Abstract
Identification of the genomic regions most intimately associated with non-Hodgkin's lymphoma (NHL) pathogenesis is confounded by the genetic heterogeneity of human populations. We hypothesize that the restricted genetic variation of purebred dogs, combined with the contrasting architecture of the human and canine karyotypes, will increase the penetrance of fundamental NHL-associated chromosomal aberrations in both species. We surveyed non-random aneuploidy in 150 canine NHL cases, revealing limited genomic instability compared to their human counterparts and no evidence for CDKN2A/B deletion in canine B-cell NHL. ‘Genomic recoding’ of canine NHL data into a ‘virtual human’ chromosome format showed remarkably few regions of copy number aberration (CNA) shared between both species; restricted to regions of dog chromosomes 13 and 31, and human chromosomes 8 and 21. Our data suggest that gene discovery in NHL may be enhanced through comparative studies exploiting the less complex association between CNAs and tumor pathogenesis in canine patients.
Keywords: comparative genomic hybridization (CGH), canine, lymphoma, chromosome, microarray
Introduction
Non-Hodgkin's lymphoma (NHL) is among the most clinically and histopathologically diverse of all human cancers, characterized by an extensive range of morphologic and biologic subtypes that present challenges to diagnosis, clinical management and prediction of outcome. The identification of non-random DNA copy number aberrations (CNAs) in a wide range of human cancers has led to expanding interest in their utility as molecular markers for more comprehensive subclassification of distinct tumor subtypes and for development of therapeutic protocols targeted to specific mechanistic pathways (for example [1,2]). The identification of robust and clinically informative molecular indicators is, however, confounded by the extensive heterogeneity of human populations, within which the highly diverse and complex disease phenotypes observed are likely to have developed from a combination of multiple genetic risk factors, each with relatively weak penetrance. This is supported by numerous reports demonstrating that the extensive phenotypic variation in NHL is borne out by a high level of molecular heterogeneity (reviewed in [3]). The CNAs and associated genes that are most fundamentally associated with the disease pathogenesis have therefore remained largely elusive.
This challenge may be alleviated in part through parallel investigations of clinically and histologically comparable tumors in more genetically homogenous populations. The purebred domestic dog (Canis familiaris, CFA) represents a powerful spontaneous model for investigation of heritable risk and genetic factors that are clinically predictive in NHL, which accounts for up to 24% of all canine malignancies (for example [4]). As with its counterpart in human patients (hNHL), canine NHL (cNHL) displays extensive clinical and histological diversity and a growing need for more robust and informative subclassification. In the domestic dog, however, the likely confounding effects of genomic heterogeneity are limited as a consequence of the strategic development of distinct and genetically isolated populations in the form of >350 discrete and intensively regulated breeds, each exhibiting restricted genetic and phenotypic diversity (reviewed in [5] and others). Thus, detection of the genetic factors that are most intimately associated with tumor predisposition and pathogenesis may be more penetrant and in turn, more evident, in these relatively homogenous purebred canine populations as compared to human patients with the equivalent disease.
The relative incidence of cNHL is approximately five-fold greater than that of hNHL [4,6]. This is due in part to the significantly increased prevalence of naturally-occurring lymphoma in many of the most popular family-owned dog breeds, whose distinct temporal, geographical and phylogenetic origins are well described [7,8]. Furthermore, there is striking variation in the relative prevalence of B- and T-cell-derived lymphoproliferative disease in different breeds [9]. For example, the incidence of B-cell lymphoma (BCL) versus T-cell lymphoma (TCL) in the Labrador Retriever is similar to that observed when considering all dog breeds as one common population (65% BCL:35% TCL). The Golden Retriever, however, shows approximately equal incidence of both BCL and TCL (54% BCL: 46% TCL), while in the Boxer the majority of NHL cases comprise TCL (estimated to be as high as 65%) [9-11]. This breed-related susceptibility, combined with evidence for familial clusters of cNHL in purebred dogs [12,13] is strongly suggestive of an intimate link between NHL pathogenesis and genetic factors [4,11]. The unique characteristics of cNHL populations thus offer alternative means to identify the subset of shared genomic aberrations that are likely to be most intrinsic to tumor pathogenesis, using the dog to ‘filter out the background noise’ that continues to confound hNHL studies. Furthermore while the human genome is organized into 23 pairs of metacentric, submetacentric and acrocentric chromosomes ranging from 247Mb to 47Mb in size, the 39 chromosome pairs comprising the domestic dog karyotype are almost exclusively acrocentric/telocentric, ranging in size from just 125Mb to 26Mb [14]. The vastly contrasting architecture of the human and domestic dog karyotypes allows the two genomes to be evaluated in the context of the 200+ evolutionarily conserved chromosome segments from which they are both configured [15]. We propose that this will aid identification of those cytogenetic aberrations that represent intrinsic features of the disease, rather than a consequence of species-specific structural genomic organization.
We present the first genome-wide survey of tumor-associated CNAs in cNHL through array-based comparative genomic hybridization (aCGH) analysis of 150 naturally occurring cases in purebred dogs. The study population targeted three of the six most popular dog breeds within the US [16], the Golden Retriever, Labrador Retriever and Boxer, due to their elevated predisposition to NHL and contrasting incidence of BCL and TCL subtypes [7]. Canine aCGH data were ‘recoded’ into human genome format, enabling direct comparison with existing data from human patients. This comparison provided evidence for a marked reduction in genomic instability in canine NHL as compared to the human counterpart, and we discuss these findings in context with the structural configuration of their genomes. These data highlight the limited subset of CNAs that are evolutionarily conserved between human and canine NHL, which may harbor disease-associated genes and hereditary risk factors that have remained beyond the limits of detection when evaluated solely within human populations.
Materials and Methods
Histopathology Evaluation of the Canine Lymphoma Cohort
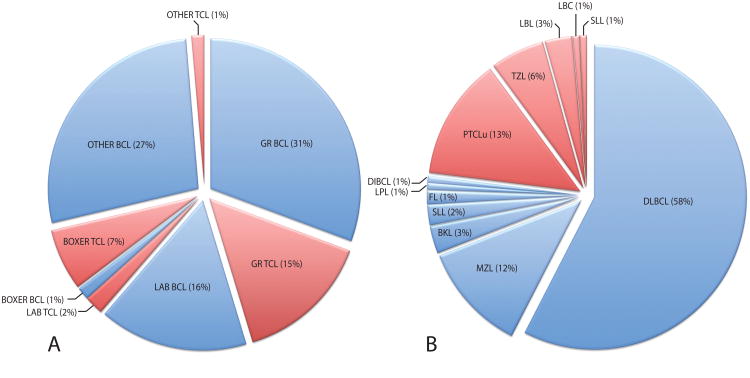
Clinical specimens were recruited between 2003 and 2009 from community or institutional veterinary practices, from client-owned dogs with clinical signs consistent with lymphoma. Specimens were acquired prior to initiation of therapy and under approved protocols with informed client consent. Representative lymph node biopsies were formalin-fixed and paraffin-embedded for histological evaluation. A diagnosis of lymphoma was confirmed by examination of hematoxylin and eosin (H&E)-stained sections in conjunction with conventional immunohistochemical techniques using CD3 and CD79a markers. Standard morphological features were used to define the histological subtype, grade and mitotic index of the tumor based on the World Health Organization (WHO) classification system for human lymphoid neoplasia [17], which has been shown to be translatable to the canine counterpart with a high degree of accuracy and reproducibility [18]. Histological specimens were reviewed independently by two or more board-certified veterinary pathologists, one of whom (VEV) was common to all cases, to maximize consistency. Patient details and histological classifications of all 150 cases are provided as supplementary online materials (SOM table I) and are summarized in figure 1. Overall 71.3% of the 150 cases evaluated were derived from the three target breeds, the Golden Retriever (GR, 68 cases), Labrador Retriever (LR, 27 cases) and Boxer (12 cases) (figure 1a). The remaining 43 (28.7%) cases were represented by 18 ‘other’ breeds, with between one and five individuals per breed. The cohort comprised 113 BCL tumors (75.3% of cases) and 37 TCL tumors (24.7% of cases). The distribution of cases by histological subtype is shown in figure 1b (insufficient tissue availability precluded definitive subtype classification of 11 cases). Overall 67.7% were high-grade tumors, 26.6% were low-grade tumors and the remaining cases were of intermediate grade. The average age at diagnosis across all cases was 7.5 years (median = 7 years, range 2 – 14 years, nine cases were of unspecified age).
Figure 1.
Distribution of cNHL cases by a) breed and tumor phenotype, and b) by histological subtype. Data for cBCL cases are denoted by blue shading, and for cTCL cases by red shading. A Fisher's exact test identified a highly significant association (p < 0.001) between breed and tumor phenotype. When all cases were stratified according to immunophenotype, the frequency of different histological subtypes showed no significant association with breed (p < 1.00 for BCL, p < 0.768 for TCL).
