Abstract
The chromatin remodeler BRAHMA (BRM) is a Trithorax Group (TrxG) protein that antagonizes the functions of Polycomb Group (PcG) proteins in fly and mammals. Recent studies also implicate such a role for Arabidopsis (Arabidopsis thaliana) BRM but the molecular mechanisms underlying the antagonism are unclear. To understand the interplay between BRM and PcG during plant development, we performed a genome-wide analysis of trimethylated histone H3 lysine 27 (H3K27me3) in brm mutant seedlings by chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq). Increased H3K27me3 deposition at several hundred genes was observed in brm mutants and this increase was partially supressed by removal of the H3K27 methyltransferase CURLY LEAF (CLF) or SWINGER (SWN). ChIP experiments demonstrated that BRM directly binds to a subset of the genes and prevents the inappropriate association and/or activity of PcG proteins at these loci. Together, these results indicate a crucial role of BRM in restricting the inappropriate activity of PcG during plant development. The key flowering repressor gene SHORT VEGETATIVE PHASE (SVP) is such a BRM target. In brm mutants, elevated PcG occupancy at SVP accompanies a dramatic increase in H3K27me3 levels at this locus and a concomitant reduction of SVP expression. Further, our gain- and loss-of-function genetic evidence establishes that BRM controls flowering time by directly activating SVP expression. This work reveals a genome-wide functional interplay between BRM and PcG and provides new insights into the impacts of these proteins in plant growth and development.
Author Summary
In flowering plants, the proper transition from vegetative growth to flowering is critical for their reproductive success and must be controlled precisely. Multiple genes have been shown to regulate the floral transition in response to environmental and endogenous cues. Among them is SHORT VEGETATIVE PHASE (SVP), a key flowering repressor gene in Arabidopsis. SVP is highly expressed during the vegetative phase to promote growth, but the mechanism by which the high expression level of SVP is maintained remains unknown. Here, we report a genome-wide study to examine the functional interplay between the BRM chromatin remodeler and the PcG proteins that catalyze trimethylation of lysine 27 on histone H3 (H3K27me3), a histone mark normally associated with transcriptionally repressed genes. We identify BRM as a direct upstream activator of SVP. BRM acts to keep the levels of H3K27me3 low at the SVP locus by inhibiting the binding and activities of the PcG proteins. Thus, our work identifies a previously unknown mechanism in regulation of flowering time and demonstrates the power of genome-wide approaches in dissecting regulatory networks controlling plant development.
Introduction
Plant development takes place in distinct phases, each of which is characterized by the activation of a particular set of genes and the repression of others. Precise control of gene expression in each phase is crucial for proper growth and development. The transition from the vegetative to the reproductive phase is controlled precisely by multiple genetic pathways in response to environmental and endogenous signals [1–4]. In Arabidopsis, a repressor complex that consists of two MADS box transcription factors, FLOWERING LOCUS C (FLC) and SVP, serves as a negative regulator of flowering time by directly repressing the expression of the floral pathway integrators FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1) [1,5,6]. SVP is highly expressed during the vegetative phase [5,7], but is down-regulated during the floral transition by the autonomous and gibberellin (GA) pathways [5], which results in the de-repression of FT and SOC1 to promote flowering. Despite its key role in controlling flowering time, the mechanisms by which SVP expression is regulated are still unknown. Particularly, no positive regulator(s) of SVP expression in the vegetative phase have been identified.
Polycomb Group (PcG) proteins are epigenetic repressors that maintain the repressed state of genes in cells where the genes should be inactive [8–11]. PcG proteins repress genes through combined activities of at least two multi-protein complexes known as Polycomb Repressive Complex 1 (PRC1) and PRC2 [8]. PRC2 is involved in the establishment and maintenance of the repressed chromatin state, by introducing the H3K27me3 mark. Subsequently, PRC1 binds to the H3K27me3 mark and compacts the chromatin, resulting in the repressed state of PcG target genes. In Arabidopsis, at least three forms of PRC2 complexes exist and each of them acts at specific developmental phases [12–15]. CLF and SWINGER (SWN) are two putative H3K27 methyltransferases and act redundantly during the vegetative and reproductive stages [16]. Several thousands of Arabidopsis genes were reported to carry the H3K27me3 mark in young seedlings [17–19]. A fraction of PcG target genes was found to carry the H3K27me3 mark specifically in either the shoot apical meristem or leaf cells [18], suggesting dynamic regulation of H3K27me3 deposition. Studies have been carried out to address how PcG proteins deposit H3K27me3 to target genes [12,13,20]. It is less known, however, about the mechanisms by which PcG activities are prevented from targeting certain genes to keep these genes on at particular developmental phases.
