Abstract
We have previously demonstrated that the anti-apoptotic protein BAD is expressed in normal human breast tissue and shown that BAD inhibits expression of cyclin D1 to delay cell-cycle progression in breast cancer cells. Herein, expression of proteins in breast tissues was studied by immunohistochemistry and results were analyzed statistically to obtain semi-quantitative data. Biochemical and functional changes in BAD-overexpressing MCF7 breast cancer cells were evaluated using PCR, reporter assays, western blotting, ELISA and extracellular matrix invasion assays. Compared to normal tissues, Grade II breast cancers expressed low total/phosphorylated forms of BAD in both cytoplasmic and nuclear compartments. BAD overexpression decreased the expression of β-catenin, Sp1, and phosphorylation of STATs. BAD inhibited Ras/MEK/ERK and JNK signaling pathways, without affecting the p38 signaling pathway. Expression of the metastasis-related proteins, MMP10, VEGF, SNAIL, CXCR4, E-cadherin and TlMP2 were regulated by BAD with concomitant inhibition of extracellular matrix invasion. siRNA knockdown of BAD increased invasion and Akt/p-Akt levels. Clinical data and the results herein suggest that in addition to the effect on apoptosis, BAD conveys anti-metastatic effects and is a valuable prognostic marker in breast cancer.
Keywords: BAD, extracellular matrix invasion, breast cancer, metastasis
Introduction
Most apoptosis regulators, including BCL-2 family members, are typically localized to the intracellular membranes, cytoplasm, or mitochondria [1-4]. We previously demonstrated that the BCL-2 antagonist, BAD is localized to the nucleus, in addition to the cytoplasm in normal human breast tissue and that BAD prevents cyclin D1 transcription; hence cell cycle progression in breast cancer cells [5]. Al-Bazz et al. [6] and we [7] reported that BAD is localized to both the nucleus and cytoplasm in breast cancer tissue, suggesting a nuclear role for BAD. Other BH3 proteins, BIM and BCL-2 are also localized to the nucleus [1-3,8] suggesting that BCL-2 family proteins may have nuclear roles [9]. Our observations suggest that although BCL-2 and BAD have opposing effects in apoptosis in vitro [10,11], their cell-cycle-related functions could be comparable (see discussion).
Many clinical studies suggest that BCL-2 expression is a strong predictor of overall and disease-free survival in breast cancer patients. BCL-2 is a favorable and superior prognostic marker [12,13] independent of lymph node status, tumor size, grade, and other biomarkers including estrogen receptor a (ERa) [14]. This is in marked contrast to the majority of in vitro studies, where BCL-2 is depicted as a pro-survival or cancer-promoting factor [10,11]; however, BCL-2 has a variety of non-apoptotic functions in vitro [10,11,15-20] as does another BCL-2 family protein MCL1 [16,18,21]. BID has been demonstrated to have a role in inflammation and immunity independent of apoptosis [22]. In recent studies non-apoptotic roles of BAD were shown to include: blood glucose regulation, cooperation with p53 in the mitochondria, cell cycle regulation, and pro-survival functions [23-28]. Many of the proteins that have critical roles in apoptosis also have non-apoptotic functions, including cytochrome C, which is a key player in the intrinsic apoptosis pathway and is required for oxidative phosphorylation-linked electron transport. In addition to their well-established roles in apoptosis, functions for caspases have been described in cell-cycle entry, cell maturation, immune system function [29,30], differentiation [31], and other apoptosis-unrelated functions [32,33]. Other pro-apoptotic molecules, e.g. apoptosis inducing factor (AIF), Endo G and Omi [34,35] also have pro-survival effects [36,37].
As a continuation of our previous work on BAD in breast cancer cells [5,38], we evaluated the role of BAD in breast cancer both in vitro and in vivo. We demonstrated that BAD regulated several key molecules governing epithelial-mesenchymal transition (EMT), which thereby modulated the extra-cellular matrix invasion of breast cancer cells in vitro. This is the first demonstration of anti-invasive effects of a BCL-2 family protein in breast cancer cells.
Materials and Methods
Cell Lines and Plasmid vectors
MCF7 human breast cancer cell lines, and conditional and transient overexpression BAD constructs has been previously described [5,39,40].
Antibodies
Following antibodies were used in this study: BAD (H-168), phospho-AKT (S473), phospho-ERK1, ERK1, ERβ, Actin, Flag, phospho-c-Jun, c-Jun, phospho-JNK, JNK, phospho-p38, CXCR4, β-catenin, GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA); phospho-BAD (S136), Sp1 (Upstate, Lake Placid, NY); AKT (Cell Signaling, Danvers, MA); cyclin D1 (Novocastra Laboratories Ltd., Newcastle Upon Tyne, UK); SNAIL (Abcam, Cambridge, MA); GFP (Sigma, St. Louis, MO) and MTA3 antibody was described in [41]. Secondary antibodies: Amersham (San Diego, California) and Chemicon (Millipore, Billerica, MA).
Immunohistochemistry
Formalin-fixed and paraffin-embedded blocks of human normal (n=3) and neoplastic breast tissue (n=4) were purchased from SeraCare GCI Global Repository (West Bridgewater, MA) and breast tissue microarrays (TMAs) BR801, BR722 were purchased from US Biomax, Inc. (Rockville, MD). Slides were incubated overnight in humid chamber at 4°C with appropriate concentrations of primary antibodies. Incubation with diluent alone served as negative controls. Slides were washed and incubated with biotinylated Super Sensitive Link followed by HRP-conjugated Super Sensitive Label (BioGenex, Fremont, CA). Diaminobenzidine (Vector, Burlingame, CA) was used as the substrate. Nuclei were lightly counterstained with hematoxylin (Richard Allen Scientific, MI). Stained slides were reviewed by a certified pathologist (RLD) and the staining intensities were scored on a 0-3 scale (0=no staining, 1=mild staining, 2=moderate staining, 3=marked staining). The scores of the staining were averaged for statistical analyses.
Western Blot
Protein samples were separated on 10% SDS-PAGE and transferred to PVDF or nitrocellulose membranes. After blocking, membranes were incubated with primary antibodies, HRP-labeled secondary antibodies, and followed by ECL to detect proteins [5].
ELISA
ELISA for p-GSK3β (Ser 9/21), total GSK3β, p-AKT (Ser 473), total AKT and VEGF (Assay Gate Inc, Ijamsville, MD), and human MMP array (RayBiotech Inc. Norcross, GA) were carried out on a fee-for-service basis. E-Cadherin was quantified by PathScan ELISA Kit (cat#7886, Cell Signaling) and human STAT (CBEL-STAT-SK) ELISA kit from Ray Biotech was used to measure phospho/total STAT1, 3, and 5.
