
- 1.Oberdörster G, Oberdörster E, Oberdörster J. Nanotoxicology: an emerging discipline evolving from studies of ultrafine particles. Environ Health Perspect. 2005;113(7):823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nel A, Xia T, Madler L, Li N. Toxic potential of materials at the nano-level. Science. 2006;311(5761):622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 3.Cedervall T, Lynch I, Lindman S, et al. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci U S A. 2007;104(7):2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch I, Cedervall T, Lundqvist M, Cabaleiro-Lago C, Linse S, Dawson KA. The nanoparticle – protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century. Adv Colloid Interface Sci. 2007:134–135. 167–174. doi: 10.1016/j.cis.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 5.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. What the cell “sees” in bionanoscience. J Am Chem Soc. 2010;132(16):5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 6.Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv Drug Deliv Rev. 2009;61(6):428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karmali PP, Simberg D. Interactions of nanoparticles with plasma proteins: implication on clearance and toxicity of drug delivery systems. Expert Opin Drug Deliv. 2011;8(3):343–357. doi: 10.1517/17425247.2011.554818. [DOI] [PubMed] [Google Scholar]
- 8.Monopoli MP, Aberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nat Nanotechnol. 2012;7(12):779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 9.Lynch I, Salvati A, Dawson KA. Protein-nanoparticle interactions: what does the cell see? Nat Nanotechnol. 2009;4(9):546–547. doi: 10.1038/nnano.2009.248. [DOI] [PubMed] [Google Scholar]
- 10.Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3(1):40–47. [Google Scholar]
- 11.Dutta D, Sundaram SK, Teeguarden JG, et al. Adsorbed proteins influence the biological activity and molecular targeting of nanomaterials. Toxicol Sci. 2007;100(1):303–315. doi: 10.1093/toxsci/kfm217. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenberg MS, Friedman AE, Finkelstein JN, Oberdörster G, McGrath JL. The influence of protein adsorption on nanoparticle association with cultured endothelial cells. Biomaterials. 2009;30(4):603–610. doi: 10.1016/j.biomaterials.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 13.Johnston HJ, Semmler-Behnke M, Brown DM, Kreyling W, Tran L, Stone V. Evaluating the uptake and intracellular fate of polystyrene nanoparticles by primary and hepatocyte cell lines in vitro. Toxicol Appl Pharmacol. 2010;242(1):66–78. doi: 10.1016/j.taap.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Guarnieri D, Guaccio A, Fusco S, Netti PA. Effect of serum proteins on polystyrene nanoparticle uptake and intracellular trafficking in endothelial cells. J Nanopart Res. 2011;13(9):4295–4309. [Google Scholar]
- 15.Lesniak A, Fenaroli F, Monopoli MP, Aberg C, Dawson KA, Salvati A. Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano. 2012;6(7):5845–5857. doi: 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- 16.Petri-Fink A, Steitz B, Finka A, Salaklang J, Hofmann H. Effect of cell media on polymer coated superparamagnetic iron oxide nanoparticles (SPIONs): colloidal stability, cytotoxicity, and cellular uptake studies. Eur J Pharm Biopharm. 2008;68(1):129–137. doi: 10.1016/j.ejpb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Y, Li WX, Li QN, et al. Effects of serum proteins on intracellular uptake and cytotoxicity of carbon nanoparticles. Carbon. 2009;47(5):1351–1358. [Google Scholar]
- 18.Klinger A, Steinberg D, Kohavi D, Sela MN. Mechanism of adsorption of human albumin to titanium in vitro. J Biomed Mater Res. 1997;36(3):387–392. doi: 10.1002/(sici)1097-4636(19970905)36:3<387::aid-jbm13>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 19.Ellingsen JE. A study on the mechanism of protein adsorption to TiO2. Biomaterials. 1991;12(6):593–596. doi: 10.1016/0142-9612(91)90057-h. [DOI] [PubMed] [Google Scholar]
- 20.Yongli C, Xiufang Z, Yandao G, Nanming Z, Tingying Z, Xinqi S. Conformational changes of fibrinogen adsorption onto hydroxyapatite and titanium oxide nanoparticles. J Colloid Interface Sci. 1999;214(1):38–45. doi: 10.1006/jcis.1999.6159. [DOI] [PubMed] [Google Scholar]
- 21.Giacomelli CE, Avena MJ, DePauli CP. Adsorption of bovine serum albumin onto TiO2 particles. J Colloid Interface Sci. 1997;188(2):387–395. [Google Scholar]