SWI/SNF-type chromatin-remodeling protein complexes are thought to utilize energy from ATP hydrolysis to mobilize, disrupt or change nucleosomes to create an open chromatin structure for the access of transcriptional factors or other regulators [21,22]. The SWI2/SNF2 ATPase in Drosophila, BRM, was initially classified as a Trithorax group (TrxG) protein since it activates the transcription of homeotic genes and thus antagonizes the function of PcG during fly development [23,24]. However, recent studies indicate that it can either activate or repress target gene expression, through increasing or decreasing the accessibility of the target DNA [24–26], yet its role in the regulation of gene expression is not well understood. Although the biochemical activities of plant SWI/SNF complexes have not been examined, progress has been made to identify the plant SWI/SNF complexes through genetic and molecular analysis [27–30]. In Arabidopsis, four SWI2/SNF2 ATPases including BRM and SPLAYED (SYD), four SWI3 proteins (SWI3A to SWI3D), two SWI/SNF ASSOCIATED PROTEINS 73 (SWP73A and SWP73B), two ACTIN RELATED PROTEINS (ARP4 and ARP7), and a single SNF5 subunit termed BUSHY (BSH) were predicted subunits of SWI/SNF complexes [27]. Previous in vitro protein-protein interaction data [28,31] and a recent effort in protein complex purification followed by peptide sequencing [32] suggest that these proteins could form several SWI/SNF complexes. Subunits of Arabidopsis SWI/SNF complex(es) play crucial roles in many aspects of plant development [26,27,33–36]. The SWP73B (also called BAF60) subunit has been shown to participate in the control of flowering time [37]. The SWI3C protein is involved in gibberellin (GA) responses [38]. brm mutants show pleiotropic phenotypes, such as reduced plant size [28,39], downward curling of leaves [28,33], mild floral homeotic defects [28,34], hypersensitivity to abscisic acid [26] and early flowering [33,39,40]. Efforts have been made to understand the reason why brm mutants show an early flowering phenotype [40], but the precise role of BRM in flowering time control is still not clear.
Although the roles of PcG proteins and BRM during plant development have been investigated individually, how their activities are coordinated is poorly understood. Interestingly, a recent report in Arabidopsis showed that loss of BRM activity led to the increased H3K27me3 at two floral homeotic genes [34], suggesting the antagonistic relationship between BRM and PcG. However, the current model is solely based on the characterization of a few identified targets of BRM, and it remains unknown to what extent BRM is required for antagonizing PcG function in plant. Also the precise mechanism by which BRM antagonizes PcG activity during plant development remains unclear. Finally, whether or not plant BRM might work synergistically with PcG proteins is completely unknown. To address these questions, we have performed a genome-wide analysis of H3K27me3 in brm mutant seedlings by chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq). We identify several hundred genes that show increased levels of H3K27me3 upon loss of BRM activity, demonstrating the critical role of BRM in preventing genes from H3K27me3-mediated repression in plant cells. We further show that there is inappropriate invasion of PcG proteins. Finally, by taking advantage of our genome-wide data, we uncover a role for BRM in repressing flowering by activating directly the expression of SVP, thus providing an explanation for the early flowering phenotype of brm mutants. Together, our results demonstrate that BRM is essential for proper H3K27me3 distribution in the genome and thus plant development.
Results
Loss of BRM Activity Leads to the Gain of H3K27me3 at Hundreds of Genes
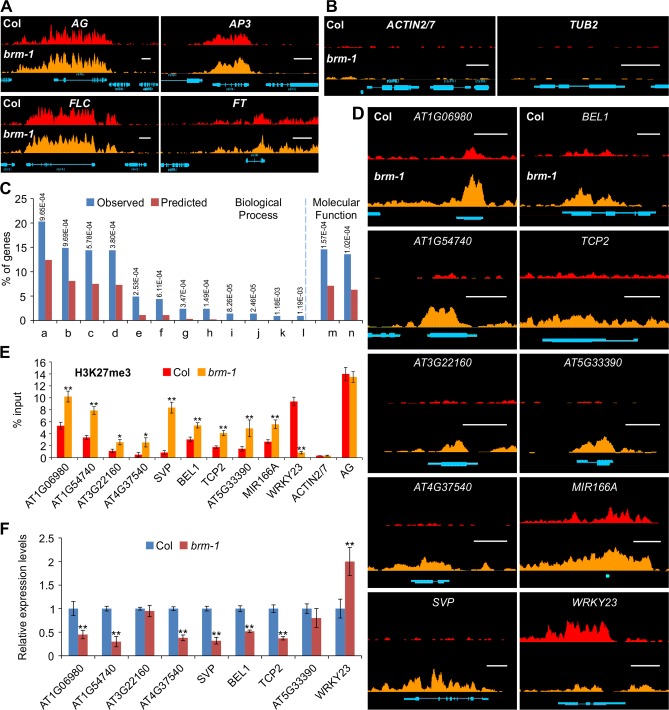
To examine whether BRM affects the patterns of H3K27me3 deposition and distribution in a genome-wide scale, we performed ChIP-seq with anti-H3K27me3 antibodies in wild-type Col and brm-1, a null allele with a T-DNA insertion [28]. Two independent biological DNA samples were generated and used for sequencing. We mapped the reads to the Arabidopsis genome and identified H3K27me3-enriched regions in both wild-type and brm mutants. Only H3K27me3-enriched regions identified in both biological replicates were chosen for further data analysis. In 14-day-old wild-type Col seedlings, we identified 5,591 regions, corresponding to 7,230 genes, which were marked by H3K27me3 (S1 Dataset). H3K27me3 target genes identified in our study cover more than 95% (6,322 out of 6634) of those reported in a previous ChIP-seq analysis [19]. Furthermore, in both Col and the brm-1 mutant, the patterns of H3K27me3 at several well-characterized H3K27me3 target genes, such as AGAMOUS (AG), APETALA3 (AP3), FLC and FT, are very similar to those reported by Lu et al [19] (Fig. 1A). In contrast, no H3K27me3 signals at two highly expressed genes, ACTIN2/7 and TUB2, were observed (Fig. 1B), demonstrating the quality and reliability of our ChIP-seq data.