PCR
Total RNA was extracted using RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was synthesized using random hexamers (Applied Biosystems: Carlsbad, CA) and M-MuLV reverse transcriptase (New England BioLabs, Inc, Inswich, MA). The quantitative real-time PCR was carried out using following primers (SNAIL: QT00010010, Actin: QT01680475, Qiagen) and iQ SYBR Green RT Supermix and iCycler MyiQ Real Time PCR system (Bio-Rad, Hercules, CA). The results were normalized to endogenous control (actin). Fold changes were calculated in relation to reference control cDNA using the method described in [42]. Semi-quantitative reverse transcriptase-PCR was used to measure β-catenin and GAPDH mRNA as previously described [5].
Flow Cytometry
CXCR4 surface expression was measured using a Flowcellect flow cytometry kit (Millipore Billerica, MA). Cells were fixed in paraformaldehyde, washed and stained with mouse anti-human CXCR4 at 4°C. Cells were stained with Phycoerythrin conjugated secondary antibody and analyzed by flow cytometry.
Invasion Assay
Cells were incubated in the presence or absence of tetracycline in starving media for 24hrs. Quiescent cells were placed on the upper wells of a Matrigel-coated invasion chambers (BD Biosciences, Bedford, MA). Bottom wells contained serum. The chambers were incubated for 24hrs at 37°C and 5% CO2. Cells on the upper surface were removed and the cells invading the underside of the membrane were fixed and stained with propidium iodide and quantified. The average of n=3 (biological replicates) with 2 technical replicates for each experiment is shown as described in [43].
BAD siRNA transfection
MCF7 cells were transfected with BAD siRNA (Santa Cruz) and control scrambled siRNA (Sigma Aldrich) using Lipofectamine (Life Technologies). Cells were serum-starved 24h following transfection and subjected to invasion assays and western blot analysis.
Statistical analysis
The scored intensities from IHC data from normal and neoplastic breast tissue were analyzed using SAS software (Cary, NC). The BR801 tissue array had both normal and matching neoplastic breast tissue from same patients, so a randomized block design, blocking on patient (effectively a paired t-test) was used to analyze BAD, p-BAD, AKT, p-AKT, ERK1, and p-ERK1 expression levels. Markers were not paired, and a one-way model was used to analyze the data. Responses were categorical scores, but the measure of mean response was desired, so analysis of variance with rank transformation was used for the non-normally distributed scores. Means of untransformed data are reported, and Fisher's protected LSD was used to compare means. Simple Spearman rank correlations were calculated among all staining scores for each marker on the same tissue sample, and correlations were compared using Fisher's z-transformation.
Results
BAD and p-BAD is down-regulated in neoplastic breast epithelium
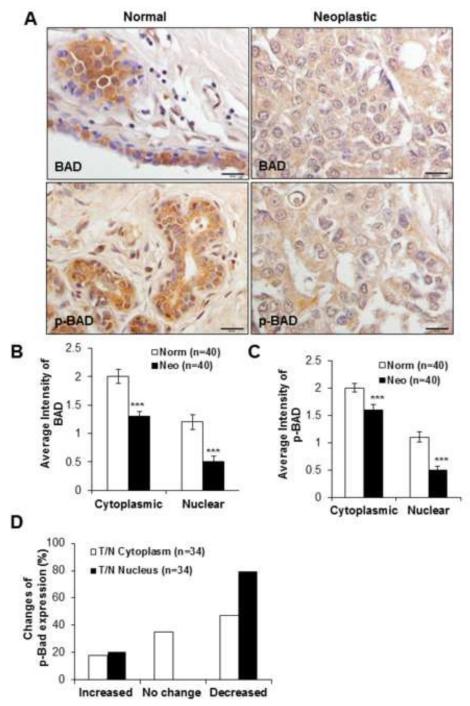
Breast tissue microarrays (TMA) were analyzed for BAD, phospho-BAD (p-BAD) by immunohistochemistry. Significantly less staining intensities for BAD and p-BAD were found in cancer than in normal breast tissue (p<0.01) and in both cases the expression of BAD in the cytoplasm exceeded that of nuclei (Figures 1A-C). In Grade II cancers (n=34) a decreased expression of p-BAD was found in 47% of cytoplasm and 80% of nuclei (Figure 1D) compared to normal tissue (p<0.001, Fischer’s exact test). These results demonstrated that BAD and p-BAD expressions are decreased both in the cytoplasm and nuclei of human breast cancer tissues.
Figure 1. BAD and p-BAD expressions in normal and neoplastic breast tissues by IHC.
(A) Down-regulation of BAD and p-BAD expressions are shown in neoplastic and normal breast tissue. Objective 40X, scale bar 50μm. (B) Semi-quantitative evaluation of IHC staining intensities of BAD and (C) p-BAD in normal and neoplastic breast tissues with the average intensities of cytoplasmic and nuclear expression of BAD and p-BAD. (D) Percentage changes of p-BAD expression in Grade II tumors compared to normal tissue differed between cytoplasm and nucleus (n=34). T=tumor, N=normal; difference significant at ***p<0.001 by Fisher’s exact test. Values represent the mean ± S.D. *p<0.05, ***p<0.001 by Student’s t-test.
BAD regulates functions associated with invasiveness in breast cancer
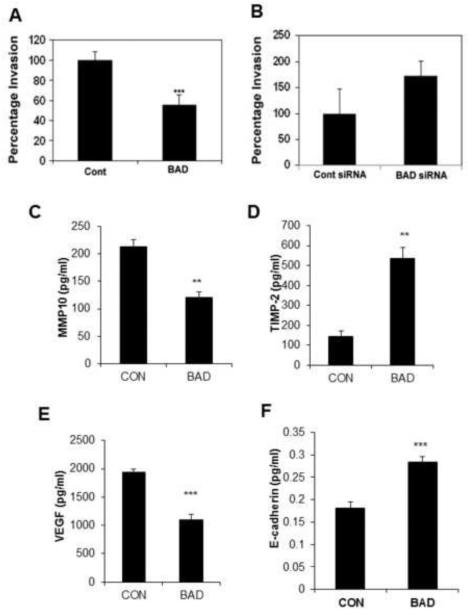
Cyclin D1 and c-Jun promote breast cancer cell invasion [44-46] and BAD inhibits both these proteins [5]; therefore, we measured the effects of BAD on cancer cell invasion and on the expression of proteins that regulate metastasis. Overexpression of BAD significantly reduced MCF7 cell invasion through Matrigel by approximately 50% (Figure 2A). As predicted, partial silencing of BAD by siRNA led to a 70% increase of MCF7 cell invasion through Matrigel (Figure 2B). The relative BAD expression in the overexpressing or knockdown cells were confirmed by western blotting analysis.
Figure 2. BAD regulates functions associated with invasiveness.
(A) Invasion by MCF7 cells was measured following BAD induction for 72 hrs. (B) Invasion by MCF7 cells was measured following silencing BAD by siRNA for 72 hrs. (C) MMP10, (D) TIMP2, (E) VEGF, and (F) E-cadherin were measured by ELISA in cell lysates of control and BAD induced cells (n=3). Values represent the mean ± S.E. **p<0.01, ***p<0.001 by Student’s t-test.