Microarray-Based Detection of Tumor-Associated CNAs
DNA was isolated from representative tumor tissue using a Qiagen DNeasy Kit (Qiagen, Valencia, CA). aCGH analysis was performed using a custom microarray comprising cytogenetically-mapped bacterial artificial chromosome (BAC) clones from the CHORI-82 dog library (http://bacpac.chori.org, BACPAC Resources, Children's Hospital Oakland Research Institute, Oakland, CA). The array comprises clones distributed at approximately 1Mb intervals throughout each dog autosome and the X chromosome (mean interval 1.10Mb, range 0.28Mb to 3.28Mb), in addition to clones representing canine orthologues of 53 human genes associated with a range of cancers [19]. Locations of BAC clones are denoted herein according to their cytogenetic location and then their Mb position on that chromosome, based on the canFam v2.0 (May 2005, [20]) dog genome sequence assembly (for example, CFA 1q11; 3.2Mb). Canine aCGH analysis was performed as described previously [19]. Microarrays were scanned at 5μm resolution using an Agilent G2565CA scanner and image data were processed using Feature Extraction version 10.7 (Agilent Technologies, Santa Clara, CA). Signal intensity data were block normalized, and arrayed loci with confidence estimates < 0.5 and/or replicates with standard deviation > 0.3 were excluded from further analysis. Data visualization and statistical analysis were performed using Genomic Workbench version 6.0 (Agilent Technologies) using the ADM1 calling algorithm with a threshold of four and a fuzzy zero correction. Parameters for genomic copy number gains and losses were set at log2 tumor DNA signal intensity:reference DNA signal intensity of +/− 0.2. Recurrent CNAs were defined as those identified in ≥20% of cases analyzed. Correlation between individual CNAs and signalment/diagnostic parameters was explored by converting log2 ratio data to ordinal values (gain/normal/loss) and performing Mantel-Haenszel Chi square tests to identify significant associations (p < 0.05). A Bonferroni correction was applied by deriving a p-value threshold for significance based on the number of arrayed genomic loci assessed in these comparisons, in order to correct for multiple testing and to control family-wise error rates, resulting in a cut-off for statistical significance at p < 2.181 × 10−5. Predictive modeling was performed for statistically significant regions using logistic regression and multiple regression analysis. Statistical analysis was performed with Stata v.11 (www.stata.com).
‘Genomic Recoding’ of cNHL Data and Comparison with hNHL data
Canine aCGH data were recoded into ‘virtual’ human genome format in order to facilitate direct visual comparison of cytogenetic profiles of human and canine NHL. Briefly, the positional co-ordinates of each arrayed canine clone were defined according to the nucleotide location of paired BAC end sequence data within the canine genome sequence assembly (http://genome.ucsc.edu, canFam v2.0, May 2005, [20]). These data were imported into the Liftover Batch Coordinate Conversion Tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver), using default settings to establish the orthologous nucleotide sequence coordinates for arrayed canine BAC clones within the human genome sequence assembly (February 2009, GRCh37/hg19). Tumor:reference signal intensity data at each arrayed locus for each cNHL case were then replotted according to these ‘virtual’ human chromosome locations.
Raw datasets were retrieved for prior BAC-array CGH studies of hNHL [21-23] (Masao Seto, Masao Nakagawa and Ichiro Takeuchi, pers. comm.), which utilized a 1.3Mb-resolution BAC-array platform comparable to that used in the present study for cNHL. These studies reported cytogenetic profiling data for 46 human nodal diffuse large B-cell lymphoma (DLBCL) cases [21,22] and 29 human nodal peripheral T-cell lymphomas (unspecified, PTCLu) cases [23], respectively and are consistent with additional studies using similar analysis platforms and techniques [21,24,25]. Raw log2 test:reference signal intensity data [21-23] were processed using the same parameters used for canine cNHL data. Human and canine data were then plotted on a common axis according to the location of arrayed BAC sequences within the GRCh37/hg19 human genome sequence assembly, enabling direct visual comparison of aCGH data for the same tumor type in both species.
Extended aCGH Analysis of Selected cNHL Cases at Higher Resolution
Five cNHL cases from the cohort were analyzed further using a ∼180,000-feature canine oligonucleotide CGH microarray (Agilent Technologies), comprising repeat-masked ∼60-mer oligonucleotides distributed at approximately 13kb intervals throughout the dog genome sequence assembly. Tumor and reference DNA samples were labeled independently with Cyanine3-dUTP and Cyanine5-dUTP, respectively, using the Genomic DNA Enzymatic Labeling Kit (Agilent Technologies). Oligonucleotide-array CGH (oaCGH) analysis was performed following the manufacturer's recommendations. Arrays were scanned at 3μm resolution using an Agilent G2565CA scanner and image data were processed using Feature Extraction version 10.7 and Genomic Workbench version 6.0 (Agilent Technologies). Data were filtered to exclude probes exhibiting non-uniform hybridization or signal saturation and were normalized using the centralization algorithm with a threshold of six. The ADM2 algorithm was used to define CNAs using a ‘three probes minimum’ filter and a threshold of six with a fuzzy zero correction.
Results
Genome-Wide Overview of Recurrent CNAs in cNHL
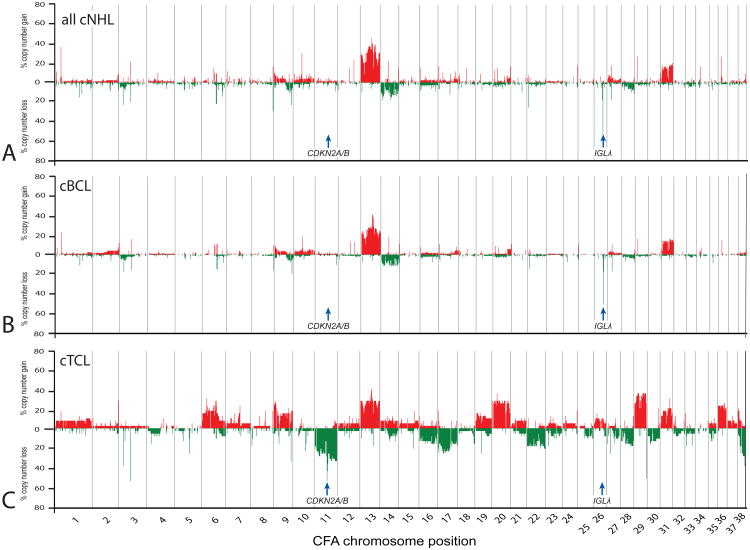
Data analysis proceeded through successively focused evaluation, commencing with a genome-wide overview of CNAs within all cNHL cases studied, followed by locus-specific comparisons at the level of tumor phenotype and histological subtype, and finally by breed. The global level of genomic instability in cNHL was markedly less extensive than that observed in similar studies of canine osteosarcomas (Angstadt et al., submitted) and histiocytic tumors (Hedan et al, manuscript in preparation), and was more comparable to that of canine intracranial tumors [26], with a limited proportion of the genome involved in highly recurrent copy number imbalance, few apparent homozygous deletions and no high level amplifications. Figure 2a presents a genome-wide penetrance plot summarizing the frequency and distribution of DNA copy aberrations at ∼1Mb intervals within the cohort of 150 cNHL cases. These data show that aneuploidy in cNHL was restricted predominantly to copy number increase along the length of CFA 13 and 31. No chromosomes showed a combination of both recurrent gains and losses among the cohort, indicating that individual CNAs were primarily unidirectional. Data for all 150 tumors were stratified into cBCL cases (n = 113) and cTCL cases (n = 37) to summarize the relationship between immunophenotype and genome-wide DNA copy number imbalance. Figure 2b shows that recurrent CNAs in cBCL consisted primarily of gains along CFA 13 and 31 and partial loss of CFA 26, with the remainder of the genome showing minimal evidence for imbalance. cTCL showed a higher frequency and wider distribution of recurrent CNAs (figure 2c), including gains of CFA 6, 9, 13, 20, 29, 31 and 36, and losses of CFA 11, 17, 22, 28 and 38, the majority of which extended along most of the length of the chromosome. These data indicate that while gains of CFA 13 and 31 are shared features of both phenotypes, the majority of the genomic imbalance observed within the cNHL cohort (figure 2a) is driven by cTCL. Regions of recurrent CNA in cBCL and cTCL are detailed in SOM table II and example data from BAC-array CGH analysis are provided in SOM figure 1.
Figure 2.
Summary of recurrent genomic copy number imbalances in a) all 150 cNHL cases, b) the 113 cBCL cases and c) the 37 cTCL cases. Each arrayed BAC clone is plotted against its chromosomal location along the x-axis. The y-axis indicates the percentage of the corresponding subset of the cNHL cohort that show demonstrated either copy number gain (shown in red above the midline) or copy number loss (shown in green below the midline) at each arrayed locus. Genes of specific interest are annotated and are discussed in the text. These data demonstrate the elevated degree of genomic instability associated with cTCL as compared to cBCL, and reveals two immunophenotype-associated chromosome copy number aberrations, located on CFA 11 and CFA 26.
Identification of CNAs Associated with Tumor Immunophenotype
Bonferroni-corrected chi-square analysis of aCGH data from of the entire cohort of 150 cNHL cases identified 134 arrayed BAC clones representing genomic regions that showed a significant difference in DNA copy number status in cBCL compared to cTCL (SOM table III). These loci were distributed across 23 different chromosomes, of which 11 were represented by a single BAC clone, consistent with a CNA ∼2Mb in size. The most extensive region of significant association encompassed 45 clones distributed along the length of CFA 11, extending from ∼9.5Mb below the centromere down to the most telomeric clone at 77.0Mb. Deletion of this ∼67.5Mb region was associated significantly with cTCL and peaked at the CDKN2A/B tumor suppressor gene locus at CFA 11q16; 44.3Mb (p < 2.90 × 10−15). Copy number loss of CDKN2A/B occurred in 20/36 cTCL cases (55.6%), and was more frequent in high-grade cTCL (10/15 cases) versus low-grade cTCL (2/6 cases), but was not evident in any of the 111 cBCL cases scored at this locus. Also associated significantly with cTCL was copy number gain of a 39.2Mb region spanning the length of CFA 29 (3.2 - 42.4Mb), as well as losses along CFA 17 and 38, and gains along CFA 20 and 36 (SOM table III).
