Abstract
The HIV-1 envelope (Env) mediates viral entry into host cells. To enable the direct imaging of conformational dynamics within Env, we introduced fluorophores into variable regions of the glycoprotein gp120 subunit and measured single-molecule fluorescence resonance energy transfer within the context of native trimers on the surface of HIV-1 virions. Our observations revealed unliganded HIV-1 Env to be intrinsically dynamic, transitioning between three distinct prefusion conformations, whose relative occupancies were remodeled by receptor CD4 and antibody binding. The distinct properties of neutralization-sensitive and neutralization-resistant HIV-1 isolates support a dynamics-based mechanism of immune evasion and ligand recognition.
The HIV-1 envelope (Env) spike is a membrane-fusion machine that mediates viral entry into cells (1). HIV-1 Env, composed of three gp120 glycoproteins and three gp41 subunits, evades recognition by antibodies by favoring a neutralization-resistant ground-state conformation in which N-linked glycans cover most of the surface (2–5). Interaction with the CD4 receptor causes structural rearrangements in gp120, which lead to formation of a co-receptor–binding site (1,2,6,7). These rearrangements include movement of the variable loops 1and 2(V1/V2) from their apical position in the unliganded trimer, to the trimer periphery (8–10). Subsequent interactions with the co-receptor trigger additional Env remodeling, with gp41 rearranging into a stable six-helix bundle that facilitates fusion between viral and cellular membranes. Although static images of HIV-1 Env in various conformations have been obtained (6–9,11–17), direct measurement of the dynamic features of the Env trimer has been lacking.
To enable real-time observations of conformational transitions in the native HIV-1 Env spike on the surface of virions, we used single-molecule fluorescence resonance energy transfer (smFRET) (18). Short peptides were introduced that permit enzymatically targeted incorporation of fluorophores (19,20) into the V1 loop and one of the following: the V4 loop, the E loop, or the V5 loop of gp120. The attachment of donor and acceptor fluorophores at these sites provided distinct reference points for observing movements of the V1 loop via time-dependent changes in FRET efficiency (Fig. 1). Dually tagged viruses were engineered for the neutralization-sensitive HIV-1NL4-3 laboratory-adapted strain and the neutralization-resistant tier-2 primary isolate HIV-1JR-FL. Dually tagged HIV-1NL4-3 viruses were isolated that met a series of functional criteria (21) (figs. S1 to S3). Insertions were then made in homologous positions in the HIV-1JR-FL Env that had little effect on infectivity (fig. S1B). Both HIV-1NL4-3 and HIV-1JR-FL tolerated the Q3 peptide (20, 22) (GQQQLG) in the V1 loop and the A1 peptide (19) (GDSLDMLEWSLM) in the V4 loop (V1-Q3/V4-A1) (fig. S4). HIV-1NL4-3 also tolerated the Q3 peptide in the V1 loop and the A1 peptide in the V5 loop (V1-Q3/V5-A1).
Fig. 1. Single-molecule FRET imaging indicates conformational dynamics of HIV-1 Env.
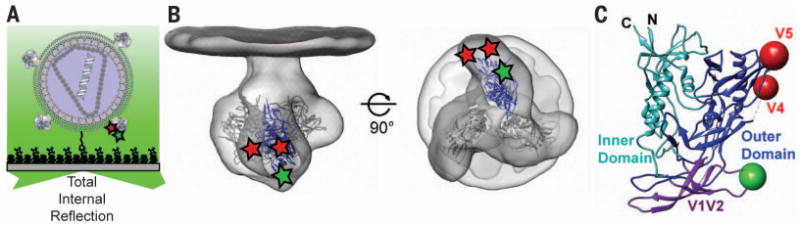
(A) Experimental design. Native HIV-1 virions containing a single dually labeled Env molecule among native trimers were immobilized on the surface of passivated quartz microscope slides and imaged via TIRF microscopy. (B) Approximate position of the organic dyes in the variable loops V1 (green; Cy3B or Cy3) and V4 or V5 (red; Cy5(4S)COT or Alexa Fluor 647) of gp120 of HIV-1 Env. The Env structure, with the ribbon structure of the gp120 trimer, is adapted from PDB accession code 4NCO (6–8, 11, 45). (C) Ribbon structures of the gp120 inner domain (cyan), outer domain (blue), and V1/V2 region (purple) are highlighted.