Figure 1. Loss-of-function of BRM results in changes of H3K27me3 distribution over several hundred endogenous genes.
(A) ChIP-seq data for the well-known H3K27me3 target genes AG, AP3, FLC and FT from wild type Col (red; top) and brm-1 (orange; bottom). Gene structures are shown underneath each panel. Scale bars, 1Kb. The plants used were 14-day-old seedlings. (B) ChIP-seq data showing no H3K27me3 signal at two constitutively expressed genes ACTIN2/7 and TUB2 in wild-type Col (red; top) and brm-1 (orange; bottom). Gene structures are shown underneath each panel. Scale bars, 1Kb. (C) Gene Ontology (GO) analysis of the genes showing increased H3K27me3 levels in brm-1. Numbers on the top are P values (hypergeometric test) for GO category enrichment generated by comparing the percentage of the corresponding categories in the genes that showed increased H3K27me3 levels with those in the whole genome. (a) Regulation of biological process;(b) Regulation of metabolic process; (c) Regulation of macromolecule metabolic process;(d) Regulation of gene expression; (e) Response to auxin stimulus; (f) Tissue development; (g) Gene silencing by miRNA; (h) Meristem maintenance; (i) Meristem determinacy; (j) Floral meristem determinacy; (k) Leaf shaping; (l) Maintenance of floral meristem identity; (m) Transcription regulator activity; (n) Transcripiton factor activity. (D) ChIP-seq data showing changes in H3K27me3 levels at 10 selected genes in brm-1. Nine of them showed an increase and one showed a decrease in H3K27me3 levels. Data for the wild-type Col are shown in red at the top, and brm-1 is shown in orange at the bottom. Gene structures are shown underneath each panel. Scale bars, 1Kb. (E) ChIP-qPCR validation using independent samples. Data are shown as percentage of input. ACTIN2/7 and AG were used as control loci that exhibited no change in H3K27me3 deposition. Error bars indicate standard deviations from three biological replicates.*: P < 0.05; **: P < 0.01. (F) Expression analysis of selected genes by qRT-PCR. The expression of each gene was normalized to that of GAPDH, and the expression level in Col was set to 1. Error bars indicate standard deviations from three biological replicates.*: P < 0.05; **: P < 0.01.
Compared to wild-type, we identified 276 genes at which H3K27me3 levels changed more than twofold in the brm-1 mutant (see the Materials and Methods section for details). Out of the 276 genes, 258 (93.5%) genes showed more than a twofold increase in H3K27me3 in brm-1, while only 18 (6.5%) genes showed more than a twofold reduction in H3K27me3 in brm-1 (S2 Dataset). Our genome-wide data show that BRM mainly acts to antagonize PRC2 activity during vegetative development, which is consistent with its expected role as a TrxG protein. However, the decreased H3K27me3 at a smaller set of genes in brm mutant suggests that BRM could also promote PcG activity at certain loci.
We performed a Gene Ontology (GO) analysis for the genes showing increased H3K27me3 deposition using the BINGO software [41]. In the classification of biological processes, these genes were highly enriched in “regulation of metabolic process” (P = 9.69E-4) and “regulation of gene expression” (P = 3.8E-4; Fig. 1C), and in terms of molecular function, the most enriched category observed was “transcription regulator activity” (P = 1.57E-4). Thus, BRM is involved in a wide spectrum of cellular processes such as gene expression regulation and metabolism through preventing PcG proteins from H3K27me3 deposition. To validate our ChIP-seq data, we randomly chose a subset of genes and performed ChIP followed by quantitative PCR (ChIP-qPCR) using independent chromatin samples. We confirmed the changes in H3K27me3 levels at all 10 selected genes in brm-1 (Fig. 1D and 1E). We did not detect any marked changes at the PcG non-targets ACTIN2/7 or the PcG target AG (Fig. 1E).
Next, we asked whether the elevated H3K27me3 levels in the brm-1 mutant caused down-regulation of the corresponding genes. We measured the expression levels of several selected genes that showed increased H3K27me3 levels in brm-1 by quantitative Reverse Transcription-PCR (qRT-PCR) and observed decreased expression for most but not all of them in brm-1 (Fig. 1F). Interestingly, we also found increased expression of WRKY23 (Fig. 1F), a gene with decreased H3K27me3 levels in brm-1 (Fig. 1D and 1E). These data indicate that there is a positive correlation between increased H3K27me3 levels and decreased gene expression in brm-1 and also suggest that increased H3K27me3 deposition alone in brm might not be sufficient for gene repression at some target loci.