The reduced level of aneuploidy in cBCL relative to cTCL (figure 2) was supported by the identification of few regions of association between tumor phenotype and DNA copy number status within cBCL. In contrast to the recurrent losses along the length of CFA 11 in cTCL, the CNA associated most significantly with cBCL was highly recurrent deletion of a discrete ∼2Mb region on CFA 26q24:28.6–30.4Mb (peaking at 30.4Mb, deleted in 69.8% of cBCL cases scored [74/106 cases], p < 2.59 × 10−14). This deletion encompasses the canine immunoglobulin lambda locus (IGLλ) and flanked both proximally and distally by normal copy number. Deletion of this region was not evident in any cTCL cases.
Since deletions of CFA 11q16; 44.3Mb and CFA 26q24; 30.4Mb were restricted to cTCL and cBCL, respectively, in the present study, we assessed their potential value as molecular indicators of tumor immunophenotype in cNHL. Predictive modeling by regression analysis showed that when assessed in combination, determination of the copy number status of these two regions alone predicts the cBCL phenotype with 93.8% accuracy, and predicts the cTCL phenotype with 59.5% accuracy, with an overall accuracy of 85.3%. Full details of this analysis are provided in SOM Appendix 1.
Evidence for CNAs Associated with Histological Subtype
Based on the extensive variation observed between the genomic profiles of cBCL and cTCL, all subsequent comparisons were stratified by tumor phenotype. Within each tumor phenotype we assessed whether genomic copy number aberrations were significantly associated with histological subtype. The two most highly represented cBCL subtypes, DLBCL (80 cases) and marginal zone lymphoma (MZL, 16 cases), showed negligible variation in DNA copy number status, with no significant differences in their genomic profiles. cTCL subtypes also showed highly conserved profiles; however four loci (two on CFA 29 and one each on CFA 7 and X) showed a significantly elevated incidence of copy number gain in PTCLu (n = 18 cases) as compared to T-zone lymphoma (TZL, n = 8 cases). These regions are defined in SOM table IV. There was no evidence for significant differences in CNA status based on tumor grade.
Evidence for CNAs Associated with Breed
We investigated evidence for genomic regions whose copy number status was associated significantly with the breed of the patient, which might be indicative of heritable risk of disease within a consistent genetic background. This analysis was restricted to the three target breeds and was stratified by phenotype. No significant correlations were found within cBCL, and the disproportionate distribution of high p-values suggested that cBCL in different breeds show highly conserved genomic copy number status (SOM table V). In contrast, cTCL showed a skew towards low p-values, suggesting that the cytogenetic profiles of tumors of this phenotype may be more strongly influenced by the genetic background of the patient as compared to cBCL. Seven individual loci (on CFA 6, 12, 20 and 31) showed highly significant association with breed, each demonstrating an elevated incidence of copy number gain in Boxers with cTCL (SOM table V).
Comparative analysis of cNHL and hNHL by genomic recoding
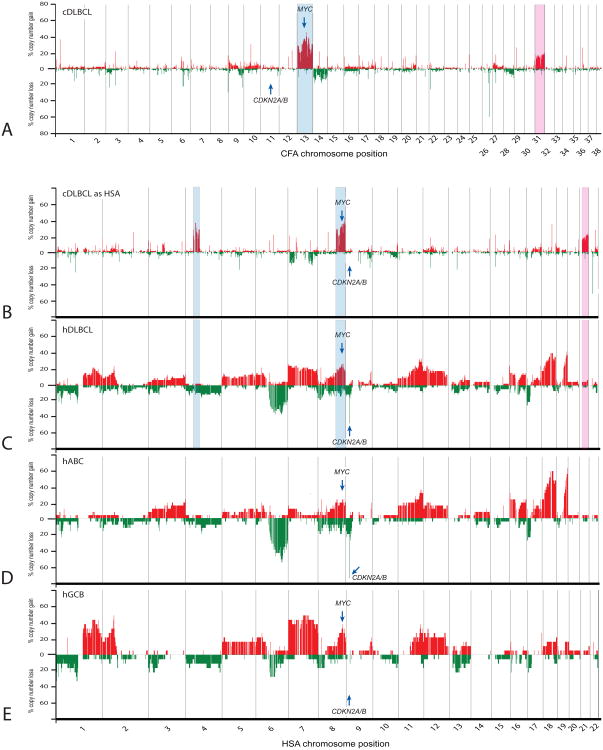
Of the 2143 arrayed canine BAC clones evaluated using the Liftover Batch Coordinate Conversion Tool, 2083 (97.2%) were converted successfully to a unique orthologous position in the human genome sequence assembly. Canine sequence data for 42 clones intersected with multiple regions of human sequence and for the remaining 18 clones no orthologous human chromosome sequence could be identified. These 60 clones were excluded from subsequent genomic recoding and comparative evaluation. With DLBCL being the most highly represented and investigated form of NHL in both species, and representing more than half of the cohort in the present study, we focused on this subtype as the basis for extended assessment of evolutionarily conserved CNAs. Figure 3a presents, in conventional canine genome format, a genome-wide penetrance plot of BAC CGH data for the 80 cDLBCL cases from the present study, demonstrating the limited aneuploidy associated with the histological subtype in canine patients. The most extensive and highly recurrent CNAs, gains of CFA 13 and 31, are highlighted in blue and pink, respectively. Recurrent gain of the MYC oncogene and absence of deletion of the CDKN2A/B tumor suppressor gene locus are also indicated, determined by the copy number status of arrayed canine BAC clones encompassing these genes [19]. Figure 3b presents the same cDLBCL data from figure 3a, now recoded into ‘virtual human genome format’ to facilitate cross-species comparisons. The human genomic regions orthologous to CFA 13 and 31 are highlighted, demonstrating that if CNAs of these chromosomal regions were conserved in hDLBCL patients, they would manifest as gains of HSA 8qdist/4pprox-qprox (blue shading) and HSA 21 (pink shading), respectively. Figure 3c shows the genome-wide penetrance plot for 46 hDLBCL cases that were evaluated using a 1.3Mb-resolution human BAC aCGH platform [21,22], demonstrating a wide range of aneuploidy distributed across the human karyotype. Comparison of these data with figure 3b reveals that hDLBCL exhibits a substantially elevated incidence of detectable genomic imbalance compared to cDLBCL, and unlike its canine counterpart, presents with recurrent CDKN2A/B deletion. Comparison of the highlighted chromosome regions in figures 3b and 3c show that while copy number increase of HSA 4pprox-qprox is infrequent in hDLBCL there is evidence for shared gain of CFA13q/HSA8q (including MYC) in both species and, to a lesser degree, of CFA31q/HSA21q.
Figure 3.
Comparison of recurrent genomic copy number imbalances in canine and human BCL. a) summarizes the genome-wide incidence of copy number gain and loss in the 80 cDLBCL cases from the present study, according to their dog chromosomal location. Gains of CFA 13 and CFA 31 are highlighted in blue and pink, respectively. In b) the same canine data are replotted according to the equivalent location of arrayed loci on the corresponding human chromosome. When recoded into orthologous human genome co-ordinates, gain of CFA 13 now manifests as partial gains on HSA 4 and 8 (highlighted in blue) and gain of CFA 31 manifests as gain of HSA 21 (highlighted in pink). Figure c) shows data in a comparable format derived from a prior study of 46 hDLBCL cases [22]. In d) and e) these 46 hDLBCL cases are segregated to compare the cytogenetic profiles of the 28 hABC-DLBCL cases and the 18 hGCB-DLBCL cases, respectively. These images demonstrate the apparently reduced degree of genomic imbalance detected in cBCL compared to its human counterpart, and reveals evidence for shared aberrations (including gain of MYC) as well as aberrations that are not shared (such as deletion of CDKN2A/B).
Takeuchi et al [22] classified all 46 hDLBCL cases evaluated by aCGH into one of two molecularly distinct variants, the activated B cell-like [ABC] form (28 cases) and the germinal center B cell-like [GCB] form (18 cases). Figures 3d and 3e, respectively present data from these 46 human cases now subdivided according to these variants. To our knowledge there is no firm evidence thus far to indicate whether these variants exist in cDLBCL. We therefore investigated whether cDLBCL data exhibited shared CNAs consistent with these tumor subtypes. Based on the lack of CDKN2A/B deletion in cDLBCL (equivalent to loss of HSA 9p21, a characteristic of hABC-DLBCL), and the conservation in cDLBCL of key CNAs associated with the GCB form in human patients (gain of MYC at CFA 13q13/HSA 8q24, and gain of CFA 31/HSA 21) [21,25], the present study provides preliminary support for an elevated representation of the GCB-form of cDLBCL.