- 22.Wassell DTH, Embery G. Adsorption of bovine serum albumin on to titanium powder. Biomaterials. 1996;17(9):859–864. doi: 10.1016/0142-9612(96)83280-7. [DOI] [PubMed] [Google Scholar]
- 23.Zaqout MS, Sumizawa T, Igisu H, Higashi T, Myojo T. Binding of human serum proteins to titanium dioxide particles in vitro. J Occup Health. 2011;53(2):75–83. doi: 10.1539/joh.l10034. [DOI] [PubMed] [Google Scholar]
- 24.Ruh H, Kühl B, Brenner-Weiss G, Hopf C, Diabaté S, Weiss C. Identification of serum proteins bound to industrial nanomaterials. Toxicol Lett. 2012;208(1):41–50. doi: 10.1016/j.toxlet.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Allouni ZE, Høl PJ, Cauqui MA, Gjerdet NR, Cimpan MR. Role of physicochemical characteristics in the uptake of TiO2 nanoparticles by fibroblasts. Toxicol In Vitro. 2012;26(3):469–479. doi: 10.1016/j.tiv.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 26.Boukha Z, Fitian L, López-Haro M, et al. Influence of the calcination temperature on the nano-structural properties, surface basicity, and catalytic behavior of alumina-supported lanthana samples. J Catalysis. 2010;272(1):121–130. [Google Scholar]
- 27.Allouni ZE, Cimpan MR, Høl PJ, Skodvin T, Gjerdet NR. Agglomeration and sedimentation of TiO2 nanoparticles in cell culture medium. Colloids Surf B Biointerfaces. 2009;68(1):83–87. doi: 10.1016/j.colsurfb.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Bradford MM. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Arvizo RR, Giri K, Moyano D, et al. Identifying new therapeutic targets via modulation of protein corona formation by engineered nanoparticles. PLoS One. 2012;7(3):e33650. doi: 10.1371/journal.pone.0033650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tkachenko NH, Yaremko ZM, Bellmann C. Effect of 1-1-charged ions on aggregative stability and electrical surface properties of aqueous suspensions of titanium dioxide. Colloid Surface A Physicochem Engineer Aspect. 2006;279(1):10–19. doi: 10.1016/j.jcis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Vainrub A, Pustovyy O, Vodyanoy V. Resolution of 90 nm (lambda/5) in an optical transmission microscope with an annular condenser. Opt Lett. 2006;31(19):2855–2857. doi: 10.1364/ol.31.002855. [DOI] [PubMed] [Google Scholar]
- 32.Nagaveni K, Hegde MS, Ravishankar N, Subbanna GN, Madras G. Synthesis and structure of nanocrystalline TiO2 with lower band gap showing high photocatalytic activity. Langmuir. 2004;20(7):2900–2907. doi: 10.1021/la035777v. [DOI] [PubMed] [Google Scholar]
- 33.Wu L, Yu JC, Wang XC, Zhang LZ, Yu JG. Characterization of mesoporous nanocrystalline TiO2 photocatalysts synthesized via a sol-solvothermal process at a low temperature. J Solid State Chem. 2005;178(1):321–328. [Google Scholar]
- 34.Horie M, Nishio K, Fujita K, et al. Protein adsorption of ultrafine metal oxide and its influence on cytotoxicity toward cultured cells. Chem Res Toxicol. 2009;22(3):543–553. doi: 10.1021/tx800289z. [DOI] [PubMed] [Google Scholar]
- 35.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci U S A. 2008;105(38):14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng ZJ, Mortimer G, Schiller T, Musumeci A, Martin D, Minchin RF. Differential plasma protein binding to metal oxide nanoparticles. Nanotechnology. 2009;20(45):455101. doi: 10.1088/0957-4484/20/45/455101. [DOI] [PubMed] [Google Scholar]
- 37.Landsiedel R, Ma-Hock L, Kroll A, et al. Testing metal-oxide nanomaterials for human safety. Adv Mater. 2010;22(24):2601–2627. doi: 10.1002/adma.200902658. [DOI] [PubMed] [Google Scholar]
- 38.Taurozzi JS, Hackley VA, Wiesner MR. A standardised approach for the dispersion of titanium dioxide nanoparticles in biological media. Nanotoxicology. 2013;7(4):389–401. doi: 10.3109/17435390.2012.665506. [DOI] [PubMed] [Google Scholar]
- 39.Vamanu CI, Høl PJ, Allouni ZE, Elsayed S, Gjerdet NR. Formation of potential titanium antigens based on protein binding to titanium dioxide nanoparticles. Int J Nanomedicine. 2008;3(1):69–74. doi: 10.2147/ijn.s1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rezwan K, Meier LP, Gauckler LJ. Lysozyme and bovine serum albumin adsorption on uncoated silica and AlOOH-coated silica particles: the influence of positively and negatively charged oxide surface coatings. Biomaterials. 2005;26(21):4351–4357. doi: 10.1016/j.biomaterials.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 41.Patil S, Sandberg A, Heckert E, Self W, Seal S. Protein adsorption and cellular uptake of cerium oxide nanoparticles as a function of zeta potential. Biomaterials. 