Abstract
Next generation sequencing (NGS) technologies have been applied extensively in many areas of genetics and genomics research. A fundamental problem when comes to analyzing NGS data is mapping short sequencing reads back to the reference genome. Most of existing software packages rely on a single uniform reference genome and do not automatically take into the consideration of genetic variants. On the other hand, large proportions of incorrectly mapped reads affect the correct interpretation of the NGS experimental results. As an example, Degner et al. showed that detecting allele-specific expression from RNA sequencing data was biased toward the reference allele. In this study, we developed a method that utilize DirectX 11 enabled graphics processing unit (GPU)’s parallel computing power to produces a personalized diploid reference genome based on all known genetic variants of that particular individual. We show that using such a personalized diploid reference genome can improve mapping accuracy and significantly reduce the bias toward reference allele in allele-specific expression analysis. Our method can be applied to any individual that has genotype information obtained either from array-based genotyping or resequencing. Besides the reference genome, no additional changes to alignment algorithm are needed for performing read mapping therefore one can utilize any of the existing read mapping tools and achieve the improved read mapping result. C++ and GPU compute shader source code of the software program is available at: http://code.google.com/p/diploid-mapping/downloads/list.
Keywords: Allele specific expression, RNA-sequencing, read mapping, reference genome, single nucleotide polymorphism, GPU programming
I. INTRODUCTION
For diploid eukaryotic organisms, the maternally and paternally derived copies of most genes are expressed at similar levels. However, for some genes, the two alleles of an individual are expressed at different rates. This phenomenon is termed allele-specific expression (ASE). In recent years, much and increasing effort has been made to identify ASE genes since they present unique opportunities to study cis-regulatory variation [1–6]
The newly emerged next generation sequencing (NGS) technologies have been increasingly recognized as an important and powerful tool for identifying ASE genes genome-wide, which improves our understanding about cisregulatory variation. To identify ASE, one can conduct RNA sequencing (RNA-Seq) experiment [7, 8] to map all generated reads to the reference genome for all exonic SNPs that are known to be heterozygous, and then quantify the magnitude of expression of each allele by counting the number of times each allele is observed in reads that mapped to that locus. Despite its simplicity, systematic bias for read mapping may affect the accuracy of identifying ASE genes. This has been pointed out recently by Degner et al. [9]
Mapping short reads onto the reference genome is a fundamental problem in analyzing next generation sequencing (NGS) data and has been an area of intensive research in the past years. A wealth of successful software programs have been developed and enjoyed wide-spread usage in many different NGS applications such as MAQ [10], SOAP [11], BOWTIE [12], BWA [13], BFAST [14], mrFAST and mrsFAST [15]. The details of these algorithms and plenty of other commonly-used read mapping software can be found in an excellent review paper [16].
Almost all of the existing read-mapping software rely on a universal reference genome–the National Center for Biotechnology Information (NCBI) human reference genome [17] which is derived from a small number of anonymous donors. Although carefully annotated and maintained, this single reference genome cannot represent all the variants found in the general population. We know that each individual possess a unique set of genetic variants in hundreds of thousands that differ from the universal reference genome that distinguish him or her from others. Such wide-spread genetic variants compounded with nonignorable sequencing errors and short read length caused a large proportion of reads unmapped or mapped to incorrect genomic locations. These mapping errors affect the interpretation of the NGS experimental results. As an example, Degner et al. showed that detecting allele-specific expression (ASE) from RNA sequencing data was biased toward the reference alleles because reads containing alternative alleles have less probability to align than reads that contains the reference allele. Therefore genes with a large amount of alternative alleles may be underestimated [9].
To reduce the impact of these genetic variants, Dewey et al. proposed to use ethnically concordant major allele reference genome sequence for read mapping [18]. Using estimated allele frequency data from the 1000 genome project [19], the authors developed three ethnically-specific major allele references for European, African and East Asian. When applied to four individuals from a nuclear family, Dewey et al. reported increased number of reads that mapped uniquely to the major allele reference genome than to the NCBI reference genome.
While much improvement is achieved using reference genomes that tailored toward the ethnical groups, it is important to note that there are still plenty of genetic variations at the individual level within each ethnical group. With the efforts such as the international HapMap project [20], the 1000 Genome project [19] and many others, we have accumulated and cataloged millions of known genetic variants, most in the form of single nucleotide polymorphisms (SNPs). In the past five years, the cost of array-based genotyping has declined sharply. As a result, for individuals or cell lines that we want to mn RNA-Seq on, the genotypes of almost all common SNPs (minor allele frequency greater than 5%) are already known. In light of this, we believe that such information, whenever available should be incorporated into the process of read mapping.
In this study, we propose a novel method that utilizes all known genetic variant information of a particular individual and combine it with the NCBI reference genome to produce a "personalized" and diploid reference genome. We showed that mapping against this personalized diploid reference genome will improve mapping accuracy and significantly reduce the bias toward reference alleles in allele-specific expression analysis. Our method can be applied to any individual whose genotype is known either from array-based genotyping or resequencing. Besides the reference genome, no additional change to alignment algorithm is required for performing read mapping therefore one can continue using any of the existing read mapping tools they like and achieve the improved read mapping result.