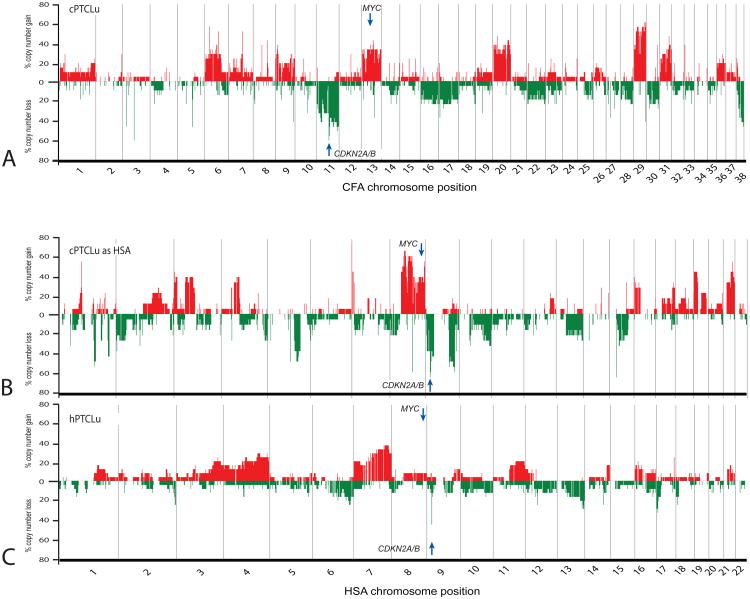
A similar assessment was performed for the most common cTCL subtype within the present study, PTCLu (48.6% of all cTCL cases evaluated). Figure 4 shows a genome-wide penetrance plot for 18 cPTCL cases in canine (figure 4a) and in recoded human genome format (figure 4b). These data recapitulate the elevated degree of aneuploidy associated with cTCL as compared to cBCL (compare figures 2b and 2c), and demonstrate recurrent gain of MYC and deletion of CDKN2A/B in cPTCLu. Figure 4c presents data for 29 hPTCLu cases from a prior BAC-aCGH study [23]. Compared to observations from DLBCL in figure 3, comparison of figures 4b and 4c show a more comparable degree of genome-wide aneuploidy in PTCLu in both species. Furthermore, unlike in DLBCL, PTCLu tumors exhibit conserved deletion of CFA 11/HSA 9p, peaking in incidence at the CDKN2A/B locus. In support of our hypothesis, the penetrance of CDKN2A/B deletion relative to the copy number status of its flanking regions, was markedly elevated in canine patients. Figure 4 indicates additional CNAs in hPTCLu that are conserved in the dog counterpart, including loss of HSA10q and 13q, and gain of HSA 1q, 3, 4, 8q (including MYC), 16p and 17q. In contrast, deletions of HSA 6q, 12q and 16q and gains of HSA 7 and 11 did not appear to be conserved in the canine counterpart.
Figure 4.
Comparison of recurrent genomic copy number imbalances in canine and human TCL. a) summarizes the genome-wide incidence of copy number gain and loss in the 18 cPTCLu cases from the present study, according to their dog chromosomal location. In b) the same data are recoded into human chromosome format. c) shows data in a comparable format derived from a prior study of 29 hPTCLu cases [23]. This comparison reveals that PTCLu is associated with extensive genomic instablility in both species, and identifies regions of evolutionarily shared imbalance, including deletion of CDKN2A/B.
Extended Analysis of cDLBCL by Oligonucleotide aCGH (oaCGH)
Five high-grade cDLBCL cases from the cNHL cohort were analyzed further by oaCGH analysis to assess evidence for the presence of additional CNAs that lay below the detection limits of the BAC array platform. These cases were selected as representative examples of the BAC aCGH profiles identified within this most highly abundant histological subtype evaluated in the study. The aCGH profiles from both array platforms for each case are compared in SOM figure 1, showing that the limited aneuploidy identified by BAC aCGH was recapitulated by the higher-resolution oaCGH platform, which also verified the balanced copy number status of CDKN2A/B in these cases (SOM figure 2). A total of 22 CNAs were shared by two or more of the five DLBCL cases evaluated by high-resolution oaCGH, of which two represented full length gains of CFA 13 and 31 (SOM table VI). The remaining 20 shared CNAs were distributed across 12 different chromosomes and were each < 8Mb in size, including eight regions that were < 0.5Mb and a further four that were < 65kb.
Discussion
Genomic Recoding Facilitates Direct Comparison of Cross-Species aCGH data
We proposed that genome-wide comparison of CNAs in canine and human NHL would aid definition of shared karyotypic features that are intimately involved in disease pathogenesis. Genomic recoding of canine aCGH data and direct comparison with existing human datasets (figure 3) supported our hypothesis that much of the complexity evident in cytogenetic profiling of hDLBCL may be associated with the greater degree of natural genomic heterogeneity inherent to human populations, masking the genetic factors that are most fundamentally associated with tumor pathogenesis. The similarity between the array platforms and analysis techniques used, coupled with shared histological classification criteria, suggest that, at the chromosomal level, the limited aneuploidy associated with cDLBCL is sufficient to result in a comparable phenotype to that of hDLBCL, which presents with considerably more extensive genomic instability. The conserved copy number increase of two orthologous subchromosomal regions in human and canine patients, despite their extensively divergent genome architecture, now highlights CFA 13q11-q21.1 (3.4-40.9Mb)/HSA 8q22-qtel (99.2-145.7Mb) and CFA 31q11-qtel (14.2-39.9Mb)/HSA 21q11-qtel (15.6-44.9Mb) with elevated evidence for harboring genes that are fundamentally involved in tumor pathogenesis, including the MYC oncogene. Further interrogation of these regions identifies additional genes that have been associated previously with human lymphoproliferative disease including OLIG2 (oligodendrocyte lineage transcription factor 2, HSA 21q22,1), ERG (v-ets erythroblastosis virus E26 oncogene like, avian, HSA 21q22.3), RUNX1 (runt-related transcription factor 1, HSA 21q22.3), CHIC2 (cysteine-rich hydrophobic domain 2, HSA 4q11-q12), KDR (vascular endothelial growth factor receptor 2, HSA 4q11-q12) and KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, HSA 4q12) [27-29] and others. Extended analysis using high-resolution oaCGH suggested that the limited complexity apparent in cDLBCL remained evident even at 50-fold higher resolution, but also identified several microdeletions/amplifications in cNHL that could not be detected using BAC aCGH analysis (SOM table VI), and which will be pursued in future studies. Despite its devastating impact, there are remarkably few comprehensive reports of high-resolution cytogenetic profiling in defined hNHL cohorts beyond the most common histological subtypes; thus it remains to be seen whether these ‘microaberrations’ are also evident in human patients, and what their biological relevance may be.
Gene expression profiling and immunohistochemical studies have shown previously that hDLBCL comprises at least two common and molecularly distinct variants (reviewed in [30]). While they share extensive overlap in histology, the GCB subtype is typically less aggressive and associated with a more favorable prognosis compared to ABC tumors; thus subclassification may be of clinically predictive value. Furthermore, previous studies have shown that ABC-DLBCL and GCB-DLBCL tumors in human patients exhibit variation in their genomic profiles, with a subset of CNAs (summarized in figures 3d and 3e) being strongly associated with one or other subtype [21,22,24,31-38]. It is not yet clear whether these distinct variants exist in cDLBCL. Although data from the present study provided preliminary support for CNAs consistent with the GCB form, the limited aneuploidy detected in cDLBCL precludes more detailed conclusions.
Comprehensive gene expression profiling and high-resolution aCGH analysis of a common cohort may be required in order to provide more robust and comprehensive evidence for molecular variants in cDLBCL. Figure 3 does, however, suggest that the extensive variation in the cytogenetic profiles of canine and human DLBCL cohorts is not attributable solely to differential representation of ABC and GCB variants. The potential value of discrimination between ABC-DLBCL and GCB-DLBCL in the purebred dog is supported further by knowledge that the frequency and distribution of these variants, and their associated chromosome rearrangements and gene dysregulation events, varies greatly between different human populations and ethnic backgrounds [39].
Immunoglobulin Gene Deletions in cBCL
Subchromosomal deletion of CFA 26 was a hallmark event in cBCL that does not appear to be conserved in hBCL. Although several cancer-associated loci (NF2, BCR, PTEN) are located within CFA26qdist, our prior studies using metaphase chromosome-based CGH [40] suggested that extensive imbalances (>5-10Mb in size) of CFA 26 were not highly recurrent in cNHL. The increased resolution capabilities of the aCGH analysis platforms used in the present study now define a highly recurrent deletion of <2Mb within CFA 26q24: 28.7-30.4Mb that was restricted to cBCL, encompassing the canine IGLλ gene. Deletions of immunoglobulin light chain loci are common events in hBCL, arising from clonal gene rearrangement during formation of a functional B-cell receptor in mature lymphocytes. Using BAC aCGH in hBCL Tagawa et al [21] identified a high frequency of localized deletion of the immunoglobulin light chain kappa locus (IGLk, HSA 2p11), while there was no evidence for recurrent deletion of IGLλ (HSA 22q11). This is contrary to the findings of the present study of cBCL, in which we observed a high incidence of deletion encompassing IGLλ (74/106 cases scored) and limited evidence for recurrent IGLk deletion (16/103 cases scored, CFA 17q16: 40.7Mb). CGH analysis revealed that the common deletion encompassing the canine IGLλ region lay close to the limit of detection of our ∼1Mb-resolution BAC-array platform; thus these contrasting findings may simply reflect the specific nature and genomic coverage of the arrayed targets in this and prior studies of hBCL by BAC-aCGH. The pattern of IGLλ versus IGLk deletion in cBCL was, however, supported by subsequent application of high-resolution oaCGH analysis to a subset of the cohort. While in humans the ratio of light chain usage in immunoglobulin formation is relatively balanced (60% IGLk, 40% IGLλ) [41], prior studies in the domestic dog have attributed approximately 90% of light chain usage to IGLλ [42]. Our aCGH data support these findings, suggesting that clonal rearrangement of immunoglobulin light chains is more likely to manifest as recurrent deletion of IGLk in hNHL and of IGLλ in cNHL. The region corresponding to the immunoglobulin heavy chain (IGH) locus (CFA 8q33.3; 76Mb) is minimally annotated in the canFam2 genome assembly, with an apparent gap in coverage spanning 0.3Mb that encompasses the predicted location of the dog IGH locus. The limited annotation of this chromosome region and the absence of BAC clones encompassing the dog IGH locus precludes complete analysis of the structural and numerical status of this region at the present time.