Although MCF7 cells are considered to be minimally invasive, this cell line was established from a pleural effusion [47] attesting to their invasiveness in vivo. We have previously used RNA interference to demonstrate that BAD regulates cyclin D1 and c-jun in MCF7 cells [5]. In these cells, a decrease in MMP10 (Figure 2C) was found coupled with increased secretion of the MMP inhibitor TIMP-2 (Figure 2D) that correlates with improved survival in breast cancer patients [48]. In addition, the production of pro-angiogenic VEGF, which correlates with increased cancer metastasis [49] was significantly reduced by the expression of BAD (Figure 2E). Increased E-cadherin expression has been shown to correlate with better prognosis in various cancer types. Overexpression of BAD induced E-cadherin (Figure 2F), which correlated with reduced extracellular matrix invasion in MCF7 cells (Figure 2A). Other proteins measured by the MMP array, as described in Material and Methods section in detail, were unchanged by BAD overexpression (data not shown).
BAD regulates β-catenin, STAT and AKT activities
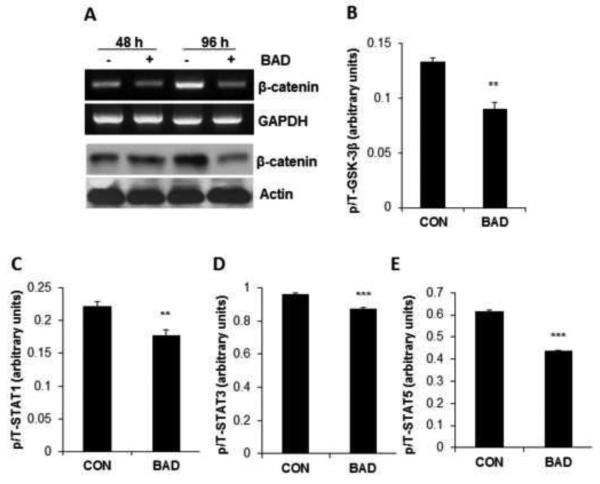
Previously, we demonstrated by overexpression, as well by RNA interference that BAD suppresses cyclin D1 via repression of the cyclin D1 promoter and BAD reduces cyclin D1 abundance induced in response to estradiol or serum via a negative regulation of c-Jun [5]. In the present study, we investigated whether BAD could regulate β-catenin, another inducer of cyclin D1 [50] in breast cancer cells. A decrease in β-catenin mRNA and protein was observed simultaneously (Figure 3A). Phosphorylation of β-catenin by GSK-3β induces its degradation and GSK-3β is inactivated by AKT/PKB phosphorylation [51]. We measured the effects of BAD on GSK-3β phosphorylation, and a significant decrease in p-GSK-3β was found in BAD over-expressing cells (Figure 3B) without a change in total GSK-3β (Supplemental Figure 1B) indicative of GSK-3β activation. STAT family proteins regulate the cell cycle in breast cancer cells [52,53]. ELISA analyses showed a significant down-regulation of both phosphorylated and total STAT1 and STAT3 in BAD over-expressing cells (Supplemental Figures 2A and 2B) resulting in a reduced p/T ratio (Figures 3C and 3D). In contrast, a decrease only in p-STAT5 but not in total STAT5 (Supplemental Figure 2C) was noted with reduced p/T ratio STAT5 (Figure 3E).
Figure 3. BAD controls expression of β-catenin signaling pathway.
(A) BAD was induced in MCF7 cells for 48 or 96 hrs. β-catenin mRNA was measured by RT-PCR with GAPDH expression measured as control. β-catenin protein was measured by western blot with actin as loading control. (B-E) Phospho and total forms of GSK-3β, STAT1, STAT3 and STAT5 were measured by ELISA. Phospho/total (p/T) ratio of each protein in control vs. BAD-overexpressing cells are shown (n=3). Values represent the mean ± S.E. **p<0.01, ***p<0.001 by Student’s t-test.
BAD regulates EMT-related proteins
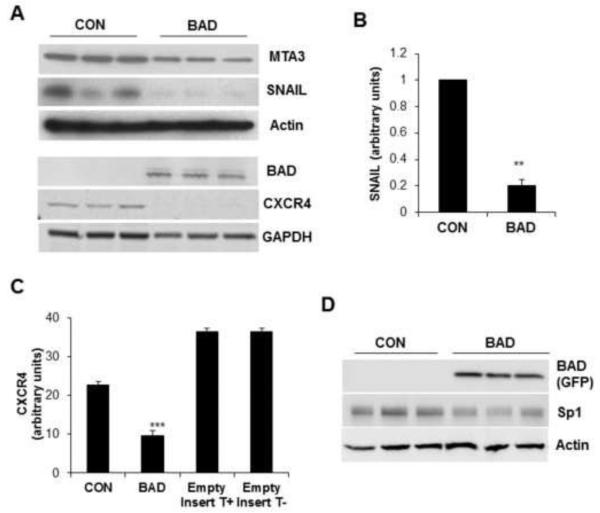
SNAIL, a well characterized inducer of epithelial to mesenchymal transition (EMT) [54], was inhibited by BAD as demonstrated by mRNA and protein expression (Figures 4A and 4B). Further, the expression of MTA3, an upstream regulator of SNAIL [41], was also blocked by BAD (Figure 4A). This observation together with the increased E-cadherin expression in these cells (Figure 2F), suggested that the transcriptional repression of the E-cadherin promoter exerted by SNAIL [54] may be antagonized by BAD. CXCL12/SDF1, a c-Jun target and its receptor CXCR4, play significant roles in breast cancer metastasis [46]. Our results demonstrated a significant down-regulation of CXCR4 by BAD (Figure 4A and 4C). In addition, Sp1, a transcription factor required for EMT, was inhibited by BAD as shown in Figure 4D.
Figure 4. BAD regulates EMT-related proteins.
(A) Expression of MTA3, SNAIL, and CXCR4 were assayed by WB in control and conditionally BAD induced MCF7 cells (n=3). Expression of actin and GAPDH are shown as protein loading controls. (B) Relative SNAIL mRNA expression was determined by qPCR in 3 individual experiments with comparable results; representative results from one experiment are shown. (C) Flow cytometric analysis of CXCR4 in control and conditionally BAD-induced MCF7 cells. MCF7 cells containing the tetracycline regulator, but not the BAD insert, were used as a control to show lack of effect of tetracycline by itself. Experiment was repeated 4 times with comparable results. Values represent the mean ± S.E. **p<0.01, ***p<0.001 by Student’s t-test. (D) Expression of Sp1 was assayed by WB in control and conditionally BAD induced MCF7 cells (n=3). Actin was used as loading control.