II. METHODS
The goal of this project is to construct a personalized diploid reference genome using known genetic variants of an individual to reduce ASE bias. This reference genome can then be used for mapping reads generated from any sequencing assay conducted on this individual to improve the read mapping accuracy. There is no need to modify the read mapping software. Since genotypes are increasingly available and readily available, we believe incorporating such information in the read mapping step is important and beneficial. We have developed a software package available for public download that is able to achieve this goal conveniently.
A. Constructing personalized, diploid reference genome
In this study, we only consider SNPs, However our method can handle indels in a similar fashion. For better comparison with existing research results, we download universal NCBI reference genome (hg18.fa) from NCBI, which was used by Degner et al. [9], although our method can be applied to any version of universal reference genome including hg19.fa. To add alternative alleles, we go through each genotype stored in the individual’s genotype file (usually in the VCF format) in parallel. A typical DirectX11 enabled graphics processing unit (GPU) usually has thousands of “Stream Processors” running on gigahertz level frequency, which is very suitable to perform such large amount of parallel computation. For a SNP that is homozygous wild type allele (identical to the reference allele), no action is taken; for a SNPs that is homozygous mutant allele, we edit the corresponding nucleotide in the reference genome sequence file; for a heterozygous SNP, we add a “mini chromosome” that is w ≥ 2k − 1 bp in length where k is the read length and w can be specified by users. When w > 2k − 1 indels can be better detected. Suggested value of w is 2k − 1 + 2m, where m is the maximum mismatches allowed during reads mapping. bwa, for example, sets the default value of m to 2 when the read length is 35 bp. The sequence of this “mini chromosome” is identical to the corresponding reference genome except at the middle position in which the alternative allele of that SNP is placed in. We name these “mini chromosomes” in a way such that their genomic locations can be easily identified.
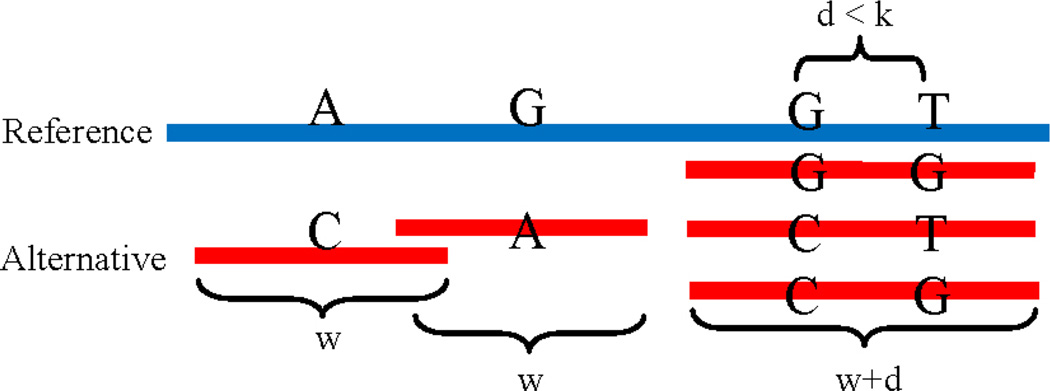
Admittedly, adding these “mini chromosomes” may result in additional multiple mapping, however, with careful bookkeeping, such multiple-mapping incidences can be resolved post-hoc. If two SNPs are located near each other, i.e., with distance of d bp, where d < k, we use a slightly longer “mini chromosome”, (w + d) bp that cover both SNPs, and adding “mini chromosomes” with all possible combinations of covered SNPs (see Figure 1). More than two nearby SNPs can be handled in similar fashion. After this step, the personalized diploid reference genome contains tens of thousands of such mini chromosomes.
Figure 1.
Examples of generated “mini chromosomes” at heterozygous SNPs. Most alternative chromosomes have the length of w. Small portion of heterozygous SNPs that are close to each other will be merged into longer ones.
We choose not to simply add another set of whole chromosomes consist with all the alternative alleles due to the following three reasons: First, currently there is a limit of how large the reference genome can be handled by many existing read mapping software. Many mapping software have strict limitation on the length of the total reference sequence (mostly, 4G bp) because the data structure unsigned integer is defined in compilers as a 32-bit number (232 = 4G). Second, most of the genomic regions are homozygous, so it is not resource-efficient, and lead to many more multiple mapping incidences.
Our program can also accept an optional command line argument indicating the individual’s gender. When this argument is set, for female individuals, we will exclude chromosome Y from the personalized reference genome and chromosome X is treated the same as any other autosome.