Unlike hDLBCL, most other forms of hBCL have received little attention at the genomic level, in part due to the relative paucity of these subtypes in human populations. Nodal marginal zone lymphoma is infrequent in human patients but is the most common indolent BCL subtype in canine populations [18,43]. Conversely classical Burkitt's like lymphoma, while relatively common in people, is considered not to exist in the dog, and both the variant Burkitt's-like lymphoma and follicular lymphoma also are rare in canine populations [18]. Mantle cell lymphoma is infrequent in both species. A comparative approach would provide an opportunity to combine these limited canine and human clinical resources into more extensive and synergistic datasets, increasing the power to define clinically informative molecular indicators for less common BCL subtypes.
Comparative Analysis of Canine and Human TCL
Similar opportunities are presented by virtue of the elevated incidence of TCL in canine versus human populations, and their contrasting relative incidence of specific TCL subtypes [4]. Data from the present study revealed more complex and extensive genomic instability in cTCL as compared to cBCL, consistent with their human counterparts (for example, [44]). Cytogenetic investigations of hTCL have focused largely on extranodal forms due to their relative abundance over nodal hTCL [6,45]. Histological classification of nodal hTCL is complex and somewhat controversial, and there is limited knowledge of genomic characteristics that are highly associated with individual subtypes. Anaplastic large cell lymphoma (ALCL) is among the more common hTCL subtypes (∼15% of all hTCL cases [45]) and has received moderate cytogenetic characterization, but is uncommon in the dog (<2% of cTCL, [18]). Conversely T-zone lymphoma (TZL) is common in the dog (representing 37% of all cTCL [18] and 22% of cTCL cases in the present study) but is so infrequent in human populations [4,45] that virtually no information exists regarding their genomic characteristics. Moreover the TZL subtype was eliminated from the most recent WHO classification of human lymphoproliferative disease due to extensive biologic and microscopic overlap with other indolent forms of hTCL [46], confounding direct correlation between human and canine tumors. The most promising candidates for comparative studies are the ‘peripheral T-cell lymphomas- unspecified’ (PTCLu), which represent ∼ 60-70% of hTCL cases in Western countries [47] and ∼ 40-50% of cTCL [18]. In both species PTCLu comprises a histologically, clinically and molecularly heterogeneous group of tumors whose characteristics do not fit within any defined entity in the WHO Classification system. This heterogeneity is recapitulated in the typically chaotic and variable cytogenetic profiles of hPTCLu [23,48-50], which was consistent with our observations for cPTCLu in the present study (figure 4). Unlike in DLBCL, canine and human PTCLu exhibit shared deletion of the CDKN2A/B tumor suppressor gene, a CNA that was more common in high-grade cTCL versus low-grade cTCL. This was consistent with our prior functional studies of high-grade cTCL [4,9,40,51], which identified a consequent increase in the proportion of tumor cells showing hyperphosphorylation of Rb, which is regulated by p16. Low-grade/indolent cNHL rarely showed inactivation of the p16/Rb pathway, although Rb phosphorylation was also observed infrequently in high-grade cBCL. We suggested that this may be a consequence of upregulation of MYC in cBCL through gain of CFA 13 or IGH/MYC fusion rather than disruption of the p16 pathway [4,51], which correlates with the absence of CDKN2A/B deletion in cBCL in the present study. Furthermore, the absence of detectable CDKN2A/B deletion in cBCL invites further examination of the functional role of this tumor suppressor gene in the pathogenesis of the clinical and histological counterpart in human patients.
Figure 4 suggests that there is greater conservation in the genomic profiles of canine and human TCL as compared to BCL, although this is likely to be at least in part a consequence of the elevated level of aneuploidy in cTCL. Interestingly the pattern of genomic gain and loss along the length of several chromosomes within the cohort of human patients was also remarkably similar in recoded canine PTCLu data, but the relative incidence of these imbalances was elevated in the canine cohort. For example, both species present with recurrent loss of HSA 8p compared to gain of HSA 8q, an elevated incidence of gain of HSA 9qdist compared to HSA 9qprox and a greater incidence of HSA 13q gains towards the telomeric region of this chromosome, but these aberrations are more frequent within the recoded data for the canine NHL population (figure 4b and c). This adds support to the hypothesis that tumor-associated CNAs may be more penetrant in canine populations.
A region of deletion of HSA 5q (peaking at 5q21) has been strongly associated with hTCL and the hPTCLu subtype in particular, with a possible correlation with longer survival in European populations [48]. There is evidence for conservation of this aberration in our cTCL cohort (figure 4b, consistent with deletion of CFA 11). Interestingly, however, figure 4c shows that this CNA was not evident in the study of hPTCLu in Japanese patients by Nakagawa et al. [23]. This may be attributable to the contrasting geographical and/racial distribution of the hPTCLu cohorts [23] or to differential exposure of these populations to pathogenic viruses (reviewed in [52]). This represents an example of the likely confounding effects of genetic heterogeneity in human populations that may be overcome by studies of purebred dogs for which genetically isolated populations are readily available for focused study.
Figure 4 shows that one of the most common CNAs in hPCTL, gain of HSA 7, was not conserved in cPTCLu. While gain of HSA 7q is frequently observed in hPTCL, this aberration is also common in a wide range of other human neoplasms, and there is evidence that it may represent a secondary abnormality acquired during tumor progression [49]. Similar circumstances may apply to gain of CFA 13, which was by far the most frequent aberration detected in the present study, resulting in copy number increase of the MYC oncogene (CFA 13q13) in 36.7% of the entire cNHL cohort (55/150 cases). This gene has received considerable attention in hBCL patients due to the high frequency of aberrant fusion events with immunoglobulin gene loci, resulting in transcriptional dysregulation and constitutive expression of MYC (reviewed in [37,53-55]). Conventional aCGH analysis is considered blind to chromosome aberrations that do not result in net DNA gain or loss, and therefore cannot detect balanced translocations. It is becoming increasingly evident, however, that many translocation events described in human cancers involve deletions and amplifications at sites of chromosome fission and fusion [56]. Moreover, we have shown previously that MYC-IGH fusion occurs in cNHL, and have detected constitutive MYC expression both in cDLBCL and cBKL [57]. aCGH data for ten cBCL cases overlapping with this prior study showed incomplete correlation between the DNA copy number status and transcriptional status of MYC (data not shown). This is consistent with findings in canine osteosarcoma [58] and lymphoid tumor cell lines [59], suggesting that multiple, complex mechanisms are involved in transcriptional control of this oncogene, and correlating with observations in human cBCL [60]. Interestingly, however, while MYC copy number increase is associated with BCL in human patients, in the present study there was no significant difference in the incidence of MYC gain when comparing cBCL (38.9% of cases) and cTCL (29.7% of cases) (p < 0.334). The profile of gain along the full length of CFA 13 was strikingly similar in both cNHL phenotypes (figure 2), peaking in incidence at 39.7Mb in cBCL and at 41.7Mb in cTCL (CFA 13q21.1), more than 10Mb distal to the MYC locus (CFA 13q13: 28.2Mb). These observations suggest that copy number gain of this oncogene is not the primary driving force for the recurrent gain in copy number of CFA 13 in cNHL, and may be a consequence rather than a causative factor. Moreover it is apparent from a range of parallel studies that gain of CFA 13 is common to a spectrum of other canine cancers, including osteosarcoma, glial tumors, histiocytic sarcomas and prostate tumors [26,51,61-63 and our unpublished observations}. Our observations of CFA 13 gain in canine cancer patients therefore closely resemble those of HSA 7 gain in human patients, and suggest that cross-species analyses may aid with identification of recurrent CNAs that are more intimately associated with disease progression rather than initiation, and which may not be as fundamentally involved in tumor pathogenesis as their frequency may suggest.