BAD regulates ERK and AKT signaling pathways
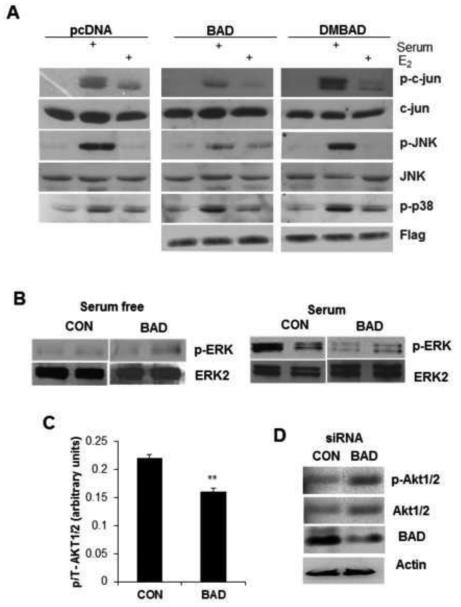
Since BAD interacts selectively with un-phosphorylated c-Jun [5], we investigated whether BAD could also regulate signal pathways that phosphorylate c-Jun. We hypothesized that BAD may inhibit the Ras-MEKK-MEK-ERK and JNK pathways selectively [5]. c-Jun is phosphorylated and activated by JNK, ERK, and a variety of other kinases [55]. As shown in Figure 5A, BAD decreased the phosphorylation of c-Jun and JNK without affecting the phosphorylation of p38, showing a degree of specificity. The inhibitory effects of BAD on the activation of JNK/c-Jun by E2 are broadly similar to that of serum. The BAD double mutant, in which both serine residues are mutated to alanine (DMBAD S75A/S99A), completely reversed the effect of BAD (Figure 5A). Inhibition of ERK by conditional over-expression of BAD is demonstrated in Figure 5B.
Figure 5. BAD regulates ERK and AKT pathways.
(A) MCF7 cells were transiently transfected with indicated plasmid vectors and were growth arrested with 10nM ICI and 0.1% serum for 48 h prior to stimulation with 5% serum or 10 nM E2. Whole cell lysate (50 μg) of each sample was resolved using SDS-PAGE and immunoblotted with indicated antibodies. (B) After BAD induction for 72 hrs, cells were serum-starved for 24 hrs and stimulated with serum for 1 hr (experiments in duplicate). Lysates from BAD induced and control cells were probed with p-ERK and ERK antibodies. (C) Phospho and total AKT were measured by ELISA and phospho/total (p/T) ratios are shown. Values represent the mean ± S.E. **p<0.01, ***p<0.001 (n=3). (D) Induction of AKT after silencing BAD by siRNA.
BAD specifically inhibited MEK-dependent ERK1/2 activation, but not Myr-AKT-induced ERK activation (Supplemental Figure 4). We examined the effects of BAD on AKT as AKT1 promotes migration of breast cancer cells [56]. BAD induced total AKT (Supplemental Figure 1A) and reduced phospho/total (p/T) ratio. Thus BAD inhibits relative p-AKT (Figure 5C). Silencing of BAD by siRNA increased p-AKT levels (and total AKT to a lesser extent), resulting in restoring the activation of AKT (Figure 5D). Thus BAD inactivates AKT, which may activate GSK-3β (Figure 3B) causing reduction of β-catenin (Figure 3A).
As BAD inhibited ERK1/2 activation, we determined the expression pattern of the signal molecules (ERK, phosphorylated ERK, AKT and phosphorylated AKT) in breast tissue blocks by immunohistochemistry. Higher ERK and a significantly less p-ERK expression were evident in the neoplastic compared to normal cells (Supplemental Figure 3A-D). Reduced AKT and increased p-AKT were found in neoplastic cytoplasm (Supplemental Fig 3E-H).
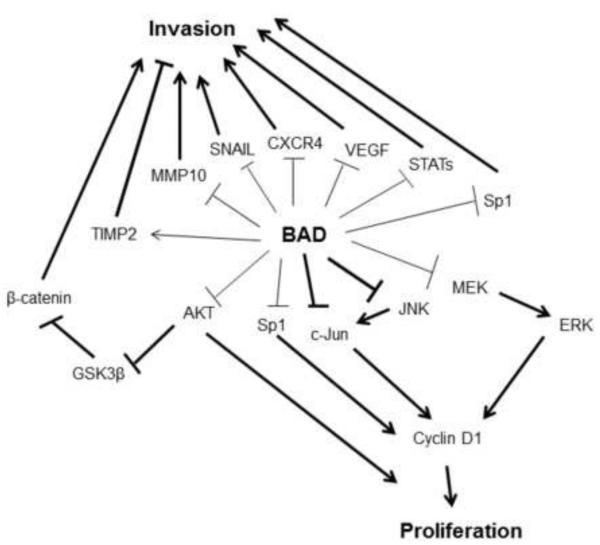
Taken together, our data suggest that in addition to the previously known cell cycle and apoptotic effects BAD also influences breast cancer cell invasion and epithelial to mesenchymal transition (Figure 6).
Figure 6. Scheme of proposed summary of BAD regulation of diverse gene expressions regulating cell invasion and proliferation in breast cancer identified in this study.
Discussion
Studies over the last decade have shown that many apoptosis regulatory proteins have additional functions unrelated to apoptosis. Previously, we demonstrated that BAD enforced a G1 phase block via inhibition of cyclin D1 synthesis in breast cancer cells. BAD is located in the cytoplasm and in the nucleus in normal breast tissue suggesting a physiological role for nuclear BAD [5]. In this report, we have extended these observations and demonstrate a novel role for BAD to inhibit breast cancer cell invasion and EMT. To our knowledge, this is the first report to demonstrate that BCL-2 family proteins have both anti-invasive and EMT-inhibiting effects in breast cancer cells. These findings may underlie the association of decreased BAD/p-BAD expression in breast cancer and the association of prolonged survival with enhanced BAD expression [6,7]. Novel findings of changes in p-BAD and relative changes in p-BAD/BAD in the nucleus vs cytoplasm in tumor vs normal (Figure 1) support a model, in which BAD could inhibit breast cancer metastasis. High BAD expression is associated with longer disease free survival, overall survival [6], and longer time to relapse in tamoxifen-treated breast cancer patients [57], and a role for BAD as a good prognostic indicator has been reported for gastric, hepatocellular and colon carcinomas [58-62]. Although in prostate cancer, BAD accelerated tumor growth in prostate cancer C4-2 xenografts [26].
Premenopausal breast carcinoma in younger women is more aggressive with a higher potential for invasion and metastasis and poor prognosis compared to postmenopausal breast carcinoma [63]. Interestingly, BCL-2 and BAD expression in premenopausal breast carcinoma was significantly lower than in post-menopausal breast carcinoma and this decrease in proteins correlated with progression from Grade I to III [64]. It is noteworthy that both BCL-2 and its antagonist BAD decreased with increasing severity of the disease.