B. Reads mapping
In this step, we perform read mapping using the personalized diploid reference genome instead of the universal NCBI reference genome. Although we use mapping software bwa v0.5.9 [13] with default parameters in this study, our pipeline scheme can accommodate any read mapping software
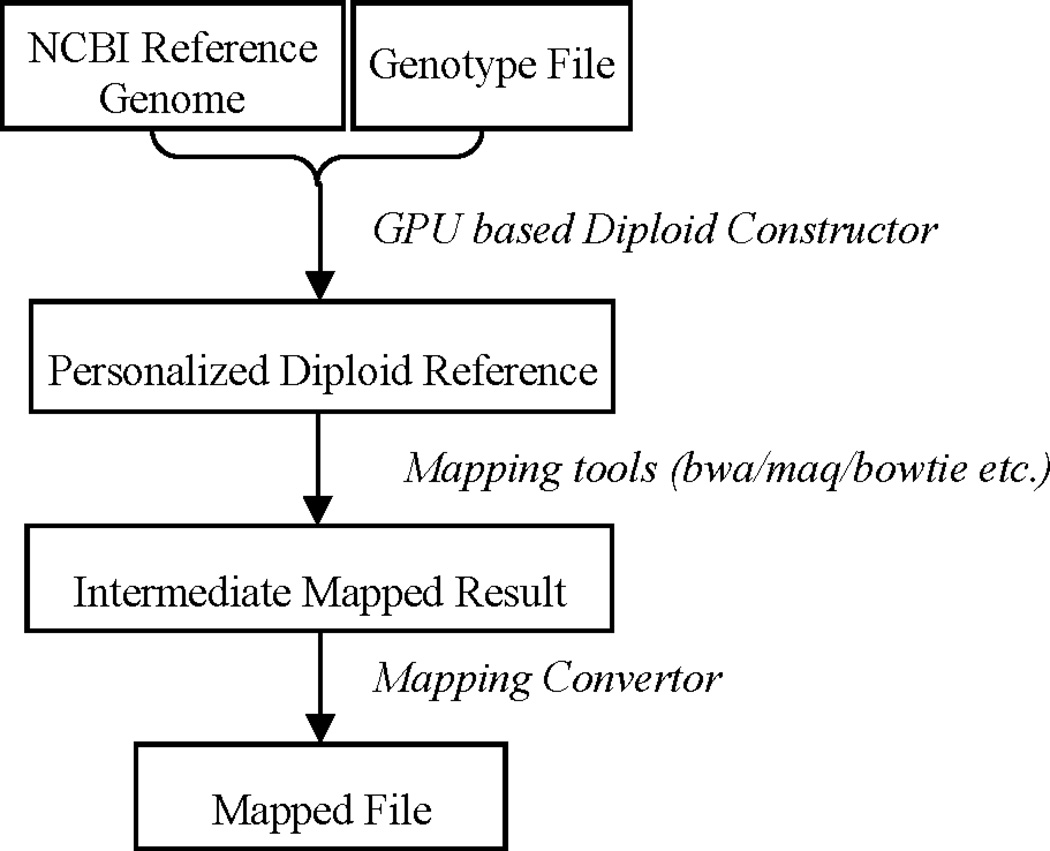
The raw output of the mapping step cannot be used directly because reads are mapped against a diploid reference genome that contains many “mini chromosomes”. We take another step to process the mapping result such that reads mapped to “mini chromosomes” are correctly interpreted as mapped to the corresponding genomic location with the alternative allele present at the middle SNP. This step contains two parts: first, recover the correct genomic mapping location, second, none-zero quality scores will be assigned according to some confidence values. Figure 2 demonstrates the whole process of our pipeline.
Figure 2.
3-step pipeline for creating personalized diploid reference genome and mapping reads against it. Diploid Constructor takes NCBI reference genome and the individual’s genotype file as input to create personalized diploid reference genome, which will then be used by multiple mapping tools to map reads. Mapping Convertor converts intermediate mapping result to regular mapped file. For example, position “chr3b.13843: 5” will be convert to “chr3:13847” (locations are 1-based).
C. An alternative method for reducing ASE bias
In addition to comparing with the common practice which is to use the universal reference genome for mapping, we also tested the masking strategy which has been used in the Degner et al. 2009 study. In this approach, all known SNP positions were “masked” prior to read-mapping. Masking was achieved by changing the nucleotide at each SNP location to one that differs from both the reference and alternative allele. The SNP locations were obtained by merging genotype files of 214 individuals defined in the 2007-03 version of the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov). In order to prevent too strong binding of the bases on both alleles to a specific masking base, we randomly choose the masking base. For example, if the nucleotides at the SNP location on the reference and alternative allele are “A” and “G”, the probability of using “C” or “T” as the mask are both 1/2.
D. Simulation studies
We conducted simulation studies to evaluate the impact on ASE bias and mapping quality when using the three competing mapping strategies: using the universal reference genome which is the status quo, using the masked universal reference genome which is introduced by Degner et al. 2009; and using the diploid personalized reference genome which we propose. In order to represent the diversity of human population and investigate its impact on the results, we selected three individuals from the HapMap panel, one Caucasian from CEPH (NA12865), one African from YRI (NA19238) and one Asian from CHB (NA18621). For each individual, we downloaded the individual’s genotype information (2007-03 version) from the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov). As Degner et al. did in their study we randomly inserted sequencing errors on reads generated. We tested three different sequencing error rates: 0, 0.01 and 0.05. When simulating reads, we choose the sequencing read length to be 35 bp and 100 bp, and then randomly sample DNA fragments across the whole diploid reference genome except chromosome X and Y. We only keep reads that cover at least one heterozygous SNP. For each of the three sequencing error rates, 2 million reads were generated. Either reference or alternative allele was selected with equal probability thus assume balanced allele specific expression. To create the masked reference genome, all SNPs identified from the 214 individuals in the International HapMap Project (genotype information obtained from the 2007-03 version) are masked. In order to increase the precision with more mapped reads, we consider SNPs located in both exons and introns.