Removal of CLF or SWN Activity in brm Background Results in a Substantial Decrease of H3K27me3 Deposition at Some Genes
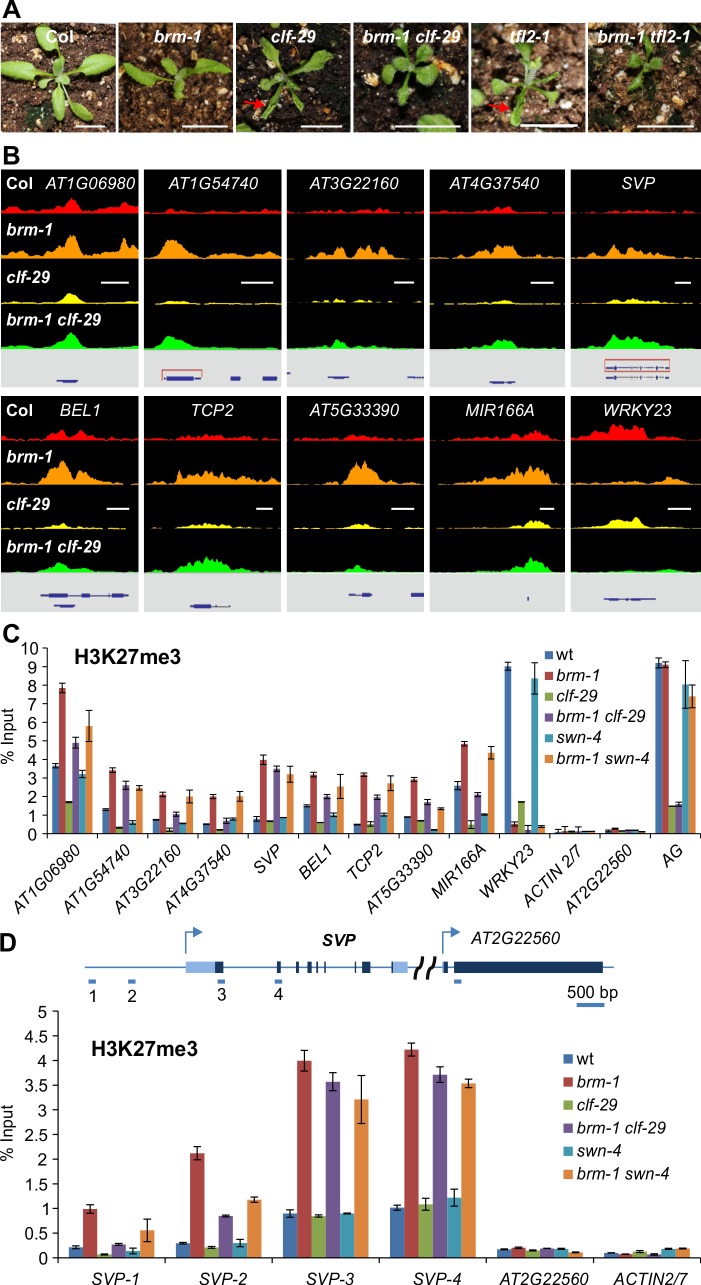
In Arabidopsis, CLF is thought to be a major H3K27 methyltransferase responsible for the deposition of H3K27me3 in tissues other than seeds [42,43]. LIKE HETEROCHROMATIN PROTEIN 1/TERMINAL FLOWER 2 (LHP1/TFL2) associates with regions with H3K27me3 across the Arabidopsis genome and was proposed to be a key component of a plant PRC1 complex [44,45]. Both clf and tfl2 single mutants showed up-ward curling of leaves (Fig. 2A) [42,46]. We reasoned that CLF might be required for the increased H3K27me3 levels at some genes in the brm-1 mutant. To test this, we first generated a brm clf double mutant to examine the genetic relationship between CLF and BRM. clf single mutants display up-wardly curled leaves while brm single mutants show down-ward curling of leaves [28]. Up-ward leaf curling in clf mutants can be caused by ectopic expression of floral homeotic genes such as AG, AP1, and AP3 [16,42]. In the brm clf double mutants, the up-ward curling of leaves was weaker than that in clf single mutants (Fig. 2A and S1 Fig.), suggesting that brm can partially suppresses clf. We also generated brm tfl2 double mutants. The leaves of the brm tfl2 double mutants showed down-ward curling as those in brm single mutants (Fig. 2A and S1 Fig.) suggesting that brm suppresses tfl2’s phenotype of up-wardly curled leaves. These genetic data support a notion that BRM antagonizes PcG function during vegetative development. Consistent with the partially rescued up-ward leaf curling in brm clf double mutants, we found decreased ectopic expression of several floral homeotic genes such as AG, AP1, and AP3 in brm clf double mutant leaves compared to clf single mutants (S2 Fig.). Interestingly, the brm clf double mutants were also smaller in terms of overall size than either single mutant, suggesting the additive effect of the two mutations in plant development. Supporting this observation, we noticed that there were more genes mis-regulated in brm clf double mutants than either single mutant (S3 Fig.).
Figure 2. Removal of CLF or SWN activity in brm background results in a substantial decrease of H3K27me3 deposition.
(A) Loss of BRM activity partially rescues the up-wardly leaf curling phenotypes of clf-29 and tfl2-1. Scale bar: 1 cm. (B) ChIP-seq data comparing H3K27me3 levels at 10 selected genes in Col, brm-1, clf-29 and brm-1 clf-29. Data for the wild type Col are shown in red, brm-1 in orange, clf-29 in yellow, and brm-1 clf-29 in green. Gene structures are shown underneath each panel. Scale bars, 1Kb. (C) ChIP-qPCR validation of the H3K27me3 ChIP-seq data using independent samples; and ChIP-qPCR detection of H3K27me3 levels in swn-4 and brm-1 swn-4 mutants. ChIP signals are shown as percentage of input. ACTIN2/7 and AT2G22560 (a flanking gene of SVP) were used as negative control loci; and AG was used as a positive control locus. Error bars indicate standard deviations from three biological replicates. (D) Top panel: schematic representation of the genomic region covering SVP and the flanking gene AT2G22560. Dark and light blue boxes indicate exon and intron, respectively. Arrows indicate the transcription start sites. Short blue lines indicate the positions of primer pairs used. Bottom panel: ChIP-qPCR determining the levels of H3K27me3 across the SVP locus. ChIP signals are shown as percentage of input. Error bars indicate standard deviations from three biological replicates.