The mechanisms, by which BAD regulates EMT-related gene expression remains to be elucidated. BAD may regulate gene expression via several proteins, including c-Jun, cyclin D1, β-catenin, and Sp1. The transcription factor Sp1 that regulates SNAIL expression (Figure 4A) is also an activator of the cyclin D1 promoter [65].
Several proteins related to tumor cell invasiveness, EMT, and metastases were down-regulated by BAD (MTA3, SNAIL, and CXCR4 as shown in Figure 4) with concomitant inhibition of MCF7 cell invasion (Figure 2). We showed that c-Jun was inhibited by BAD [5] and c-Jun is known to promote cellular migration and invasion [45,46]. As patients with breast cancers expressing higher levels of BAD protein had a longer survival [6], and survival is directly related to metastasis, it is plausible that increased expression of BAD could be associated with a lowered metastatic potential. c-Jun as well as cyclin D1 (inhibited by BAD) are known to promote migration and invasion by breast cancer cells (vide infra). Previous studies have shown that BH3 only protein BMF may regulate anoikis, which also plays a role in invasiveness at least in vitro [66]. In this study, BAD had an opposite effect to that of BIM and BMF and decreased anoikis. A reversal or inhibition of EMT by BAD was further evident by its stimulatory effect on E-cadherin expression (Figure 4D). Increased E-cadherin expression correlates with better prognosis in many cancer types, and E-cadherin transcription is inhibited by SNAIL [54]. In addition, Sp1, a transcription factor required for EMT, was inhibited by BAD. Collectively, our data indicated a role for BAD in the reduction of cancer progression and as an inhibitor of EMT, cancer cell migration, and invasion.
In vitro data supports the a pro-invasive role for BCL-2 and its pro-survival partner BCLxL [67-70] or anti-invasive role for BCL-2 [71]. Most in vitro results suggest an anti-apoptotic role for BCL-2, yet expression correlates with improved prognosis. Increased BCL-2 and BAD expression correlate with improved outcome in breast cancer. Given the anti-invasive effects of BCL-2 in vivo, the effects of synthetic BCL-2 antagonists, should be further examined.
Conclusion
We have presented clinical data showing that p-BAD and BAD expression is decreased in breast cancer compared with normal breast tissue. BAD impedes breast cancer invasion and migration correlating with the inhibition of EMT and transcription factors that promote breast cancer cell migration. These functions are distinct from the role of BAD to promote mitochondria-mediated apoptosis.
Supplementary Material
Supplemental Figure 1: Regulation of AKT and GSK-3β by BAD. (A) Phospho-AKT and total AKT (n=4), (B) Phospho-GSK-3β and total GSK-3β (n=3) were measured by ELISA in lysates from BAD-induced or control cells. Values represent the mean ± S.E. **p <0.01, **p<0.01, ***p<0.001 by Student’s t-test compared to control.
Supplemental Figure 2: Regulation of STAT1, 3, 5 by BAD. (A-B) The activities of STAT1, phospho-STAT1 were measured in cell lysates by ELISA following induction of BAD for 72hrs. (C-F) Similar measurements of STAT3 and STAT5 in the same lysates (n=3 for each STAT). Values represent the mean ± S.E. ***p<0.001 by Student’s t-test compared to control.
Supplemental Figure 3: Immunohistochemical staining showing expression of (A and B) ERK, (C and D) phospho-ERK (p-ERK); (E and F) AKT, and (G, H) phospho-AKT (p-AKT) in normal and neoplastic breast epithelia (n=7). Magnification objective 40X, scale bar 50μm.
Supplemental Figure 4: BAD specifically inhibits MEK dependent ERK1/2 activation, but not Myr-AKT-induced ERK activation. MCF7 cells were transiently transfected with indicated plasmid vectors and were growth for 24h. Whole cell lysates were probed with p-ERK and ERK antibodies. Expression of ERK are shown as protein loading controls.
BAD and p-BAD expressions are decreased in breast cancer compared with normal breast tissue.
BAD impedes breast cancer invasion and migration.
BAD inhibits the EMT and transcription factors that promote cancer cell migration.
Invasion and migration functions of BAD are distinct from the BAD’s role in apoptosis.
Acknowledgement
This work was supported partially by NIH grant (R01CA84048, PI: Wimalasena), University of Tennessee Graduate School of Medicine, Medical Center (PI: Wimalasena), University of Tennessee Graduate School of Medicine Physician’s Medical Education and Research Foundation (R084025002, PI: Wimalasena, and R181721242, PI: Cekanova). Dr. Jay Wimalasena is thankful to undergraduate students of UT: Erica Smith, Rhett Layman, and Blair Tatge for their technical assistance.
Abbreviations
- AIF
apoptosis inducible factor
- AP-1
activator protein-1
- AKT
protein kinase B
- Apaf-1
apoptosis protease activating factor-1
- BAD
Bcl-2-associated death promoter
- BCL-2
B-cell lymphoma 2
- BCLxL
B-cell lymphoma-extra large
- BH3
Bcl-2 homology domain 3
- BRCA1
breast cancer type 1 susceptibility protein
- CDK4
cyclin-dependent kinase-4
- CXCL12/SDF1
stromal cell -derived factor-1
- CXCR4
chemokine receptor type 4
- DM
double mutant
- ECL
enhanced chemiluminescence
- EGFP
enhance GFP
- EMSA
electrophoretic mobility shift assay
- EMT
epithelial-mesenchymal transition
- ERa
estrogen receptor a
- ERβ
estrogen receptor β
- ERK
extracellular signal-regulated kinases
- FADD
Fas-associated protein with death domain
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescent protein
- GSK3β
glycogen synthase kinase 3 beta
- HER2
human epidermal growth factor receptor-2
- HIF
Hypoxia-inducible factor 1, alpha subunit
- HRP
horseradish peroxidase
- IHC
immunohistochemistry
- p
phospho
- Ras/MEK/ERK
MAPK signaling pathway
- JNK
c-Jun kinase
- MCL1
myeloid leukemia cell differentiation protein-1
- MMP10
metalloproteinase-10
- MTA3
metastasis-associated protein-3
- Rb
retinoblastoma protein
- SNP
single-nucleotide polymorphism
- Sp1
specificity protein-1
- STAT
Signal transducer and activator of transcription
- TMA
tissue microarrays
- TIMP2
metallopeptidase inhibitor 2
- TRE
transcription response elements
- VEGF
Vascular endothelial growth factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interest section: The authors have declared that no conflict of interest exists.