E. Real data studies
We analyzed two sets of RNA-Seq data: one is the one studied in Degner et al. 2009 the other is 68 individuals from Pickrell et al 2010 [21]. Just like in the simulation studies we also use three different read-mapping strategies. Here we only consider SNPs located within exons.
We analyzed two aspects of the performance of different mapping strategies: mapping bias towards reference alleles and total number of reads that are successfully mapped. For the first, at each SNP locus, we first decide the number of reads that cover the SNP; after filtering out SNPs with too shallow mapping depth (this step is optional). In this study, we use threshold of five reads. We then count the number of reads that match the reference allele and the alternative allele respectively. For the second, we want to maximize the number of RNA-seq reads that can be mapped successfully. Therefore, a mapping strategy that can produce more mapped reads with high accuracy is preferred. In the simulation study, because we know each read’s true location, we can compare the number of reads that are correctly mapped back to their true locations; for real data, because reads’ true locations are unknown, we compare the number of reads that are successfully mapped.
III. RESULTS
A. ASE bias in simulation studies
The most important statistic that measure ASE bias is the proportion of reads that mapped to the reference allele. Simulation studies showed that the ratios of reads mapped to reference alleles are very close to the theoretical value--50% for both diploid and masked genome methods, regardless of the error rate, whereas conventional method yielded upward bias towards the reference alleles and the bias increase with the error rate. This indicates that both methods yield much reduced bias at all error rates. Even assuming no sequencing error, universal reference genome method is still suffering from inherent bias. The same pattern was observed on all three HapMap samples that represent different ethnic groups. We also found that increasing read length resulted in more bias. Table 1 shows the results for individual YRI NA19238.
Table 1.
Simulation results show that universal reference genome method suffers from serious bias towards reference alleles. This bias increases dramatically with the increment of error rate. However, the masked and diploid genome methods do not have apparent bias toward either reference or alternative alleles while error rate does not have influence to the ratio. (A) Read length is 35 bp and maximum mismatch is set to 2. (B) When the read length increase to 100 bp, ASE bias will also increase considerably given the same sequencing error rate.
| A Ratio of reads mapped to reference alleles | ||
|---|---|---|
| Read length=35bp, max mismatch=2 | ||
| Error rate | Method | Ratio |
| 0% | universal | 50.3024% |
| masked | 49.9580% | |
| diploid | 50.0139% | |
| 1% | universal | 51.3741% |
| masked | 49.9367% | |
| diploid | 49.9791% | |
| 5% | universal | 61.3801% |
| masked | 49.9985% | |
| diploid | 50.0760% | |
| B Ratio of reads mapped to reference alleles | ||
|---|---|---|
| Read length=100bp, max mismatch=2 | ||
| Error rate | Method | Ratio |
| 0% | universal | 50.0615% |
| masked | 49.9968% | |
| diploid | 50.0005% | |
| 1% | universal | 55.987% |
| masked | 49.9834% | |
| diploid | 49.9947% | |
| 5% | universal | 77.1133% |
| masked | 50.058% | |
| diploid | 50.0345% | |
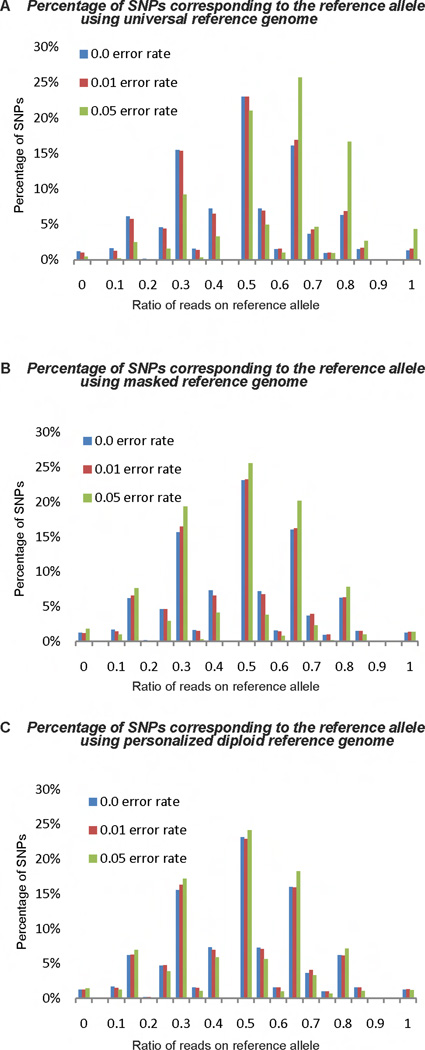
To better understand the magnitude of the bias and the impact of sequencing errors, we plotted the distribution of proportions of reference alleles obtained using different read mapping strategies (Figure 3). We found that when using universal reference genome method, the proportions of reference allele in majority of the SNPs are greater than 0.5. This asymmetry caused the mapping bias towards reference alleles. The asymmetry also increases dramatically as the error rate increases. However, neither diploid nor masked genome method shows apparent asymmetry.