To ensure that only a single fluorescently labeled gp120 molecule was present on the surface of the HIV-1 virus, plasmid encoding wild-type Env was cotransfected at a ratio of 40:1 over the dually tagged Env. Viruses were harvested and enzymatically labeled with donor (e.g., Cy3B or Cy3) and acceptor [e.g., Cy5(4S)COT or Alexa Fluor 647] fluorophores (fig. S5) (21,23). A biotin-lipid incorporated into the viral membrane, which had negligible effects on virus infectivity (fig. S7), facilitated surface immobilization on streptavidin-coated quartz microscope slides and single-molecule imaging (24).
HIV-1NL4-3 and HIV-1JR-FL virions containing a fluorescently labeled gp120 domain within the context of a native Env trimer were imaged at a time resolution of 25 frames per second (40-ms integration time) by using a prism-based total internal reflection fluorescence (TIRF) microscope (Fig. 1A). Donor and acceptor fluorescence were collected, optically separated, and recorded on electron-multiplying charge-coupled device arrays. FRET efficiencies were calculated from donor and acceptor fluorescence intensities and compiled into population histograms (Fig. 2, A and B) (21). Consistent with the probability of tagged gp120 monomer incorporation into the Env trimer (1:40 ratio to wild-type gp120) and with labeling efficiencies of 40% and 55% for the Q3 and A1 tags, respectively, ∼12% of all surface-bound virions displayed FRET, equating to 23% of all visible molecules (21) (fig. S6).
Fig. 2. Ligands remodel the conformational landscape of HIV-1 Env.
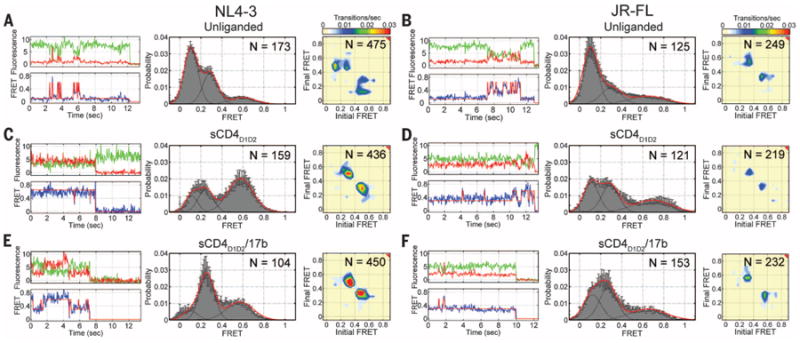
(A) (Top left) Representative fluorescence (Cy3B, donor, green; Cy5(4S) COT, acceptor, red) and (bottom left) FRET (blue) trajectories obtained from a single HIV-1NL4-3 Env on the surface of an intact virion. Idealization of FRET trajectories (red) was achieved by fitting each trace to a three-state Markov model (28). (Center) FRET trajectories from individual unliganded HIV-1NL4-3 Env proteins were compiled into a population FRET histogram and fit to the sum of three Gaussian distributions (red) with means 0.1, 0.3, and 0.6 (black). (Right) TDPs displaying the relative frequencies of observed transitions were generated from the idealization of individual FRET trajectories. (B) (Left) Fluorescence and FRET trajectories for the unliganded HIV-1JR-FL Env. (Center) FRET histogram fit to the sum of three Gaussian distributions with means 0.1, 0.3, and 0.65, and TDP. (C and D) Fluorescence and FRET trajectories, FRET histograms, and TDPs for (C) HIV-1NL4-3 Env and (D) HIV-1JR-FL Env in the presence of sCD4D1D2 (5 μM). (E and F) The same data for (E) HIV-1NL4-3 Env and (F) HIV-1JR-FL Env in the presence of sCD4D1D2 (5 μM) and 17b Fab (2 μM). The number of FRET trajectories and the number of FRET transitions are indicated on the histograms and TDPs, respectively. Histograms represent the means ± SEM determined from three independent populations of smFRET traces.