To test if CLF is required for the increased H3K27me3 levels at some genes in the brm-1 mutant, we measured genome-wide H3K27me3 levels in brm clf double mutants by ChIP-seq and compared them with those in brm single mutants. We found that removal of CLF activity led to a marked reduction of H3K27me3 levels at approximately half of the genes with increased H3K27me3 levels in brm-1 (133 out of 258; Fig. 2B; S3 Dataset), indicating the requirement for CLF activity for the increased H3K27me3 levels at some of the genes in brm mutants. We validated these results by ChIP-qPCR at selected genes (Fig. 2C). It is worth noting, however, that there was no drastic loss in H3K27me3 levels at the rest of the genes in the brm clf double mutant relative to the brm single mutant (Fig. 2B and 2C), which might be explained by the redundant SWN activity at these loci. To examine the contribution of SWN, we generated the brm-1 swn-4 double mutant. We found that H3K27me3 levels at the majority of the selected loci in the brm swn double mutant were lower than those in the brm single mutant (Fig. 2C). Furthermore, we scanned the SVP locus and included a region from the neighboring gene At2g22560, for H3K27me3 distribution in all five mutant backgrounds. As shown in Fig. 2D, the results were consistent with those in Fig. 2B and 2C, suggesting a redundant role of CLF and SWN at SVP. These observations are consistent with a scenario in which BRM acts to protect some gene loci from PcG activity in developing seedlings so that these genes stay active.
Increased Occupancy of CLF/SWN at Target Loci in brm Mutants
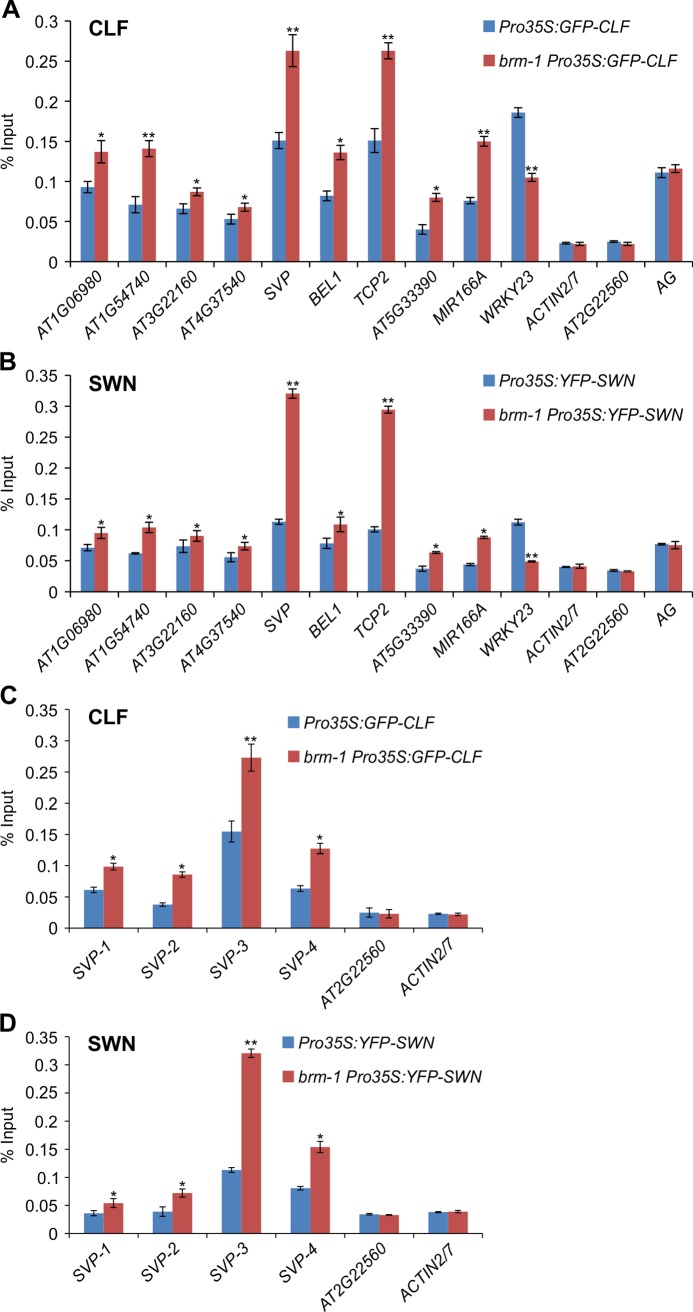
To determine if the increase in H3K27me3 levels in brm mutants was due to increased CLF/SWN presence at the loci, we first measured CLF occupancy at the loci in the brm mutant relative to wild-type using a GFP-tagged CLF line [43]. As shown in Fig. 3A, CLF occupancy was increased at all the selected genes, suggesting that, in the absence of BRM, CLF is allowed to access some inappropriate genomic regions, resulting in increased H3K27me3 levels. We then examined the involvement of SWN. For that, we generated an YFP-tagged SWN line and performed ChIP with anti-YFP antibodies to measure SWN occupancy in the brm mutant relative to wild type. We found that the occupancy of SWN was also increased at the majority of the loci when BRM was absent (Fig. 3B). Furthermore, we also scanned the SVP locus, including the region in the neighboring gene, to compare patterns of CLF/SWN occupancy between wild type and brm-1. As shown in Fig. 3C and 3D, the two proteins were markedly enriched in brm-1 across the SVP locus with a strong bias towards the transcription start site (TSS), implicating a redundant action of CLF and SWN at SVP. These observations suggest that increased CLF/SWN occupancy could contribute to the elevated levels of H3K27me3 in brm mutants. On the other hand, by comparing H3K27me3 levels and CLF/SWN occupancy at SVP relative to the control loci such as At2g22560 and ACTIN, low but significant levels of H3K27me3 (Fig. 2D) and CLF/SWN (Fig. 3C and 3D) at SVP were found in wild-type plants. This suggested that BRM prevents high levels of H3K27me3 and CLF/SWN occupancy rather than excluding them. Alternatively, it could also function to keep PcG in an inactive state. At the WRKY23 locus, CLF/SWN occupancy was reduced in the brm mutant (Fig. 3A and 3B), consistent with the decreased H3K27me3 levels observed at this locus (Fig. 1D and 1E).