Figure 3.
Distributions of reference allele proportions for SNPs tested in the simulation study (required read coverage depth > 5). The distributions spread across 0 to 1 because randomness of sampling. (A) Using universal reference genome. (B) Using masked reference genome. (C) Using personalized diploid reference genome.
B. Mapping accuracy
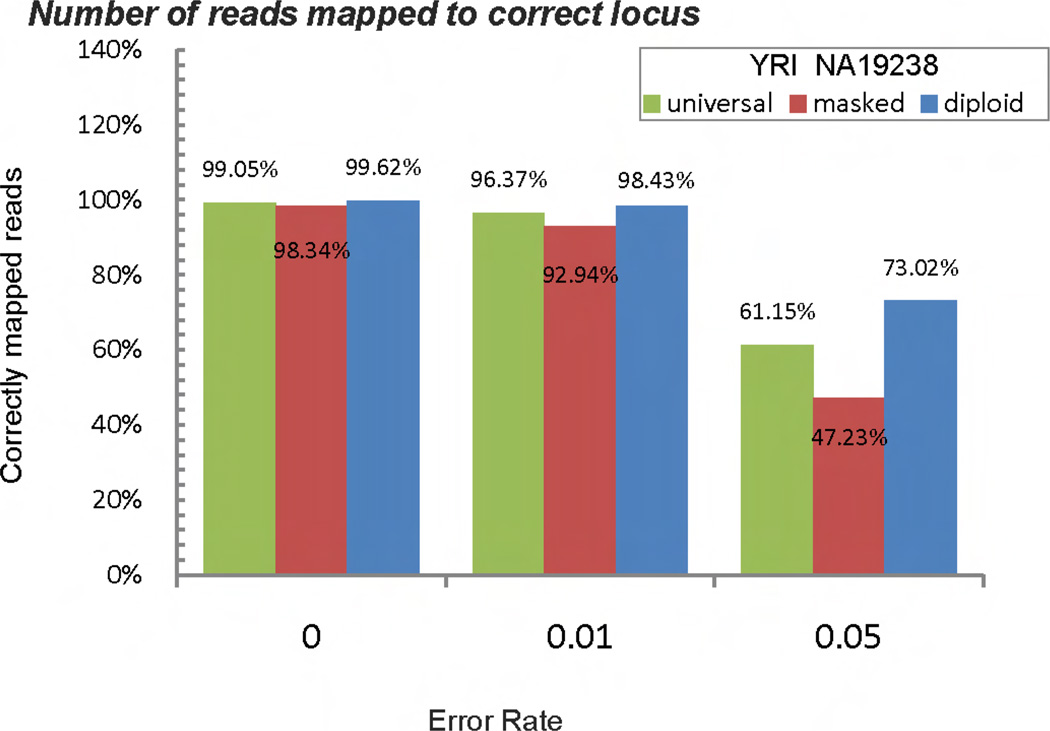
The percentage of correctly mapped reads is an important measure when evaluating mapping strategies. Although masking the reference allele in the universal reference genome reduces ASE bias, we found this strategy produces unreliable mapping result [9] and significantly lower overall mapping success rate, especially when moderate sequencing error is present. Our simulation shows that when the sequencing error rate reaches 0.05, the diploid genome method can correctly map 25% more reads compare to masked genome method. This significant improvement suggest that the diploid genome method has a higher mapping success rate overall. Figure 4 shows the mapping success rates from the three methods when mapping 2 million reads with different error rates.
Figure 4.
Simulation results of individual YRI NA19238 show that diploid genome method can improve mapping quality. The diploid genome method shows the highest correctness of mapping results among three methods. Universal reference genome method, although has mapping bias, shows better mapping quality than masked genome method.