References
- 1.Krajewski S, Tanaka S, Takayama S, Schibler MJ, Fenton W, Reed JC. Investigation of the subcellular distribution of the bcl-2 oncoprotein: residence in the nuclear envelope, endoplasmic reticulum, and outer mitochondrial membranes. Cancer Res. 1993;53:4701–4714. [PubMed] [Google Scholar]
- 2.Strasser A, O'Connor L, Dixit VM. Apoptosis signaling. Annu Rev Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- 3.Akao Y, Otsuki Y, Kataoka S, Ito Y, Tsujimoto Y. Multiple subcellular localization of bcl-2: detection in nuclear outer membrane, endoplasmic reticulum membrane, and mitochondrial membranes. Cancer Res. 1994;54:2468–2471. [PubMed] [Google Scholar]
- 4.Yang Q, Sakurai T, Jing X, Utsunomiya H, Shan L, Nakamura Y, Nakamura M, Oura S, Suzuma T, Yoshimura G, Umemura T, Kokawa Y, Kakudo K. Expression of Bcl-2, but not Bax, correlates with estrogen receptor status and tumor proliferation in invasive breast carcinoma. Pathol Int. 1999;49:775–780. doi: 10.1046/j.1440-1827.1999.00942.x. [DOI] [PubMed] [Google Scholar]
- 5.Fernando R, Foster JS, Bible A, Strom A, Pestell RG, Rao M, Saxton A, Baek SJ, Yamaguchi K, Donnell R, Cekanova M, Wimalasena J. Breast cancer cell proliferation is inhibited by BAD: regulation of cyclin D1. J Biol Chem. 2007;282:28864–28873. doi: 10.1074/jbc.M700785200. [DOI] [PubMed] [Google Scholar]
- 6.Al-Bazz YO, Underwood JC, Brown BL, Dobson PR. Prognostic significance of Akt, phospho-Akt and BAD expression in primary breast cancer. Eur J Cancer. 2009;45:694–704. doi: 10.1016/j.ejca.2008.11.044. [DOI] [PubMed] [Google Scholar]
- 7.Fernando R, Cekanova M, Worraratphoka J, Sukhthankar M, Siriwardhana N, Moustaid-Moussa N, Baek SJ, Donnel R, Strom A, Wang Q, Zou M, Wimalasena J. BAD is a multifunctional protein in breast cancer cells. Cancer Res; Proceedings of the 101st Annual Meeting of the American Association for Cancer Research; Washington, DC Philadelphia (PA): AACR; Apr 17-21, 2010. (2010) Abstract nr 1046. [Google Scholar]
- 8.Massaad CA, Portier BP, Taglialatela G. Inhibition of transcription factor activity by nuclear compartment-associated Bcl-2. J Biol Chem. 2004;279:54470–54478. doi: 10.1074/jbc.M407659200. [DOI] [PubMed] [Google Scholar]
- 9.Choi DH, Kim S, Rimm DL, Carter D, Haffty BG. Immunohistochemical biomarkers in patients with early-onset breast carcinoma by tissue microarray. Cancer J. 2005;11:404–411. doi: 10.1097/00130404-200509000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Huang DC, O'Reilly LA, Strasser A, Cory S. The anti-apoptosis function of Bcl-2 can be genetically separated from its inhibitory effect on cell cycle entry. EMBO J. 1997;16:4628–4638. doi: 10.1093/emboj/16.15.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 12.Park SH, Kim H, Song BJ. Down regulation of bcl2 expression in invasive ductal carcinomas is both estrogen- and progesterone-receptor dependent and associated with poor prognostic factors. Pathol Oncol Res. 2002;8:26–30. doi: 10.1007/BF03033697. [DOI] [PubMed] [Google Scholar]
- 13.Castiglione F, Sarotto I, Fontana V, Destefanis M, Venturino A, Ferro S, Cardaropoli S, Orengo MA, Porcile G. Bcl2, p53 and clinical outcome in a series of 138 operable breast cancer patients. Anticancer Res. 1999;19:4555–4563. [PubMed] [Google Scholar]
- 14.Callagy GM, Webber MJ, Pharoah PD, Caldas C. Meta-analysis confirms BCL2 is an independent prognostic marker in breast cancer. BMC Cancer. 2008;8:153. doi: 10.1186/1471-2407-8-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chattopadhyay A, Chiang CW, Yang E. BAD/BCL-[X(L)] heterodimerization leads to bypass of G0/G1 arrest. Oncogene. 2001;20:4507–4518. doi: 10.1038/sj.onc.1204584. [DOI] [PubMed] [Google Scholar]
- 16.Crescenzi E, Palumbo G, Brady HJ. Bcl-2 activates a programme of premature senescence in human carcinoma cells. Biochem J. 2003;375:263–274. doi: 10.1042/BJ20030868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linette GP, Li Y, Roth K, Korsmeyer SJ. Cross talk between cell death and cell cycle progression: BCL-2 regulates NFAT-mediated activation. Proc Natl Acad Sci U S A. 1996;93:9545–9552. doi: 10.1073/pnas.93.18.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trisciuoglio D, Gabellini C, Desideri M, Ziparo E, Zupi G, Del Bufalo D. Bcl-2 regulates HIF-1alpha protein stabilization in hypoxic melanoma cells via the molecular chaperone HSP90. PLoS ONE. 2010;5:e11772. doi: 10.1371/journal.pone.0011772. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Vairo G, Innes KM, Adams JM. Bcl-2 has a cell cycle inhibitory function separable from its enhancement of cell survival. Oncogene. 1996;13:1511–1519. [PubMed] [Google Scholar]
- 20.Vairo G, Soos TJ, Upton TM, Zalvide J, DeCaprio JA, Ewen ME, Koff A, Adams JM. Bcl-2 retards cell cycle entry through p27(Kip1), pRB relative p130, and altered E2F regulation. Mol Cell Biol. 2000;20:4745–4753. doi: 10.1128/mcb.20.13.4745-4753.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujise K, Zhang D, Liu J, Yeh ET. Regulation of apoptosis and cell cycle progression by MCL1. Differential role of proliferating cell nuclear antigen. J Biol Chem. 2000;275:39458–39465. doi: 10.1074/jbc.M006626200. [DOI] [PubMed] [Google Scholar]
- 22.Yeretssian G, Correa RG, Doiron K, Fitzgerald P, Dillon CP, Green DR, Reed JC, Saleh M. Non-apoptotic role of BID in inflammation and innate immunity. Nature. 2011;474:96–99. doi: 10.1038/nature09982. [DOI] [PubMed] [Google Scholar]
- 23.Jiang P, Du W, Heese K, Wu M. The Bad guy cooperates with good cop p53: Bad is transcriptionally up-regulated by p53 and forms a Bad/p53 complex at the mitochondria to induce apoptosis. Mol Cell Biol. 2006;26:9071–9082. doi: 10.1128/MCB.01025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danial NN. BAD: undertaker by night, candyman by day. Oncogene. 2008;27(Suppl 1):S53–70. doi: 10.1038/onc.2009.44. [DOI] [PubMed] [Google Scholar]