In earlier studies we reported the identification of evolutionarily conserved chromosome aberrations that are shared by human and canine counterparts of the same spontaneous malignancy. These included MYC-IGH fusion in NHL, BCR-ABL fusion in chronic myelogenous leukemia, PTEN deletion in osteosarcoma, MYC gain in glioma and osteosarcoma, and deletion of RB in chronic lymphocytic leukemia [9,26,40,57,59,61, Angstadt et al. submitted]. The present study indicates that while the aCGH profiles of hNHL and cNHL share common features, there is limited conservation of the hallmark CNAs of each species. These findings are reminiscent of our prior report of intracranial tumors in the dog, in which we determined that canine meningiomas show negligible evidence for deletion of the NF2 tumor suppressor gene that is widely regarded as a hallmark of human meningiomas, while canine gliomas do not appear to present with CNAs orthologous to the deletions of HSA 1p and 19q that are characteristic of human glioma patients [26]. We suggested that the apparent contrast between the extensive histological and clinical similarity of canine and human tumors versus the lack of conservation in structural chromosome rearrangements might be related to their vastly contrasting genome architecture. In figures 3 and 4 it is apparent from our ‘genomic recoding’ approach that conversion of canine to human chromosome co-ordinates has the effect of visually ‘disrupting’ the genome into smaller defined fragments at the subchromosomal level, resulting in a series of peaks and troughs in the resulting CNA penetrance plots. This fragmentation reflects the highly contrasting structural organization of the 200+ evolutionarily conserved chromosome segments shared by the human and canine genomes as a result of evolution from a common ancestral karyotype [14,15]. For example, CFA 11, recurrently deleted in cTCL, is orthologous to two regions (located on HSA 5 and 9) that show recurrent deletion in hTCL patients. Conversely gains of CFA 13 and 29 in cTCL correspond to neighboring syntenic chromosome segments forming the distal half of HSA 8q, which is frequently gained in hTCL (reviewed in [44]). Comparison of genome organization in phylogenetically diverse species has shown that almost 20% of chromosome breakpoints were reused during mammalian evolution, many of which represent centromeric regions and are associated with higher than average gene density. The locations of antigen receptor genes and many other cancer-associated chromosome breakages correspond closely to evolutionary breakpoints, suggesting that they may be inherently fragile [64]. Characterization of the genes disrupted by cancer-associated translocation events has been notoriously challenging using traditional cytogenetic approaches [56]. Thus cross-species genomics analysis offers exciting opportunities for expediting gene discovery by focused evaluation of genes located at evolutionary breakpoints.
Despite the apparently limited aneuploidy detected in cNHL, interrogation of recurrent CNAs identified in this study identified an extensive catalogue of chromosomal regions and genes presenting with recurrent DNA copy number imbalance in cNHL. Within these are key genes that have been associated previously with a range of human malignancies, while others highlight pertinent targets for focused evaluation in future studies. While conversion of our cDLBCL aCGH data into human genome format indicated a limited degree of global conservation in their cytogenetic profiles, a subset of common, evolutionarily conserved aberrations are evident. We propose that targeted analysis of these conserved CNAs will accelerate progress in defining the fundamental underlying molecular pathogenesis of these cancers in both species, and manipulating this knowledge to improve outcome. Recent advances in development of immunotherapeutic strategies that are effective across species [65] have shown great promise for a ‘one medicine’ approach to NHL. High-resolution cross-species cytogenomic analysis coupled with gene expression profiling of a common case series is now required in order to define and explore the most biologically relevant genomic parallels, to establish their functional role in tumor initiation and progression, and to evaluate their potential for targeted therapy in both human and canine patients.
Supplementary Material
Supplementary Online Materials: SOM Table I: Signalment and histopathological data for the canine lymphoma cohort. Cases are listed according to tumor immunophenotype and then by histological subtype and tumor grade (HG/IG/LG = high/intermediate/low grade). M = male, F = female. ‘nd’ indicates data not determined.
Breed abbreviations are: Australian Shepherd Dog (ASD), Border Collie (BC), Beagle (BEAG), Bernese Mountain Dog (BMD), Boxer (BOX), Briard (BRI), Chesapeake Bay Retriever (CBR), Cocker Spaniel (CS), Doberman (DOB), English Bulldog (EBD), English Bull Terrier (EBT), Flat Coated Retriever (FCR), Great Dane (GD), Golden Retriever (GR), German Shepherd Dog (GSD), Labrador Retriever (LAB), Mastiff (MAST), Rottweiler (ROTT), Standard Poodle (STP), West Highland White Terrier (WHWT), Yorkshire Terrier (YT).
Histological subtype abbreviations are: Burkitt's-like lymphoma (BKL), diffuse intermediate B-cell lymphoma (DIBCL), diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), lymphoblastic convoluted lymphoma (LBC), lymphoblastic lymphoma (LBL), lymphoplasmacytoid lymphoma (LPL), marginal zone lymphoma (MZL), peripheral T-cell lymphoma unspecified (PTCLu), small lymphocytic lymphoma (SLL), T-zone lymphoma (TZL).
SOM Table II: Genomic location of recurrent CNAs in cNHL. Regions showing CNA in 20% or more of the cohort (table 2a = cBCL, table 2b = cTCL) are listed according to the chromosomal location of the corresponding arrayed BAC clone. Each region is then defined by the frequency of DNA copy number gain or loss within the total number of cases scored at that locus. The orthologous region in the human genome is also shown alongside a description of gene sequences encoded within that region. Genes shown in bold type are members of the Canine Gene Census list [27] and have been shown previously to be associated with human malignancies. Due to the extensive number of CNAs identified in cTCL (encompassing >1000 genes), SOM table IIb is restricted only to genes defined in the Cancer Gene Census list.
SOM Table III: Genomic regions with copy number status significantly associated with immunophenotype in cNHL (p < 2.181 × 10−5). Regions are defined by the location of the corresponding arrayed clone in the dog genome assembly, which is shown against the orthologous location in the human genome. Loci are listed in descending order according to the Bonferroni-corrected p-value for significance at each locus. nd indicates data not determined, indicating regions that could not be mapped to a unique orthologous location in the human genome. Genes shown in bold type are members of the Canine Gene Census list [27].
SOM Table IV: Genomic regions with copy number status significantly associated with tumor subtype within cTCL (cPTCLu versus cTZL, p < 2.181 × 10−5). The significance of each association is shown according to a Bonferroni-corrected p-value to correct for multiple testing. Clones are listed according to their location in the dog genome assembly, and the corresponding location in the human genome is also shown. Genes located within these regions are listed.
SOM Table V: Association between DNA copy number status and breed, stratified by phenotype. A) cBCL showed a skew towards large p-values and no significant associations. B) cTCL showed a skew towards small p-values, and seven loci showed significant association with copy number gain in Boxers. The chromosome locations (canine and human) and Bonferroni-corrected p-values (p < 2.181 × 10−5) for these regions are shown. Genes located within these regions are listed.
SOM Table VI: Common aberration analysis of five cDLBCL cases evaluated by oligonucleotide aCGH revealed 22 regions of CNA shared by two or more cases. The chromosome location and nucleotide span of each shared CNA is indicated, in addition to the nature of the CNA (gain or loss).
SOM figure 1: Comparison of BAC-array CGH and oligonucleotide-array CGH analysis of five cDLBCL cases and one cPTCLu case from Golden Retrievers. The elevated resolution capabilities of the latter aid definition of the genomic boundaries of common CNAs and identify subchromosomal CNAs that were not detectable by BAC aCGH. Overall, however, oaCGH analysis supported our observations from BAC-array analysis that showed limited genomic instability in cDLBCL as compared to its human equivalent, comprised predominantly of highly recurrent copy number gain or loss of a small subset of entire chromosomes or extensive chromosome regions. This in turn suggests that our observations are not a consequence of the resolution limitations of BAC-array CGH analysis.
SOM figure 2: Magnified view of oaCGH data along CFA 11 for the five cDLBCL cases shown in SOM figure 1. The CFA 11 ideogram is shown below each oaCGH profile, and the arrow denotes the location of the CDKN2A/B locus at CFA 11q16, which shows a balanced copy number in each cDLBCL case. Data for these five cDLBCL cases are compiled in the final panel (blue trace) along with a cPTCLu case (red trace) for which CDKN2A/B deletion was identified previously by BAC aCGH (SOM figure 1, verifying the ability of the oaCGH platform to detect copy number loss of this locus.
SOM Appendix 1: Predictive modeling analysis of genomic regions associated with tumor phenotype.
Acknowledgments
This study was supported by funds from the National Institutes of Health (R01CA112211, awarded to MB) and the American Kennel Club Canine Health Foundation (CHF615, awarded to MB and JM). We gratefully acknowledge the expertise of Sandra Horton and the NCSU Histopathology laboratory in the preparation and diagnostic evaluation of clinical specimens for this study. We acknowledge the assistance of Drs Andrea Angstadt and Dahlia Nielsen with bioinformatics analysis. We thank Drs Masao Seto, Masao Nakagawa and Ichiro Takeuchi (Aichi Cancer Center Research Institute, Nagoya, Japan) for assistance with retrieval of raw aCGH data from prior studies of human lymphoma patients.
Footnotes
Declaration of Interests: The authors report no declarations of interest.