C. Real data analysis on ASE bias and mapped reads
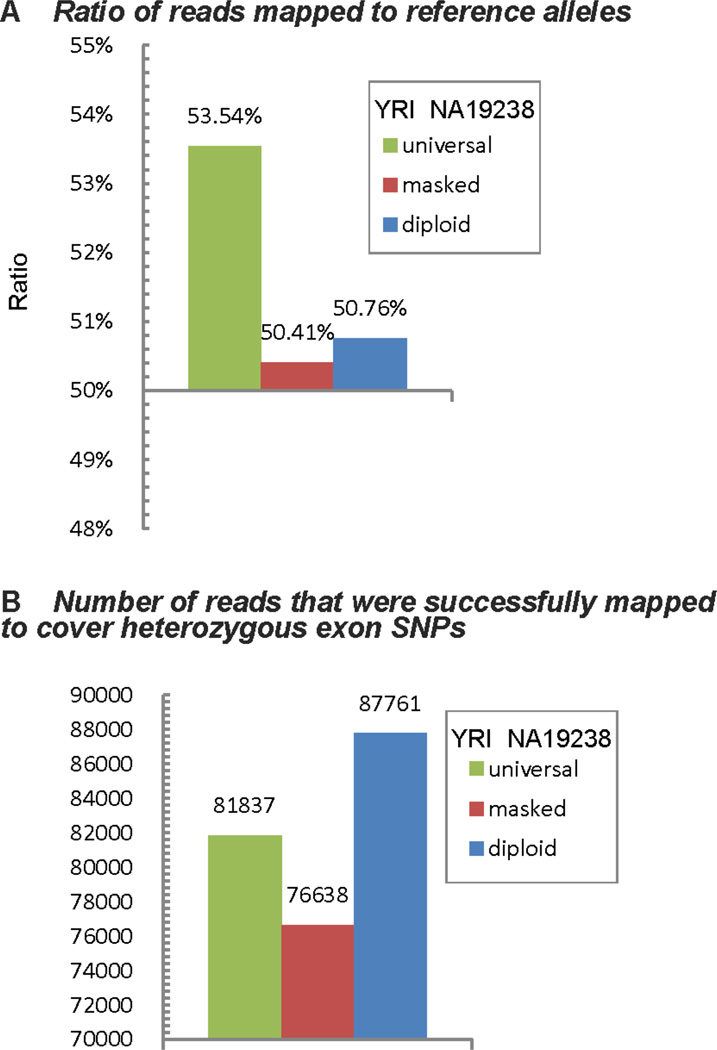
We reanalyzed the real RNA-Seq data presented in Degner et al. 2009 to compare the levels of ASE detection bias resulted from different mapping strategies. We also compared the number of reads that were successfully mapped to cover heterozygous exon SNPs using the three read mapping strategies. Figure 5 shows the results. From the figure, we observe the same pattern as in the simulated data: using either masked reference genome or the personalized diploid reference genome vastly reduce ASE bias while our method resulted in much higher success rate of read mapping than using the masked reference genome.
Figure 5.
(A) Results derived from running 3 different methods on real data show that diploid genome method can effectively reduce bias towards reference alleles. Although masked genome method can also reduce such bias, it might be over-reduced because of its unreliable mapping result. (B) Number of reads that cover heterozygous exon SNPs. This figure shows that masked genome method loses 12.70/0 of successfully mapped reads compare to diploid genome method. Therefore, it can be inferred that masked genome method is problematic.
D. More real data results
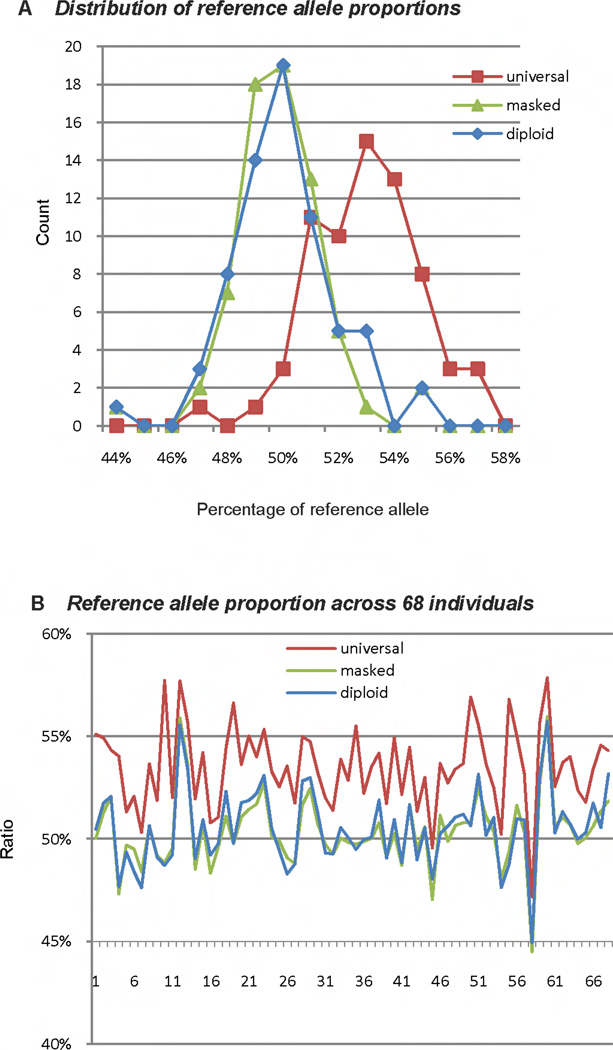
To verify that our new method can reduce ASE bias in general population, i.e., individuals whose genotype is known, we conducted experiments on a set of individuals with real data from a recent study of Pickrell et al 2010 [21]. We download the entire dataset from http://eqtl.uchicago.edu/RNA_Seq_data/, and then select 68 individuals whose genotype can be found in the 2007-03 version of the International HapMap Project. Figure 6 shows the distribution of the ratio of mapped reference allele. Again we observe using masked reference genome or personalized diploid reference genome reduce bias towards reference allele.
Figure 6.