- 25.Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, Gygi SP, Korsmeyer SJ. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- 26.Smith AJ, Karpova Y, D'Agostino R, Jr., Willingham M, Kulik G. Expression of the Bcl-2 protein BAD promotes prostate cancer growth. PLoS ONE. 2009;4:e6224. doi: 10.1371/journal.pone.0006224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craik AC, Veldhoen RA, Czernick M, Buckland TW, Kyselytzia K, Ghosh S, Lai R, Damaraju S, Underhill DA, Mackey JR, Goping IS. The BH3-only protein Bad confers breast cancer taxane sensitivity through a nonapoptotic mechanism. Oncogene. 2010;29:5381–5391. doi: 10.1038/onc.2010.272. [DOI] [PubMed] [Google Scholar]
- 28.Seo SY, Chen YB, Ivanovska I, Ranger AM, Hong SJ, Dawson VL, Korsmeyer SJ, Bellows DS, Fannjiang Y, Hardwick JM. BAD is a pro-survival factor prior to activation of its pro-apoptotic function. J Biol Chem. 2004;279:42240–42249. doi: 10.1074/jbc.M406775200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nhan TQ, Liles WC, Schwartz SM. Physiological functions of caspases beyond cell death. Am J Pathol. 2006;169:729–737. doi: 10.2353/ajpath.2006.060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM, Dale JK, Puck J, Davis J, Hall CG, Skoda-Smith S, Atkinson TP, Straus SE, Lenardo MJ. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- 31.Miura M, Xiao-Dong C, Allen MR, Yanming B, Gronthos S, Byoung-Moo S, Lakhani S, Flavell RA, Feng X-H, Robey PG, Young M, Shi S. A crucial role of caspase-3 in osteogenic differentiation of bone marrow stromal stem cells. Journal of Clinical Investigation. 2004;114:1704–1713. doi: 10.1172/JCI20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galluzzi L, Joza N, Tasdemir E, Maiuri MC, Hengartner M, Abrams JM, Tavernarakis N, Penninger J, Madeo F, Kroemer G. No death without life: vital functions of apoptotic effectors. Cell Death Differ. 2008;15:1113–1123. doi: 10.1038/cdd.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saleh M, Vaillancourt JP, Graham RK, Huyck M, Srinivasula SM, Alnemri ES, Steinberg MH, Nolan V, Baldwin CT, Hotchkiss RS, Buchman TG, Zehnbauer BA, Hayden MR, Farrer LA, Roy S, Nicholson DW. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 2004;429:75–79. doi: 10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- 34.Vahsen N, Cande C, Briere JJ, Benit P, Joza N, Larochette N, Mastroberardino PG, Pequignot MO, Casares N, Lazar V, Feraud O, Debili N, Wissing S, Engelhardt S, Madeo F, Piacentini M, Penninger JM, Schagger H, Rustin P, Kroemer G. AIF deficiency compromises oxidative phosphorylation. EMBO J. 2004;23:4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang KJ, Ku CC, Lehman IR. Endonuclease G: a role for the enzyme in recombination and cellular proliferation. Proc Natl Acad Sci U S A. 2006;103:8995–9000. doi: 10.1073/pnas.0603445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Rosenberg S, Wang H, Imtiyaz HZ, Hou YJ, Zhang J. Conditional Fas-associated death domain protein (FADD): GFP knockout mice reveal FADD is dispensable in thymic development but essential in peripheral T cell homeostasis. J Immunol. 2005;175:3033–3044. doi: 10.4049/jimmunol.175.5.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeh WC, Pompa JL, McCurrach ME, Shu HB, Elia AJ, Shahinian A, Ng M, Wakeham A, Khoo W, Mitchell K, El-Deiry WS, Lowe SW, Goeddel DV, Mak TW. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science. 1998;279:1954–1958. doi: 10.1126/science.279.5358.1954. [DOI] [PubMed] [Google Scholar]
- 38.Fernando RI, Wimalasena J. Estradiol abrogates apoptosis in breast cancer cells through inactivation of BAD: Ras-dependent nongenomic pathways requiring signaling through ERK and Akt. Mol Biol Cell. 2004;15:3266–3284. doi: 10.1091/mbc.E03-11-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polzien L, Baljuls A, Rennefahrt UEE, Fischer A, Schmitz W, Zahedi RP, Sickmann A, Metz R, Albert S, Benz R, Hekman M, Rapp UR. Identification of Novel in Vivo Phosphorylation Sites of the Human Proapoptotic Protein BAD: PORE-FORMING ACTIVITY OF BAD IS REGULATED BY PHOSPHORYLATION. Journal of Biological Chemistry. 2009;284:28004–28020. doi: 10.1074/jbc.M109.010702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 41.Fujita N, Kajita M, Taysavang P, Wade PA. Hormonal regulation of metastasis-associated protein 3 transcription in breast cancer cells. Mol Endocrinol. 2004;18:2937–2949. doi: 10.1210/me.2004-0258. [DOI] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Castillo-Pichardo L, Martinez-Montemayor MM, Martinez JE, Wall KM, Cubano LA, Dharmawardhane S. Inhibition of mammary tumor growth and metastases to bone and liver by dietary grape polyphenols. Clin Exp Metastasis. 2009;26:505–516. doi: 10.1007/s10585-009-9250-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Jiao X, Wang C, Ju X, Lu Y, Yuan L, Lisanti MP, Katiyar S, Pestell RG. Cyclin D1 induction of cellular migration requires p27(KIP1) Cancer Res. 2006;66:9986–9994. doi: 10.1158/0008-5472.CAN-06-1596. [DOI] [PubMed] [Google Scholar]
- 45.Jiao X, Katiyar S, Willmarth NE, Liu M, Ma X, Flomenberg N, Lisanti MP, Pestell RG. c-Jun induces mammary epithelial cellular invasion and breast cancer stem cell expansion. J Biol Chem. 2010;285:8218–8226. doi: 10.1074/jbc.M110.100792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katiyar S, Jiao X, Wagner E, Lisanti MP, Pestell RG. Somatic excision demonstrates that c-Jun induces cellular migration and invasion through induction of stem cell factor. Mol Cell Biol. 2007;27:1356–1369. doi: 10.1128/MCB.01061-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soule HD, Vazguez J, Long A, Albert S, Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst. 1973;51:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 48.Nakopoulou L, Katsarou S, Giannopoulou I, Alexandrou P, Tsirmpa I, Panayotopoulou E, Mavrommatis J, Keramopoulos A. Correlation of tissue inhibitor of metalloproteinase-2 with proliferative activity and patients' survival in breast cancer. Mod Pathol. 2002;15:26–34. doi: 10.1038/modpathol.3880486. [DOI] [PubMed] [Google Scholar]