References
- 1.Bernheim A. Cytogenomics of cancers: From chromosome to sequence. Mol Oncol. doi: 10.1016/j.molonc.2010.06.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan WJ. Pathogenesis of diffuse large B cell lymphoma. Int J Hematol. 2010 doi: 10.1007/s12185-010-0602-0. Published online: 29 June 2010. [DOI] [PubMed] [Google Scholar]
- 3.Jevremovic D, Viswanatha DS. Molecular diagnosis of hematopoietic and lymphoid neoplasms. Hematol Oncol Clin North Am. 2009;23:903–933. doi: 10.1016/j.hoc.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Modiano JF, Breen M, Valli VE, Wojcieszyn JW, Cutter GR. Predictive value of p16 or Rb inactivation in a model of naturally occurring canine non-Hodgkin's lymphoma. Leukemia. 2007;21:184–187. doi: 10.1038/sj.leu.2404392. [DOI] [PubMed] [Google Scholar]
- 5.Shearin A, Ostrander EA. Leading the way: canine models of genomics and disease. Disease models and mechanisms. 2010;3:27–34. doi: 10.1242/dmm.004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morton LM, Wang SS, Devesa SS, Hartge P, Weisenburger DD, Linet MS. Lymphoma incidence patterns by WHO subtype in the United States, 1992-2001. Blood. 2006;107:265–276. doi: 10.1182/blood-2005-06-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.American Kennel Club. The Complete Dog Book. New York: Ballantine Books; 2006. [Google Scholar]
- 8.Parker HG, Kim LV, Sutter NB, et al. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 9.Modiano JF, Breen M, Burnett RC, et al. Distinct B-cell and T-cell lymphoproliferative disease prevalence among dog breeds indicates heritable risk. Cancer Res. 2005;65:5654–5661. doi: 10.1158/0008-5472.CAN-04-4613. [DOI] [PubMed] [Google Scholar]
- 10.Lurie DM, Lucroy MD, Griffey SM, Simonson E, Madewell BR. T-cell-derived malignant lymphoma in the boxer breed. Vet Comp Oncol. 2004;2:171–175. doi: 10.1111/j.1476-5810.2004.00047.x. [DOI] [PubMed] [Google Scholar]
- 11.Pastor M, Chalvet-Monfray K, Marchal T, et al. Genetic and environmental risk indicators in canine non-Hodgkin's lymphomas: breed associations and geographic distribution of 608 cases diagnosed throughout France over 1 year. J Vet Intern Med. 2009;23:301–310. doi: 10.1111/j.1939-1676.2008.0255.x. [DOI] [PubMed] [Google Scholar]
- 12.Onions DE. A prospective survey of familial canine lymphosarcoma. J Natl Cancer Inst. 1984;72:909–912. [PubMed] [Google Scholar]
- 13.Lobetti RG. Lymphoma in 3 related Rottweilers from a single household. J S Afr Vet Assoc. 2009;80:103–105. doi: 10.4102/jsava.v80i2.180. [DOI] [PubMed] [Google Scholar]
- 14.Breen M. Canine cytogenetics--from band to basepair. Cytogenet Genome Res. 2008;120:50–60. doi: 10.1159/000118740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derrien T, Andre C, Galibert F, Hitte C. Analysis of the unassembled part of the dog genome sequence: chromosomal localization of 115 genes inferred from multispecies comparative genomics. J Hered. 2007;98:461–467. doi: 10.1093/jhered/esm027. [DOI] [PubMed] [Google Scholar]
- 16.American Kennel Club. 2009 Dog Registration Statistics. 2010 Oct 18th; < http://www.akc.org/reg/dogreg_stats.cfm>.
- 17.Jaffee E, Harris N, Stein H, Vardiman J. Pathology and Genetics of Tumors of Hematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2001. World Health Organization Classification of Tumors. [Google Scholar]
- 18.Valli VE. Veterinary pathologists achieve 80% agreement in application of WHO diagnoses to canine lymphoma. Cancer Therapy. 2008;6:221–226. [Google Scholar]
- 19.Thomas R, Duke SE, Karlsson EK, et al. A genome assembly-integrated dog 1 Mb BAC microarray: a cytogenetic resource for canine cancer studies and comparative genomic analysis. Cytogenet Genome Res. 2008;122:110–121. doi: 10.1159/000163088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindblad-Toh K, Wade CM, Mikkelsen TS, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 21.Tagawa H, Suguro M, Tsuzuki S, et al. Comparison of genome profiles for identification of distinct subgroups of diffuse large B-cell lymphoma. Blood. 2005;106:1770–1777. doi: 10.1182/blood-2005-02-0542. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi I, Tagawa H, Tsujikawa A, et al. The potential of copy number gains and losses, detected by array-based comparative genomic hybridization, for computational differential diagnosis of B-cell lymphomas and genetic regions involved in lymphomagenesis. Haematologica. 2009;94:61–69. doi: 10.3324/haematol.12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa M, Nakagawa-Oshiro A, Karnan S, et al. Array comparative genomic hybridization analysis of PTCL-U reveals a distinct subgroup with genetic alterations similar to lymphoma-type adult T-cell leukemia/lymphoma. Clin Cancer Res. 2009;15:30–38. doi: 10.1158/1078-0432.CCR-08-1808. [DOI] [PubMed] [Google Scholar]
- 24.Chen W, Houldsworth J, Olshen AB, et al. Array comparative genomic hybridization reveals genomic copy number changes associated with outcome in diffuse large B-cell lymphomas. Blood. 2006;107:2477–2485. doi: 10.1182/blood-2005-07-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oudejans JJ, van Wieringen WN, Smeets SJ, et al. Identification of genes putatively involved in the pathogenesis of diffuse large B-cell lymphomas by integrative genomics. Genes Chromosomes Cancer. 2009;48:250–260. doi: 10.1002/gcc.20632. [DOI] [PubMed] [Google Scholar]
- 26.Thomas R, Duke SE, Wang HJ, et al. ‘Putting our heads together’: insights into genomic conservation between human and canine intracranial tumors. J Neurooncol. 2009 doi: 10.1007/s11060-009-9877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. [accessed Oct. 2010];Nat Rev Cancer. 2004 4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitelman Database of Chromosome Aberrations in Cancer. 2005 Internet. Available from: http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- 29.Mitelman F, Johansson B, Mertens F. [Accessed November 2010]; [Accessed November 2010];Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2010 < http://cgap.nci.nih.gov/Chromosomes/Mitelman>.
- 30.Staudt LM, Dave S. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv Immunol. 2005;87:163–208. doi: 10.1016/S0065-2776(05)87005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 32.Monti S, Savage KJ, Kutok JL, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 33.Bea S, Colomo L, Lopez-Guillermo A, et al. Clinicopathologic significance and prognostic value of chromosomal imbalances in diffuse large B-cell lymphomas. J Clin Oncol. 2004;22:3498–3506. doi: 10.1200/JCO.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 34.Bea S, Zettl A, Wright G, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood. 2005;106:3183–3190. doi: 10.1182/blood-2005-04-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 36.Chang CC, McClintock S, Cleveland RP, et al. Immunohistochemical expression patterns of germinal center and activation B-cell markers correlate with prognosis in diffuse large B-cell lymphoma. Am J Surg Pathol. 2004;28:464–470. doi: 10.1097/00000478-200404000-00005. [DOI] [PubMed] [Google Scholar]
- 37.Hartmann EM, Ott G, Rosenwald A. Molecular biology and genetics of lymphomas. Hematol Oncol Clin North Am. 2008;22:807–823. vii. doi: 10.1016/j.hoc.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Lenz G, Wright GW, Emre NC, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Han T, Iqbal J, et al. Diffuse large B-cell lymphoma in Chinese patients: immunophenotypic and cytogenetic analyses of 124 cases. Am J Clin Pathol. 2010;133:305–313. doi: 10.1309/AJCP4H6ADGYDZMOA. [DOI] [PubMed] [Google Scholar]
- 40.Thomas R, Smith KC, Ostrander EA, Galibert F, Breen M. Chromosome aberrations in canine multicentric lymphomas detected with comparative genomic hybridisation and a panel of single locus probes. Br J Cancer. 2003;89:1530–1537. doi: 10.1038/sj.bjc.6601275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauer TR, Jr, McDermid HE, Budarf ML, Van Keuren ML, Blomberg BB. Physical location of the human immunoglobulin lambda-like genes, 14.1, 16.1, and 16.2. Immunogenetics. 1993;38:387–399. doi: 10.1007/BF00184519. [DOI] [PubMed] [Google Scholar]
- 42.Arun SS, Breuer W, Hermanns W. Immunohistochemical examination of light-chain expression (lambda/kappa ratio) in canine, feline, equine, bovine and porcine plasma cells. Zentralbl Veterinarmed A. 1996;43:573–576. doi: 10.1111/j.1439-0442.1996.tb00489.x. [DOI] [PubMed] [Google Scholar]
- 43.Novara F, Arcaini L, Merli M, et al. High-resolution genome-wide array comparative genomic hybridization in splenic marginal zone B-cell lymphoma. Hum Pathol. 2009;40:1628–1637. doi: 10.1016/j.humpath.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 44.Seto M, Honma K, Nakagawa M. Diversity of genome profiles in malignant lymphoma. Cancer Sci. 2010;101:573–578. doi: 10.1111/j.1349-7006.2009.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morton LM, Turner JJ, Cerhan JR, et al. Proposed classification of lymphoid neoplasms for epidemiologic research from the Pathology Working Group of the International Lymphoma Epidemiology Consortium (InterLymph) Blood. 2007;110:695–708. doi: 10.1182/blood-2006-11-051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seelig DM, Avery PR, Kisseberth WC, Modiano JF. T cell Lymphoproliferative Diseases. In: Weiss DJ, Wardrop KJ, editors. Schalm's Veterinary Hematology. 6th. Hoboken, NJ: Wiley-Blackwell; 2010. [Google Scholar]