The counts of individuals at each expression ratio and each individual’s reference. (A) The reference allele ratios for all individuals are in the range [44%, 58%). For diploid and masked genome method, there are 19 individuals located in the ratio region [50%, 51%), which is the peak of their curves. The universal reference genome method, however, shifted the peak to the ratio region [53%, 54%) for 15 individuals. (B) Using universal reference genome method will always produce higher reference ratio compare to diploid and masked genome method, which indicates that universal reference genome method will inevitably introduce bias towards the reference alleles.
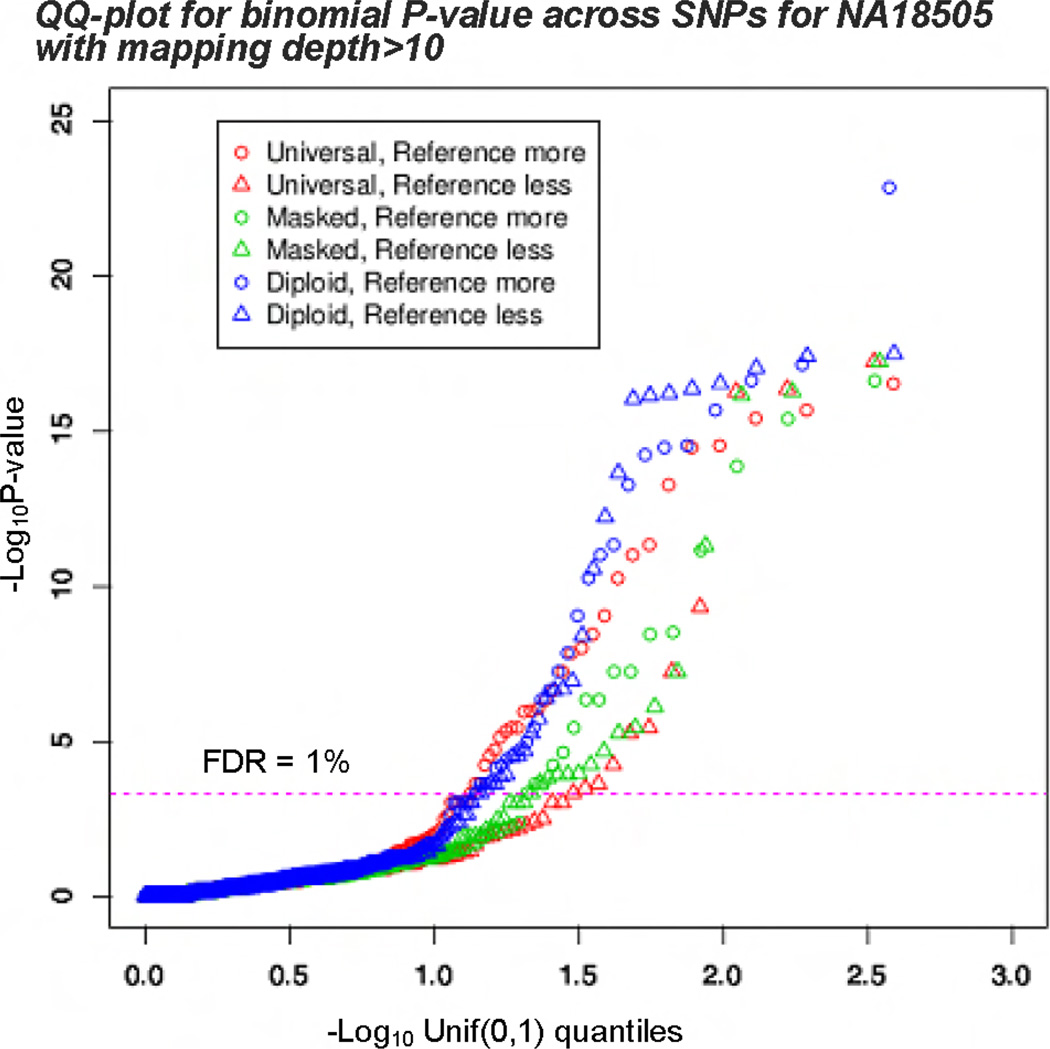
As ASE is widespread across heterozygous SNPs the P-values yield from binomial test must also display this enrichment and its impact to the expression bias. Figure 7 shows the QQ-plot of the P-values across quantiles. When using universal reference genome, we see that the two curves representing “reference more” and “reference less” respectively are far apart, indicating bias. For the other two methods: using masked reference genome or personalized diploid reference genome, the bias essentially disappeared.
Figure 7.
QQ-plots of P-values for one-sided binomial tests for heterozygous SNPs which are categorized into more or less expression for reference alleles than alternative alleles. Above the dotted line of FDR= 1%.