Figure 3. Physical occupancy of CLF and SWN at selected genes in brm mutants.
(A) Analysis of CLF occupancy at selected genes as determined by ChIP-qPCR using anti-GFP antibody in brm-1 Pro35S:GFP-CLF and Pro35S:GFP-CLF plants. (B) Analysis of SWN occupancy at selected genes as determined by ChIP-qPCR using anti-GFP antibody in brm-1 Pro35S:YFP-SWN and Pro35S:YFP-SWN plants. (C) and (D) ChIP-qPCR to determine the levels of CLF (C) and SWN (D) occupancy across the SVP locus. The primers used are the same as those in Fig. 2D. ChIP signals are shown as percentage of input. ACTIN2/7 and AT2G22560 were used as negative control loci. AG was a positive control locus. Error bars indicate standard deviations among three biological replicates. *: P < 0.05; **: P < 0.01.
Physical Occupancy of BRM at Target Loci
Next, we asked how BRM antagonizes PcG function during vegetative growth, i.e., whether it does so directly or indirectly. One of the possibilities that could explain the increased H3K27me3 deposition and PcG protein occupancy on chromatin in brm is the elevated expression level of genes encoding PcG subunits. To address this issue, we examined the expression levels of genes encoding PRC2 components, including CLF, SWN, EMBRYONIC FLOWER2 (EMF2), VERNALIZATION2 (VRN2), FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) and FERTILIZATION INDEPENDENT SEED2 (FIS2) [12] in brm mutants. The expression of these genes was not increased markedly in brm-1 compared to that in wild-type (S4 Fig.), indicating that BRM does not antagonize PcG through repressing the expression of PcG-encoding genes. We also measured histone H3 levels at selected genes, and found a slight increase in brm-1 (S5 Fig.). However, the change in H3 levels was very small and thus could not fully account for the change in H3K27me3 levels. These observations point to the possibility that BRM acts directly at the target loci to antagonize PcG proteins.
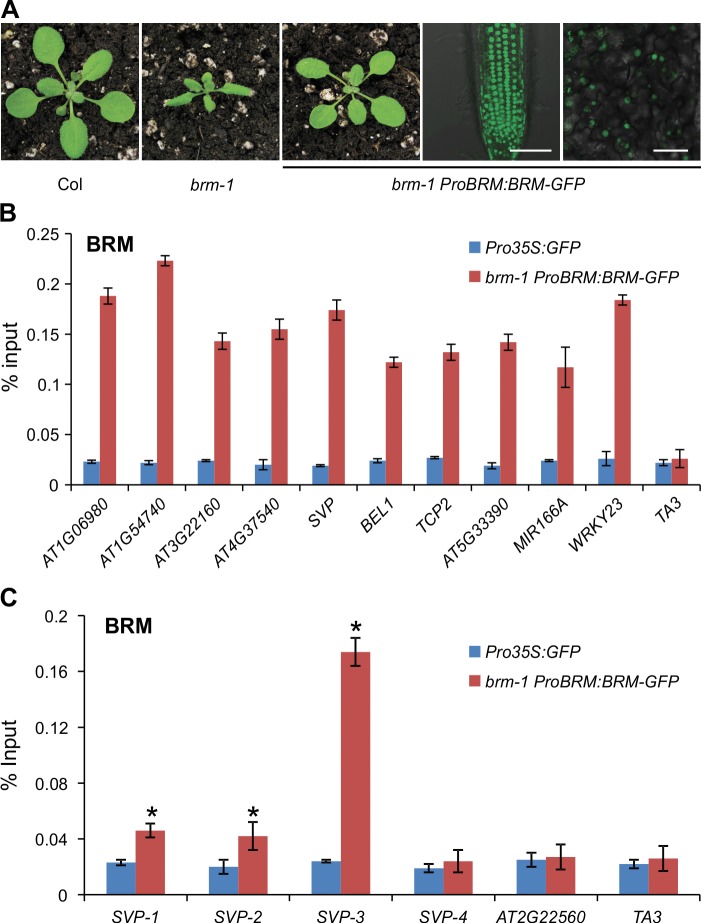
We then tested whether BRM acts directly on the affected genes by physically binding to these genes. We performed ChIP-qPCR experiments to examine BRM occupancy at the affected genes. For the ChIP assay, we used a transgenic Arabidopsis line expressing a GFP-tagged BRM transgene under the control of the BRM native promoter (ProBRM:BRM-GFP) [47]. The transgene could fully rescue the morphological defects of the brm-1 null mutant (Fig. 4A), suggesting that it is functional in vivo. ChIP was performed with anti-GFP antibodies and Pro35S:GFP plants were used as the negative control. The ChIP DNA was analyzed by qPCR to examine the enrichment of BRM at target genes. Genomic regions around the transcription start site at all examined genes were significantly enriched in the BRM-GFP ChIP (Fig. 4B). Furthermore, we scanned the SVP locus, including the negative control region in the neighboring gene, for BRM occupancy. As shown in Fig. 4C, the BRM was found to be significantly enriched at the SVP locus, particularly near the TSS. The physical association of BRM with these selected genes, in combination with increased H3K27me3 deposition and decreased expression of the genes in brm mutants, suggests that BRM acts directly at these target loci, to keep the PRC2 activity off and thus promote gene activity. Loss of BRM activity allows the access to these loci by PRC2, which turns off or decreases gene expression.