The unliganded V1-Q3/V4-A1–labeled HIV-1NL4-3 Env exhibited evidence of spontaneous reversible fluctuations between at least three conformations, each identified by a distinct FRET value: low (∼0.10), intermediate (∼0.30), and high FRET (∼0.60) (Fig. 2A). The unliganded V1-Q3/V4-A1-labeled HIV-1JR-FL Env and a HIV-1NL4-3 Env protein dually labeled in the V1 and V5 loops (V1-Q3/V5-A1) exhibited similar behavior (Fig. 2B and fig. S8). These findings, together with evidence of isotropic fluorophore tumbling (table S1), suggest that the time-dependent changes in FRET efficiency observed in each system reflect changes in interfluorophore distance. The observed transitions in the milliseconds-to-seconds time scale indicated that the HIV-1 Env trimer of both strains exhibits the intrinsic capacity to undergo large-scale conformational changes (25), which reposition the V1/V2 loop closer to the gp120 outer domain, where V4 and V5 reside.
The most populated conformation for both unliganded Env proteins exhibited low FRET (Fig. 2, A and B). This finding is consistent with prefusion HIV-1 Env predominantly occupying a closed, ground-state configuration that is conformationally masked (6, 7, 26), where the V1/V2 loops are proximal to the apex of the trimer and distal to the Env periphery (Fig. 1, B and C, and table S2). Consistent with this interpretation, mutations D368R and E370R (22) in the CD4-binding site of gp120 of HIV-1NL4-3, which inhibit formation of the bridging sheet necessary for CD4 binding (11, 27), stabilize the low-FRET state (fig. S9).
We next investigated the sequence of conformational transitions of the unliganded HIV-1 Env through the application of hidden Markov modeling (28). The smFRET trajectories from the V1-Q3/V4-A1-labeled HIV-1NL4-3 and HIV-1JR-FL Envs were fit to a three-state model containing low-, intermediate-, and high-FRET states. The well-defined peaks arranged symmetrically with respect to the diagonal axis in transition-density plots (TDPs) (Fig. 2, A and B) reflect the quality of the idealization and indicate that the system is in equilibrium. These data revealed that the neutralization-sensitive HIV-1NL4-3 Env frequently transitioned out of the low-FRET ground-state configuration into the high-FRET state, which had preferential access to the intermediate-FRET state. Direct transitions between low and intermediate FRET were seldom observed (Fig. 2A). The D368R/E370R mutant displayed a dramatic reduction in transitions out of the low-FRET state (fig. S9). This finding further verified our assignment of the low-FRET state as a ground-state conformation. In contrast, the neutralization-resistant HIV-1JR-FL Env exhibited reduced dynamics compared with HIV-1NL4-3 (Fig. 2B and table S3). This finding was most notably reflected in a substantial reduction in occupancy of the intermediate-FRET state. When transitions out of the low-FRET ground state were observed, however, HIV-1JR-FL Env, like HIV-1NL4-3, appeared to rapidly inter-convert between the high- and intermediate-FRET states. Again, direct transitions between low- and intermediate-FRET states were seldom observed. These findings indicate that both HIV-1 isolates preferentially achieve the intermediate-FRET configuration via the high-FRET state. Moreover, they reveal dynamic distinctions between the neutralization-sensitive laboratory-adapted HIV-1NL4-3 and the neutralization-resistant clinical isolate HIV-1JR-FL.