IV. DISCUSSION
ASE offers biological insights from understanding transcription regulation to disease susceptibility. Detecting ASE from RNA-Seq data has become an increasingly important topic for genetics and genomics researchers. As pointed out by Degner et al., the current read mapping strategy produces “a significant bias toward higher mapping rates of the allele in the reference sequence, compared with the alternative allele.” [9], therefore, it is of great importance to develop alternative strategy to reduce ASE bias. In this study, we proposed a novel strategy that utilizes known personal genotype information that is increasingly available in the post-genomic era. In our method, we first construct a personalized diploid reference genome using available genotype information, and then use the constructed reference genome with a regular existing read mapping software such as BWA to map reads generated from the RNA-Seq experiment. Using both simulated data and real data, we showed that our strategy can effectively reduce the ASE bias, and increase the success rate of read-mapping. We believe our method provides an attractive solution to ASE detection using RNA-Seq data.
The drawback of using the universal NCBI reference genome in read mapping has been noticed in the literature. Dewey et al. developed three ethnicity-specific major allele reference genomes for European, African and East Asian based on HapMap data and use that for read mapping. They reported improved genotyping accuracy using this synthetic reference genome [18]. In this study, we went further by constructing reference genome that is “personalized”, i.e., taking into account of known genotype information of that particular individual. With the rapid dissemination and declining cost of array-based genotyping technologies, genotypes of millions of SNPs are routinely available. Thus our method is widely applicable. Our strategy is developed independently of that of Vijaya Satya et al. 2012 [22]. Despite many similarities between the two methods, there are some notable differences: our personalized reference genome is able to accommodate indels in addition to SNP markers; we have tested our strategies on a much larger datasets to examine the population-level of the performance improvement; we have tested the performance of our method on longer read (100 bp) and found even better result in reducing ASE; we also implemented the construction of the personalized diploid reference genome using GPU compute shader code to improve the computation speed.
Using our software program, constructing a personalized diploid reference genome from a dense genotyping file only takes about 10 minutes on a commodity computer (Intel Core 2 Duo CPU, AMD Radeon HD 6900 GPU, 8GB memory), which seems a small price to pay for enhanced read mapping result.
ACKNOWLEDGMENT
This work is partially supported by NIH grant R21HG004751. We are grateful for Dr. Degner at the Jonathan Pritchard Lab for helpful discussion.
Footnotes
Conflict of Interest: none declared.
Contributor Information
Shuai Yuan, Mathematics & Computer Science Department, Emory University, 400 Dowman Drive Atlanta, GA 30322, USA, shuaiyuan@emory.edu.
Zhaohui Qin, Department of Biostatistics and Bioinformatics, Emory University, 1518 Clifton Road Atlanta, GA 30322, USA, zhaohui.qin@emory.edu.
REFERENCES
- 1.Serre D, Gurd S, Ge B, Sladek R, Sinnett D, Harmsen E, Bibikova M, Chudin E, Barker DL, Dickinson T, et al. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4(2):e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knight JC. Allele-specific gene expression uncovered. Trends Genet. 2004;20(3):113–116. doi: 10.1016/j.tig.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Milani L, Lundmark A, Nordlund J, Kiialainen A, Flaegstad T, Jonmundsson G, Kanerva J, Schmiegelow K, Gunderson KL, Lonnerholm G, et al. Allele-specific gene expression patterns in primary leukemic cells reveal regulation of gene expression by CpG site methylation. Genome Res. 2009;19(1):1–11. doi: 10.1101/gr.083931.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wittkopp PJ, Haerum BK, Clark AG. Independent effects of cisand trans-regulatory variation on gene expression in Drosophila melanogaster. Genetics. 2008;178(3):1831–1835. doi: 10.1534/genetics.107.082032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ronald J, Akey JM, Whittle J, Smith EN, Yvert G, Kruglyak L. Simultaneous genotyping, gene-expression measurement, and detection of allele-specific expression with oligonucleotide arrays. Genome Res. 2005;15(2):284–291. doi: 10.1101/gr.2850605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297(5584):1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 7.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mannnalian transcriptomes by RNASeq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degner JF, Marioni JC, Pai AA, Pickrell JK, Nkadori E, Gilad Y, Pritchard JK. Effect of read-mapping biases on detecting allelespecific expression from RNA-sequencing data. Bioinformatics. 2009;25(24):3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 12.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Bioi. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homer N, Merriman B, Nelson SF. BFAST: an alignment tool for large scale genome resequencing. PLoS One. 2009;4(11):e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hach F, Honnozdiari F, Alkan C, Birol I, Eichler EE, Sahinalp SC. mrsFAST: a cache-oblivious algorithm for short-read mapping. Nat Methods. 2010;7(8):576–577. doi: 10.1038/nmeth0810-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11(5):473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 2007. 35(Database issue):D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dewey FE, Chen R, Cordero SP, Ormond KE, Caleshu C, Karczewski KJ, Whirl-Carrillo M, Wheeler MT, Dudley JT, Byrnes JK, et al. Phased whole-genome genetic risk in a family quartet using a major allele reference sequence. PLoS Genet 2011. 7(9):e1002280. doi: 10.1371/journal.pgen.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464(7289):768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vijaya Satya R, Zavaljevski N, Reifman J. A new strategy to reduce allelic bias in RNA-Seq readmapping. Nucleic Acids Res. 2012;464(7289):768–772. doi: 10.1093/nar/gks425. [DOI] [PMC free article] [PubMed] [Google Scholar]