Figure 4. Physical occupancy of BRM at selected genes.
(A) ProBRM:BRM-GFP could complement the brm-1 phenotype. GFP signals were detected by confocal microscopy in 14-day-old brm-1 ProBRM:BRM-GFP roots and leaves, respectively. Scale bar: 50 μm. (B) BRM occupancy at selected genes as determined by ChIP using anti-GFP antibody in brm-1 ProBRM:BRM-GFP plants with Pro35S:GFP plants as control. ChIP signals are shown as percentage of input. TA3, a transposable element gene that is not targeted by BRM [34], was used as a negative control locus. (C) ChIP-qPCR to determine the occupancy of BRM across the SVP locus. ChIP signals are shown as percentage of input. The position of primer pairs used is the same as in Fig. 2D. AT2G22560, a flanking locus of SVP, was used as a negative control locus. Error bars indicate standard deviations among three biological replicates. *: P < 0.05.
BRM Positively Regulates SVP Expression
In the sections below, we present our observations to demonstrate that SVP is a main target of BRM in the control of flowering. SVP is a key negative regulator of flowering in Arabidopsis, and loss-of-function of SVP results in early flowering [5,7]. Consistent with its role in maintaining the duration of the vegetative phase, SVP is highly expressed in seedlings, but is barely detectable in inflorescence tissues [7]. We noticed initially from our ChIP-seq and ChIP-qPCR data (Fig. 1D and 1E) that H3K27me3 levels drastically increased at the SVP locus in brm-1 compared with wild-type. These data suggest that the SVP locus becomes a PRC2 target in the absence of BRM activity. The increase in H3K27me3 levels at the SVP locus in brm raises the possibility that BRM may act to keep SVP on by antagonizing PcG activity during vegetative growth. To test this hypothesis, we first extended the single time point expression analysis of SVP in brm-1 as presented in Fig. 1F by examining the expression of SVP in the brm-1 mutant spanning several developmental time points. Indeed, the expression of SVP in the brm-1 mutant was consistently lower than that in wild-type plants over a time course spanning 8 to 14 days after germination (DAG, Fig. 5A), suggesting that BRM activity is continually required for the high levels of SVP expression in seedlings. The decreased expression of SVP was unlikely due to the accelerated floral transition of brm-1 plants, since the expression of AP1, a marker gene for the vegetative-to-floral developmental transition [48,49], remained low throughout the time course (S6 Fig.).
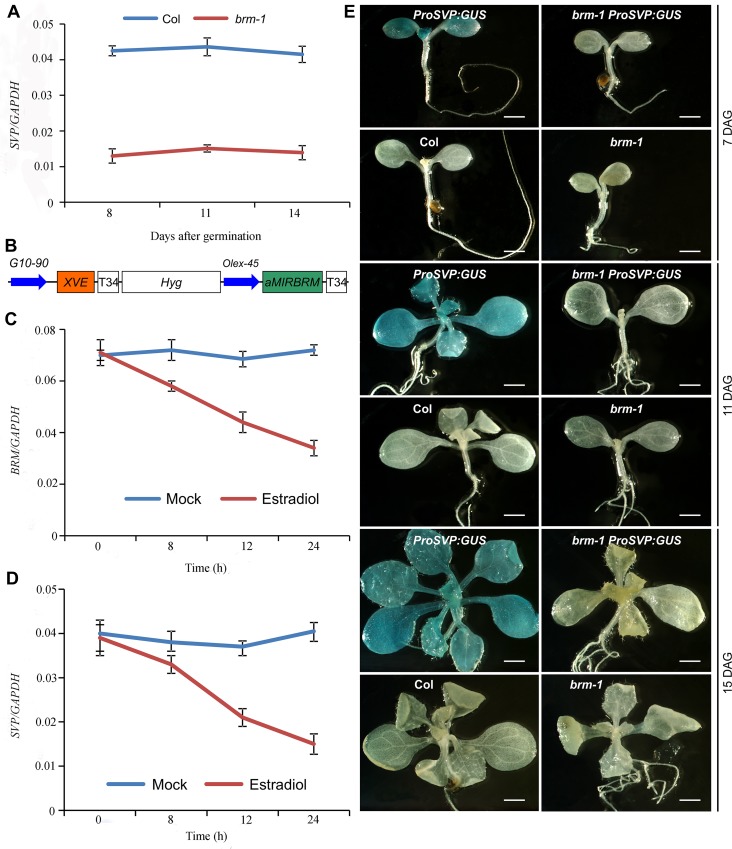
Figure 5. SVP expression is tightly controlled by BRM.