We next sought to investigate how the conformational landscape of the Env trimer responds to ligands. To measure the effects of CD4 and the co-receptor binding to Env, we used saturating concentrations of soluble CD4 (sCD4D1D2) (5 μM) (fig. S10); the CD4 mimetic small molecule JRC-II-191 (100 μM) (29); and the antigen-binding fragment (Fab) of antibody 17b (2 μM), a surrogate for the co-receptor. Antibody 17b binds Env at the co-receptor–binding site, which promotes a conformation of Env resembling the co-receptor–bound conformation (8,11,30). Whereas HIV-1NL4-3 and HIV-1JR-FL Env use distinct co-receptors (CXCR4 and CCR5, respectively), 17b binds to an epitope conserved in both strains. For HIV-1NL4-3 Env, the addition of sCD4 D1D2 stabilized the high-FRET state (Fig. 2C). The combination of sCD4D1D2 and 17b Fab stabilized the intermediate-FRET state (Fig. 2E). Likewise, the CD4-mimetic JRC-II-191 stabilized the intermediate-FRET state (fig. S11). Although saturating ligand concentrations were used (figs. S10A and S11C), TDPs indicate that the ligand-bound Env remains dynamic. In each of the ligand-saturated conditions examined, conformational sampling between high- and intermediate-FRET states predominated, whereas transitions between low and intermediate FRET were again rarely observed. For HIV-1JR-FL Env, sCD4D1D2 and sCD4D1D2/17b stabilized intermediate- and high-FRET states to similar extents (Fig. 2, D and F, and table S2). To test if this was due to incomplete sCD4D1D2 binding, the conformational equilibrium of HIV-1JR-FL Env was tested in the presence of the potent HIV-1 inhibitor, dodecameric sCD4D1D2 (sCD4D1D2-Igαtp, 0.1 mg/ml) (31). Here again, intermediate- and high-FRET states were stabilized to similar extents, and Env exhibited transitions between both states, despite the presence of a saturating ligand concentration (fig. S12).
Collectively, these observations reveal that the unliganded Env proteins of both HIV-1 isolates predominantly occupy ground-state configurations but are inherently competent to access conformations stabilized by sCD4D1D2 and sCD4D1D2/17b. These data indicate that spontaneously sampled Env conformations may serve a functional role in infectivity. For instance, this finding suggests, as has been previously speculated (32), that CD4 and 17b may facilitate the capture of preexisting Env conformations that are spontaneously sampled in the unliganded spike, rather than triggering a trimer-opening event. These observations also have implications for antibody neutralization strategies, as HIV-1NL4-3 can be readily stabilized in a single conformation by activating ligands, whereas the neutralization-resistant HIV-1JR-FL Env persistently exhibits relatively unstable, isoenergetic intermediate-and high-FRET states.
To examine directly the correlation between the occupancy of intermediate-FRET Env configurations and sensitivity to neutralization, we probed the response of both strains to 17b Fab alone (2 μM) as a function of time (Fig. 3, A and B). As expected, the neutralization-sensitive HIV-1NL4-3 Env showed a marked increase in occupancy of the intermediate-FRET state over time, whereas the neutralization-resistant HIV-1JR-FL Env showed no increase even after 60 min of incubation and only a slight increase after 90 min (Fig. 3, A to C). These experiments highlight functional distinctions of the two Env proteins and confirm that the intermediate-FRET state, which these data reveal is directly associated with co-receptor binding, is masked to a greater extent in the clinically isolated HIV-1JR-FL Env, which explains the increased resistance to 17b neutralization (Fig. 3D).
Fig. 3. Conformational masking of the co-receptor–binding site.