- 47.Rudiger T, Weisenburger DD, Anderson JR, et al. Peripheral T-cell lymphoma (excluding anaplastic large-cell lymphoma): results from the Non-Hodgkin's Lymphoma Classification Project. Ann Oncol. 2002;13:140–149. doi: 10.1093/annonc/mdf033. [DOI] [PubMed] [Google Scholar]
- 48.Zettl A, Rudiger T, Konrad MA, et al. Genomic profiling of peripheral T-cell lymphoma, unspecified, and anaplastic large T-cell lymphoma delineates novel recurrent chromosomal alterations. Am J Pathol. 2004;164:1837–1848. doi: 10.1016/S0002-9440(10)63742-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson M, Horsman DE, Weisenburger DD, et al. Cytogenetic abnormalities and clinical correlations in peripheral T-cell lymphoma. Br J Haematol. 2008;141:461–469. doi: 10.1111/j.1365-2141.2008.07042.x. [DOI] [PubMed] [Google Scholar]
- 50.Thorns C, Bastian B, Pinkel D, et al. Chromosomal aberrations in angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma unspecified: A matrix-based CGH approach. Genes Chromosomes Cancer. 2007;46:37–44. doi: 10.1002/gcc.20386. [DOI] [PubMed] [Google Scholar]
- 51.Fosmire SP, Thomas R, Jubala CM, et al. Inactivation of the p16 cyclin-dependent kinase inhibitor in high-grade canine non-Hodgkin's T-cell lymphoma. Vet Pathol. 2007;44:467–478. doi: 10.1354/vp.44-4-467. [DOI] [PubMed] [Google Scholar]
- 52.Vose JM. Peripheral T-cell non-Hodgkin's lymphoma. Hematol Oncol Clin North Am. 2008;22:997–1005. x. doi: 10.1016/j.hoc.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 53.Campbell LJ. Cytogenetics of lymphomas. Pathology. 2005;37:493–507. doi: 10.1080/00313020500368253. [DOI] [PubMed] [Google Scholar]
- 54.Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 55.Hunt KE, Reichard KK. Diffuse large B-cell lymphoma. Arch Pathol Lab Med. 2008;132:118–124. doi: 10.5858/2008-132-118-DLBL. [DOI] [PubMed] [Google Scholar]
- 56.Strefford JC, An Q, Harrison CJ. Modeling the molecular consequences of unbalanced translocations in cancer: lessons from acute lymphoblastic leukemia. Cell Cycle. 2009;8:2175–2184. doi: 10.4161/cc.8.14.9103. [DOI] [PubMed] [Google Scholar]
- 57.Breen M, Modiano JF. Evolutionarily conserved cytogenetic changes in hematological malignancies of dogs and humans - man and his best friend share more than companionship. Chromosome Res. 2008;16:145–154. doi: 10.1007/s10577-007-1212-4. [DOI] [PubMed] [Google Scholar]
- 58.Angstadt AY, Motsinger-Reif A, Thomas R, et al. Array comparative genomic hybridization and qRT-PCR analysis of canine osteosarcoma parallels its human counterpart. doi: 10.1002/gcc.20908. submitted. [DOI] [PubMed] [Google Scholar]
- 59.Seiser EL, Thomas R, Richards KL, et al. Reading Between the Lines: Molecular Characterization of Five Widely Utilized Canine Lymphoid Tumor Cell Lines. doi: 10.1111/j.1476-5829.2011.00299.x. submitted. [DOI] [PubMed] [Google Scholar]
- 60.Stasik CJ, Nitta H, Zhang W, et al. Increased MYC gene copy number correlates with increased mRNA levels in diffuse large B-cell lymphoma. Haematologica. 95:597–603. doi: 10.3324/haematol.2009.012864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomas R, Wang HJ, Tsai PC, et al. Influence of genetic background on tumor karyotypes: Evidence for breed-associated cytogenetic aberrations in canine appendicular osteosarcoma. Chromosome Res. 2009 doi: 10.1007/s10577-009-9028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunn KA, Thomas R, Binns MM, Breen M. Comparative genomic hybridization (CGH) in dogs--application to the study of a canine glial tumour cell line. Vet J. 2000;160:77–82. doi: 10.1053/tvjl.2000.0495. [DOI] [PubMed] [Google Scholar]
- 63.Winkler S, Reimann-Berg N, Escobar HM, et al. Polysomy 13 in a canine prostate carcinoma underlining its significance in the development of prostate cancer. Cancer Genet Cytogenet. 2006;169:154–158. doi: 10.1016/j.cancergencyto.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 64.Murphy WJ, Larkin DM, Everts-van der Wind A, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- 65.Balhorn RL, Skorupski KA, Hok S, Balhorn MC, Guerrero T, Rebhun RB. A selective high affinity ligand (SHAL) designed to bind to an over-expressed human antigen on non-Hodgkin's lymphoma also binds to canine B-cell lymphomas. Vet Immunol Immunopathol. doi: 10.1016/j.vetimm.2010.05.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Online Materials: SOM Table I: Signalment and histopathological data for the canine lymphoma cohort. Cases are listed according to tumor immunophenotype and then by histological subtype and tumor grade (HG/IG/LG = high/intermediate/low grade). M = male, F = female. ‘nd’ indicates data not determined.
Breed abbreviations are: Australian Shepherd Dog (ASD), Border Collie (BC), Beagle (BEAG), Bernese Mountain Dog (BMD), Boxer (BOX), Briard (BRI), Chesapeake Bay Retriever (CBR), Cocker Spaniel (CS), Doberman (DOB), English Bulldog (EBD), English Bull Terrier (EBT), Flat Coated Retriever (FCR), Great Dane (GD), Golden Retriever (GR), German Shepherd Dog (GSD), Labrador Retriever (LAB), Mastiff (MAST), Rottweiler (ROTT), Standard Poodle (STP), West Highland White Terrier (WHWT), Yorkshire Terrier (YT).
Histological subtype abbreviations are: Burkitt's-like lymphoma (BKL), diffuse intermediate B-cell lymphoma (DIBCL), diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), lymphoblastic convoluted lymphoma (LBC), lymphoblastic lymphoma (LBL), lymphoplasmacytoid lymphoma (LPL), marginal zone lymphoma (MZL), peripheral T-cell lymphoma unspecified (PTCLu), small lymphocytic lymphoma (SLL), T-zone lymphoma (TZL).
SOM Table II: Genomic location of recurrent CNAs in cNHL. Regions showing CNA in 20% or more of the cohort (table 2a = cBCL, table 2b = cTCL) are listed according to the chromosomal location of the corresponding arrayed BAC clone. Each region is then defined by the frequency of DNA copy number gain or loss within the total number of cases scored at that locus. The orthologous region in the human genome is also shown alongside a description of gene sequences encoded within that region. Genes shown in bold type are members of the Canine Gene Census list [27] and have been shown previously to be associated with human malignancies. Due to the extensive number of CNAs identified in cTCL (encompassing >1000 genes), SOM table IIb is restricted only to genes defined in the Cancer Gene Census list.
SOM Table III: Genomic regions with copy number status significantly associated with immunophenotype in cNHL (p < 2.181 × 10−5). Regions are defined by the location of the corresponding arrayed clone in the dog genome assembly, which is shown against the orthologous location in the human genome. Loci are listed in descending order according to the Bonferroni-corrected p-value for significance at each locus. nd indicates data not determined, indicating regions that could not be mapped to a unique orthologous location in the human genome. Genes shown in bold type are members of the Canine Gene Census list [27].
SOM Table IV: Genomic regions with copy number status significantly associated with tumor subtype within cTCL (cPTCLu versus cTZL, p < 2.181 × 10−5). The significance of each association is shown according to a Bonferroni-corrected p-value to correct for multiple testing. Clones are listed according to their location in the dog genome assembly, and the corresponding location in the human genome is also shown. Genes located within these regions are listed.
SOM Table V: Association between DNA copy number status and breed, stratified by phenotype. A) cBCL showed a skew towards large p-values and no significant associations. B) cTCL showed a skew towards small p-values, and seven loci showed significant association with copy number gain in Boxers. The chromosome locations (canine and human) and Bonferroni-corrected p-values (p < 2.181 × 10−5) for these regions are shown. Genes located within these regions are listed.
SOM Table VI: Common aberration analysis of five cDLBCL cases evaluated by oligonucleotide aCGH revealed 22 regions of CNA shared by two or more cases. The chromosome location and nucleotide span of each shared CNA is indicated, in addition to the nature of the CNA (gain or loss).
SOM figure 1: Comparison of BAC-array CGH and oligonucleotide-array CGH analysis of five cDLBCL cases and one cPTCLu case from Golden Retrievers. The elevated resolution capabilities of the latter aid definition of the genomic boundaries of common CNAs and identify subchromosomal CNAs that were not detectable by BAC aCGH. Overall, however, oaCGH analysis supported our observations from BAC-array analysis that showed limited genomic instability in cDLBCL as compared to its human equivalent, comprised predominantly of highly recurrent copy number gain or loss of a small subset of entire chromosomes or extensive chromosome regions. This in turn suggests that our observations are not a consequence of the resolution limitations of BAC-array CGH analysis.
SOM figure 2: Magnified view of oaCGH data along CFA 11 for the five cDLBCL cases shown in SOM figure 1. The CFA 11 ideogram is shown below each oaCGH profile, and the arrow denotes the location of the CDKN2A/B locus at CFA 11q16, which shows a balanced copy number in each cDLBCL case. Data for these five cDLBCL cases are compiled in the final panel (blue trace) along with a cPTCLu case (red trace) for which CDKN2A/B deletion was identified previously by BAC aCGH (SOM figure 1, verifying the ability of the oaCGH platform to detect copy number loss of this locus.
SOM Appendix 1: Predictive modeling analysis of genomic regions associated with tumor phenotype.