- 49.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, Pestell RG, Hung MC. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A. 2000;97:4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu D, Pan W. GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem Sci. 2010;35:161–168. doi: 10.1016/j.tibs.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 53.Matsumura I, Kitamura T, Wakao H, Tanaka H, Hashimoto K, Albanese C, Downward J, Pestell RG, Kanakura Y. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 1999;18:1367–1377. doi: 10.1093/emboj/18.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 56.Ju X, Katiyar S, Wang C, Liu M, Jiao X, Li S, Zhou J, Turner J, Lisanti MP, Russell RG, Mueller SC, Ojeifo J, Chen WS, Hay N, Pestell RG. Akt1 governs breast cancer progression in vivo. Proc Natl Acad Sci U S A. 2007;104:7438–7443. doi: 10.1073/pnas.0605874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cannings E, Kirkegaard T, Tovey SM, Dunne B, Cooke TG, Bartlett JM. Bad expression predicts outcome in patients treated with tamoxifen. Breast Cancer Res Treat. 2007;102:173–179. doi: 10.1007/s10549-006-9323-8. [DOI] [PubMed] [Google Scholar]
- 58.Galmiche A, Ezzoukhry Z, Francois C, Louandre C, Sabbagh C, Nguyen-Khac E, Descamps V, Trouillet N, Godin C, Regimbeau JM, Joly JP, Barbare JC, Duverlie G, Maziere JC, Chatelain D. BAD, a proapoptotic member of the BCL2 family, is a potential therapeutic target in hepatocellular carcinoma. Mol Cancer Res. 2010;8:1116–1125. doi: 10.1158/1541-7786.MCR-10-0029. [DOI] [PubMed] [Google Scholar]
- 59.Jeong EG, Lee SH, Kim SS, Ahn CH, Yoo NJ. Immunohistochemical analysis of phospho-BAD protein and mutational analysis of BAD gene in gastric carcinomas. APMIS. 2007;115:976–981. doi: 10.1111/j.1600-0463.2007.apm_804.x. [DOI] [PubMed] [Google Scholar]
- 60.Yoo NJ, Lee JW, Jeong EG, Soung YH, Nam SW, Lee JY, Lee SH. Expressional analysis of anti-apoptotic phospho-BAD protein and mutational analysis of pro-apoptotic BAD gene in hepatocellular carcinomas. Dig Liver Dis. 2006;38:683–687. doi: 10.1016/j.dld.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 61.Lee JW, Soung YH, Kim SY, Nam SW, Kim CJ, Cho YG, Lee JH, Kim HS, Park WS, Kim SH, Lee JY, Yoo NJ, Lee SH. Inactivating mutations of proapoptotic Bad gene in human colon cancers. Carcinogenesis. 2004;25:1371–1376. doi: 10.1093/carcin/bgh145. [DOI] [PubMed] [Google Scholar]
- 62.Sinicrope FA, Rego RL, Foster NR, Thibodeau SN, Alberts SR, Windschitl HE, Sargent DJ. Proapoptotic Bad and Bid protein expression predict survival in stages II and III colon cancers. Clin Cancer Res. 2008;14:4128–4133. doi: 10.1158/1078-0432.CCR-07-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Agrup M, Stal O, Olsen K, Wingren S. C-erbB-2 overexpression and survival in early onset breast cancer. Breast Cancer Res Treat. 2000;63:23–29. doi: 10.1023/a:1006498721508. [DOI] [PubMed] [Google Scholar]
- 64.Yu B, Sun X, Shen HY, Gao F, Fan YM, Sun ZJ. Expression of the apoptosis-related genes BCL-2 and BAD in human breast carcinoma and their associated relationship with chemosensitivity. Journal of Experimental & Clinical Cancer Research. 2010;29 doi: 10.1186/1756-9966-29-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marampon F, Casimiro MC, Fu M, Powell MJ, Popov VM, Lindsay J, Zani BM, Ciccarelli C, Watanabe G, Lee RJ, Pestell RG. Nerve Growth factor regulation of cyclin D1 in PC12 cells through a p21RAS extracellular signal-regulated kinase pathway requires cooperative interactions between Sp1 and nuclear factor-kappaB. Mol Biol Cell. 2008;19:2566–2578. doi: 10.1091/mbc.E06-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmelzle T, Mailleux AA, Overholtzer M, Carroll JS, Solimini NL, Lightcap ES, Veiby OP, Brugge JS. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc Natl Acad Sci U S A. 2007;104:3787–3792. doi: 10.1073/pnas.0700115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim H, Chung H, Kim HJ, Lee JY, Oh MY, Kim Y, Kong G. Id-1 regulates Bcl-2 and Bax expression through p53 and NF-kappaB in MCF-7 breast cancer cells. Breast Cancer Res Treat. 2008;112:287–296. doi: 10.1007/s10549-007-9871-6. [DOI] [PubMed] [Google Scholar]
- 68.Pinkas J, Martin SS, Leder P. Bcl-2-mediated cell survival promotes metastasis of EpH4 betaMEKDD mammary epithelial cells. Mol Cancer Res. 2004;2:551–556. [PubMed] [Google Scholar]
- 69.Rahman KM, Sarkar FH, Banerjee S, Wang Z, Liao DJ, Hong X, Sarkar NH. Therapeutic intervention of experimental breast cancer bone metastasis by indole-3-carbinol in SCID-human mouse model. Mol Cancer Ther. 2006;5:2747–2756. doi: 10.1158/1535-7163.MCT-06-0221. [DOI] [PubMed] [Google Scholar]
- 70.Mantena SK, Baliga MS, Katiyar SK. Grape seed proanthocyanidins induce apoptosis and inhibit metastasis of highly metastatic breast carcinoma cells. Carcinogenesis. 2006;27:1682–1691. doi: 10.1093/carcin/bgl030. [DOI] [PubMed] [Google Scholar]
- 71.Ke H, Parron VI, Reece J, Zhang JY, Akiyama SK, French JE. BCL2 inhibits cell adhesion, spreading, and motility by enhancing actin polymerization. Cell Res. 2010;20:458–469. doi: 10.1038/cr.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Regulation of AKT and GSK-3β by BAD. (A) Phospho-AKT and total AKT (n=4), (B) Phospho-GSK-3β and total GSK-3β (n=3) were measured by ELISA in lysates from BAD-induced or control cells. Values represent the mean ± S.E. **p <0.01, **p<0.01, ***p<0.001 by Student’s t-test compared to control.
Supplemental Figure 2: Regulation of STAT1, 3, 5 by BAD. (A-B) The activities of STAT1, phospho-STAT1 were measured in cell lysates by ELISA following induction of BAD for 72hrs. (C-F) Similar measurements of STAT3 and STAT5 in the same lysates (n=3 for each STAT). Values represent the mean ± S.E. ***p<0.001 by Student’s t-test compared to control.
Supplemental Figure 3: Immunohistochemical staining showing expression of (A and B) ERK, (C and D) phospho-ERK (p-ERK); (E and F) AKT, and (G, H) phospho-AKT (p-AKT) in normal and neoplastic breast epithelia (n=7). Magnification objective 40X, scale bar 50μm.
Supplemental Figure 4: BAD specifically inhibits MEK dependent ERK1/2 activation, but not Myr-AKT-induced ERK activation. MCF7 cells were transiently transfected with indicated plasmid vectors and were growth for 24h. Whole cell lysates were probed with p-ERK and ERK antibodies. Expression of ERK are shown as protein loading controls.