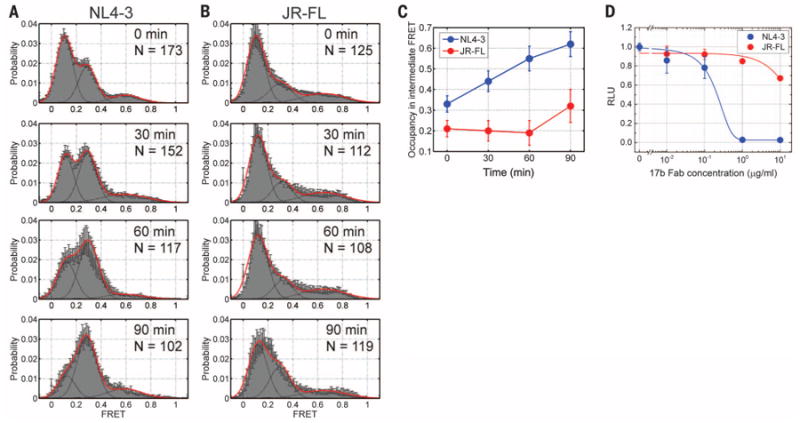
(A and B) Population FRET histograms each composed of 100 to 150 smFRET trajectories for (A) HIV-1NL4-3 Env and (B) HIV-1JR-FL Env in presence of 17b Fab (2 μM) acquired after 0, 30, 60, and 90 min of incubation. All histograms are displayed as in Fig. 2. (C) Quantification of the occupancy in the intermediate-FRET state for (blue) HIV-1NL4-3 Env and (red) HIV-1JR-FL Env. (D) Corresponding neutralization of HIV-1NL4-3 Env and HIV-1JR-FL infectivity by 17b Fab. The data are presented as the means ± SEM determined from three independent measurements.
The observed changes in the conformational landscape of HIV-1 Env in response to sCD4 and 17b allow us to assign the low- and intermediate-FRET configurations to ground-state and the CD4/17b-bound Env conformations, respectively (8,13). The assignment of the high-FRET states is less clear because the HIV-1NL4-3 and HIV-1JR-FL Env isolates differ in their responses to sCD4. Based on our observations of HIV-1NL4-3 the high-FRET state may correspond to a CD4-bound conformation. Alternatively, it may represent a previously uncharacterized structural state that is a necessary intermediate during the activation of Env by CD4.
A model of binding through capture of preexisting conformations implies that intrinsic access to activated states is required for Env function. We therefore asked if broadly neutralizing antibodies that can neutralize up to 90% of all HIV-1 isolates (33–37), some of which bind to the ground-state Env conformation (13, 37, 38), remodel the conformational landscape by stabilizing HIV-1 Env in the ground state. Surface-bound viruses from both strains were incubated with neutralizing concentrations of antibodies VRC01, PG16, PGT128, PGT145, or 2G12 (figs. S3 and S13), which recognize different HIV-1 Env epitopes (39). smFRET measurements indicated that all broadly neutralizing antibodies tested exhibited a tendency to stabilize the low-FRET ground-state conformation of both HIV-1NL4-3 and HIV-1JR-FL Env (Fig. 4). Each antibody, however, differentially affected the intermediate- and high-FRET configurations (fig. S14 and table S2). The potent small-molecule inhibitor of HIV-1 entry, BMS-626529 (40), also stabilized the low-FRET ground-state conformation for both HIV-1JR-FL and HIV-1NL4-3 to an extent comparable to the broadly neutralizing antibodies tested (Fig. 4, F and K). In contrast, the small-molecule CD4-mimetic, JRC-II-191, stabilized activated FRET states of HIV-1NL4-3 Env (fig. S11). These findings suggest that stabilization of the ground-state Env conformation is both a general and effective means of inhibiting HIV-1 entry.
Fig. 4. Ground-state stabilization by broady neutralizing antibodies and an inhibitor.