2007;28(31):4600–4607. doi: 10.1016/j.biomaterials.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alkilany AM, Nagaria PK, Hexel CR, Shaw TJ, Murphy CJ, Wyatt MD. Cellular uptake and cytotoxicity of gold nanorods: molecular origin of cytotoxicity and surface effects. Small. 2009;5(6):701–708. doi: 10.1002/smll.200801546. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Niu J, Zhang W, Zhang L, Shang E. Influence of aqueous media on the ROS-mediated toxicity of ZnO nanoparticles toward green fluorescent protein-expressing Escherichia coli under UV-365 irradiation. Langmuir. 2014;30(10):2852–2862. doi: 10.1021/la5000028. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Zhang W, Niu J, Chen Y. Mechanism of photogenerated reactive oxygen species and correlation with the antibacterial properties of engineered metal-oxide nanoparticles. ACS Nano. 2012;6(6):5164–5173. doi: 10.1021/nn300934k. [DOI] [PubMed] [Google Scholar]
- 45.Hamilton RF, Wu N, Porter D, Buford M, Wolfarth M, Holian A. Particle length-dependent titanium dioxide nanomaterials toxicity and bioactivity. Part Fibre Toxicol. 2009;6:35. doi: 10.1186/1743-8977-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iavicoli I, Leso V, Fontana L, Bergamaschi A. Toxicological effects of titanium dioxide nanoparticles: a review of in vitro mammalian studies. Eur Rev Med Pharmacol Sci. 2011;15(5):481–508. [PubMed] [Google Scholar]
- 47.Jin CY, Zhu BS, Wang XF, Lu QH. Cytotoxicity of titanium dioxide nanoparticles in mouse fibroblast cells. Chem Res Toxicol. 2008;21(9):1871–1877. doi: 10.1021/tx800179f. [DOI] [PubMed] [Google Scholar]
- 48.Long TC, Saleh N, Tilton RD, Lowry GV, Veronesi B. Titanium dioxide (P25) produces reactive oxygen species in immortalized brain microglia (BV2): implications for nanoparticle neurotoxicity. Environ Sci Technol. 2006;40(14):4346–4352. doi: 10.1021/es060589n. [DOI] [PubMed] [Google Scholar]
- 49.Vamanu CI, Cimpan MR, Høl PJ, Sørnes S, Lie SA, Gjerdet NR. Induction of cell death by TiO2 nanoparticles: studies on a human monoblastoid cell line. Toxicol In Vitro. 2008;22(7):1689–1696. doi: 10.1016/j.tiv.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Andersson PO, Lejon C, Ekstrand-Hammarström B, et al. Polymorph-and size-dependent uptake and toxicity of TiO2 nanoparticles in living lung epithelial cells. Small. 2011;7(4):514–523. doi: 10.1002/smll.201001832. [DOI] [PubMed] [Google Scholar]
- 51.Carlson C, Hussain SM, Schrand AM, et al. Unique cellular interaction of silver nanoparticles: size-dependent generation of reactive oxygen species. J Phys Chem B. 2008;112(43):13608–13619. doi: 10.1021/jp712087m. [DOI] [PubMed] [Google Scholar]
- 52.Tenzer S, Docter D, Rosfa S, et al. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS Nano. 2011;5(9):7155–7167. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- 53.Zhang S, Nelson A, Beales PA. Freezing or wrapping: the role of particle size in the mechanism of nanoparticle-biomembrane interaction. Langmuir. 2012;28(35):12831–12837. doi: 10.1021/la301771b. [DOI] [PubMed] [Google Scholar]
- 54.Muller KH, Motskin M, Philpott AJ, et al. The effect of particle agglomeration on the formation of a surface-connected compartment induced by hydroxyapatite nanoparticles in human monocyte-derived macrophages. Biomaterials. 2014;35(3):1074–1088. doi: 10.1016/j.biomaterials.2013.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kendall M, Ding P, Kendall K. Particle and nanoparticle interactions with fibrinogen: the importance of aggregation in nanotoxicology. Nanotoxicology. 2011;5(1):55–65. doi: 10.3109/17435390.2010.489724. [DOI] [PubMed] [Google Scholar]
- 56.Morrissey BW, Strömberg RR. Conformation of adsorbed blood proteins by infrared bound fraction measurements. J Colloid Interface Sci. 1974;46(1):152–164. [Google Scholar]
- 57.Fleischer CC, Payne CK. Nanoparticle-cell interactions: molecular structure of the protein corona and cellular outcomes. Acc Chem Res. 2014;47(8):2651–2659. doi: 10.1021/ar500190q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walkey CD, Chan WCW. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem Soc Rev. 2012;41(7):2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 59.Ding HM, Ma YQ. Computer simulation of the role of protein corona in cellular delivery of nanoparticles. Biomaterials. 2014;35(30):8703–8710. doi: 10.1016/j.biomaterials.2014.06.033. [DOI] [PubMed] [Google Scholar]