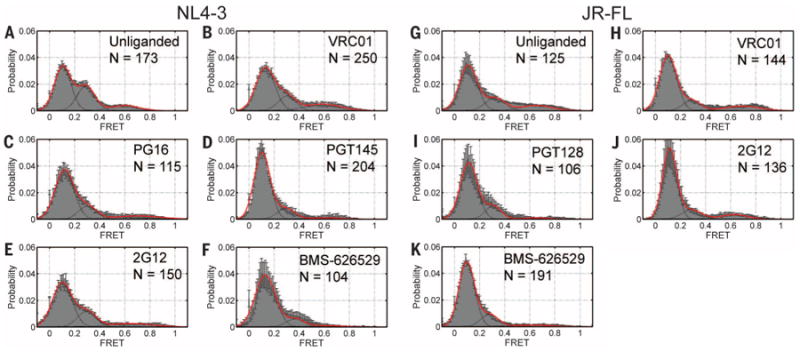
Population FRET histograms for (A) unliganded HIV-1NL4-3 Env and in the presence of broadly neutralizing antibodies (B) VRC01, (C) PG16, (D) PGT145, (E) 2G12 (all at 0.7 μM), and (F) the small molecule BMS-626529 (100 μM).The same data for (G) unliganded HIV-1JR-FL Env and in the presence of (H) VRC01, (I) PGT128, (J) 2G12, and (K) BMS-626529. Histograms are displayed as in Fig. 2.
The data presented here demonstrate that smFRET imaging can be applied to biomolecular systems as complex as HIV-1 Env on the surface of intact virions. These findings serve as an important step toward achieving smFRET imaging in living cells. HIV-1 Env from both laboratory-adapted and clinically isolated strains can spontaneously transition from a stable ground-state configuration to transient, CD4- and co-receptor–stabilized states. Thus, gp120 exhibits physical properties that typify dynamic enzymes, whose functions are regulated through the modulation of intrinsically accessible conformations (25,41). Moreover, the smFRET methodologies developed here allow for the precise delineation of conformations induced by antibodies and ligands in functional virions (42). Our analysis also directly reveals molecular and temporal events in gp120 that underlie the two-step activation of HIV-1 Env by CD4 and co-receptor. The reduced dynamics of HIV-1JR-FL Env out of the ground state, as well as the resistance to stabilization in a single conformation, reveals the mechanism of the masking of functional centers by neutralization-resistant HIV-1 isolates. Furthermore, this work suggests that stabilizing the ground state represents an effective strategy to antagonize viral fusion machines.
Supplementary Material
Acknowledgments
We thank J. Jin, L. Agosto, T. Wang, R. B. Altman, and M. R. Wasserman for assistance; C. Walsh, J. Binely, A. Trkola, and M. Krystal for reagents; and members of the Structural Biology Section, Vaccine Research Center, for critically reading the manuscript. We thank I. Wilson and J. Sodroski for encouraging us to extend our approach to a R5-tropic Env. This work was supported by NIH grants R21AI100696 to W.M. and S.C.B; P0156550 to W.M., S.C.B., and A.B.S.; and R01GM098859 to S.C.B.; by the Irvington Fellows Program of the Cancer Research Institute to J.B.M; by a fellowship from the China Scholarship Council-Yale World Scholars to X.M.; by grants from the International AIDS Vaccine Initiative's (IAVI) Neutralizing Antibody Consortium to D.R.B., W.C.K., and P.D.K.; and by funding from the NIH Intramural Research Program (Vaccine Research Center) to P.D.K IAVI's work is made possible by generous support from many donors including: the Bill & Melinda Gates Foundation and the U.S. Agency for International Development (USAID). This study is made possible by the generous support of the American people through USAID. The contents are the responsibility of the authors and do not necessarily reflect the views of USAID or the U.S. government. Reagents from the NIH are subject to nonrestrictive material transfer agreements. Patent applications pertaining to this work are the U.S. and World Application US2009/006049 and WO/2010/053583, Synthesis of JRC-II-191 (A.B.S., J.R.C.), U.S. Patent Application 13/202,351, Methods and Compositions for Altering Photopysical Properties of Fluorophores via Proximal Quenching (S.C.B., Z.Z.); U.S. Patent Application 14/373,402 Dye Compositions, Methods of Preparation, Conjugates Thereof, and Methods of Use (S.C.B., Z.Z.); and International and US Patent Application PCT/US13/42249 Reagents and Methods for Identifying Anti-HIV Compounds (S.C.B., J.B.M., W.M.). S.C.B. is a co-founder of Lumidyne Corporation.
Footnotes
Supplementary Materials: www.sciencemag.org/content/346/6210/759/suppl/DC1
Materials and Methods
References (46–56)
The data presented in this paper are tabulated in the main paper and in the supplementary materials.
References
- 1.Harrison SC. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyatt R, Sodroski J. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 3.Pantophlet R, Burton DR. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 4.Wei X, et al. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 5.Wyatt R, et al. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 6.Julien JP, et al. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyumkis D, et al. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White TA, et al. PLOS Pathog. 2010;6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu G, Liu J, Taylor KA, Roux KH. J Virol. 2011;85:2741–2750. doi: 10.1128/JVI.01612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwong PD, et al. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pancera M, et al. Proc Natl Acad Sci U S A. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran EE, et al. PLOS Pathog. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao Y, et al. Nat Struct Mol Biol. 2012;19:893–899. doi: 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris A, et al. Proc Natl Acad Sci U S A. 2011;108:11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moscoso CG, et al. Proc Natl Acad Sci U S A. 2011;108:6091–6096. doi: 10.1073/pnas.1016113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu SR, et al. Proc Natl Acad Sci U S A. 2010;107:18844–18849. doi: 10.1073/pnas.1007227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy R, Hohng S, Ha T. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Z, et al. ACS Chem Biol. 2007;2:337–346. doi: 10.1021/cb700054k. [DOI] [PubMed] [Google Scholar]
- 20.Lin CW, Ting AY. J Am Chem Soc. 2006;128:4542–4543. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Materials and Methods are available as supplementary materials on Science Online.
- 22.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 23.Zheng Q, et al. Chem Soc Rev. 2014;43:1044–1056. doi: 10.1039/c3cs60237k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee NG, Lyon LA, Le Doux JM. Nanotechnology. 2009;20:065103. doi: 10.1088/0957-4484/20/6/065103. [DOI] [PubMed] [Google Scholar]
- 25.Henzler-Wildman K, Kern D. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 26.Kwong PD, et al. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 27.Olshevsky U, et al. J Virol. 1990;64:5701–5707. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin F. Biophys J. 2004;86:1488–1501. doi: 10.1016/S0006-3495(04)74217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haim H, et al. PLOS Pathog. 2009;5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan N, et al. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arthos J, et al. J Biol Chem. 2002;277:11456–11464. doi: 10.1074/jbc.M111191200. [DOI] [PubMed] [Google Scholar]
- 32.DeVico AL. Curr HIV Res. 2007;5:561–571. doi: 10.2174/157016207782418560. [DOI] [PubMed] [Google Scholar]
- 33.Zhou T, et al. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker LM, et al. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker LM, et al. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pancera M, et al. Nat Struct Mol Biol. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Julien JP, et al. Proc Natl Acad Sci U S A. 2013;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Julien JP, et al. PLOS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwong PD, Mascola JR, Nabel GJ. Nat Rev Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- 40.Li Z, et al. Antimicrob Agents Chemother. 2013;57:4172–4180. doi: 10.1128/AAC.00513-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subsequent to the submission of this manuscript, the structure of a trimeric prefusion HIV-1 Env (43) was determined in complex with broadly neutralizing antibodies PGT122 (35) and 35O22 (44). To determine the conformational state these antibodies captured in the crystal lattice, we measured smFRET on labeled JR-FL virions in the presence of either PGT122 or 35O22 or both. These data indicated that PGT122 strongly stabilized the ground state. In contrast, 35O22 had little effect on Env conformation. HIV-1 Env complexed with both antibodies exhibited slight ground state stabilization.
- 43.Pancera M, et al. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, et al. Nature. 2014 doi: 10.1038/nature13601. [DOI] [Google Scholar]
- 45.McLellan JS, et al. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