(A) The expression of SVP is drastically decreased in developing brm-1 seedlings compared with that in Col (grown at 22°C) as determined by qRT-PCR. (B) Schematic diagram of the region between the right and left T-DNA borders of the XVE::aMIRBRM construct. The precursor of aMIRBRM was inserted behind a LexA operator sequence fused to the-45 35S minimal promoter (OLexA-45). Other components of the vector were described previously (Curtis and Grossniklaus 2003). (C) BRM expression in 7-old-day XVE::aMIRBRM transgenic seedlings mock treated or treated with 10μm β-estradiol for 0, 8, 12, and 24h, respectively. (D) SVP expression in 7-day-old XVE::aMIRBRM transgenic seedlings mock treated or treated with 10μm β-estradiol for 0, 8, 12, and 24h, respectively. The expression of each gene in A, C, and D was normalized to that of GAPDH. Error bars indicate standard deviations among three technical replicates from one representative experiment. (E) GUS activity patterns of ProSVP:GUS in Col and brm-1 backgrounds in 7, 11, and 14-DAG (days after germination) seedlings. Col and brm-1 were included as negative controls. Scale bar: 0.5 mm.
To confirm that BRM activates the expression of SVP, we generated XVE::aMIRBRM transgenic lines that harbor an inducible artificial microRNA (amiRNA) targeting BRM (Fig. 5B). As shown in Fig. 5C, BRM transcript levels in 7-day-old XVE::aMIRBRM seedlings treated with β-estradiol to induce the amiRNA were gradually decreased by approximately 50% during a 24h time course, indicating that the amiRNA was effective. SVP transcript levels showed a similar reduction kinetics in the time course (Fig. 5D). This result reveals that proper BRM activity is required for SVP expression.
To further verify that BRM activates SVP expression at the transcriptional level, we obtained a previously developed SVP promoter-GUS fusion reporter line (ProSVP:GUS) [5], and introduced it into the brm-1 background by genetic crosses (brm-1 ProSVP:GUS). As shown in Fig. 5E, GUS activity in brm-1 ProSVP:GUS was almost invisible compared to that in ProSVP:GUS at all three time points (Fig. 5E), suggesting that the promoter of SVP has no detectable activity when BRM is absent. As negative controls, we also stained Col wild-type and brm-1 mutants but saw no signals (Fig. 5E). Documented Arabidopsis gene expression data indicate a temporal and spatial overlap of the SVP and BRM expression patterns in leaves (S7 Fig.) [50], which is consistent with a role for BRM as a positive regulator of SVP in developing seedlings. These observations demonstrate a positive spatial and temporal correlation between BRM and SVP expression, and when combined with our BRM-GFP ChIP data (Fig. 4B and 4C) that showed a direct BRM binding to the SVP locus, indicate that BRM directly promotes SVP expression during vegetative development.
BRM Represses Flowering Mainly via Regulating SVP Transcription
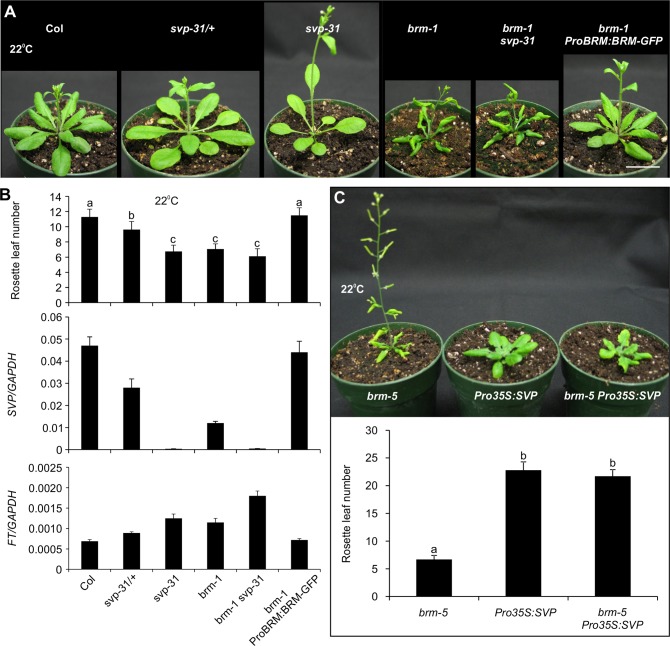
Having shown above a positive role for BRM in regulating SVP expression, we next sought to investigate whether the BRM-SVP module can largely explain the early flowering phenotype of the brm mutant. Both brm and svp single mutants show early flowering phenotypes under long-day conditions [6,7,33,39,40], but it is not known whether there is a common molecular mechanism underlying their flowering phenotypes. We first estimated the flowering time in the two mutants by counting the number of leaves at bolting (Fig. 6A and 6B, top panel). brm-1 and svp-31, a null T-DNA insertion mutant [6], flowered at roughly the same time. The svp-31 heterozygous plants flowered significantly later than their homozygous siblings but earlier than wild-type Col plants, indicating that SVP controls flowering in a dosage-dependent manner, consistent with previous observations [7]. Next, taking advantage of the dosage-dependent nature of flowering control by SVP, we quantified SVP transcript levels by qRT-PCR in the mutant plants to estimate the contribution of SVP to the flowering control by BRM (Fig. 6B, middle panel). Our qRT-PCR data confirmed that svp-31 is a null allele and the heterozygous plants accumulated approximately half the amount of SVP transcripts found in wild-type plants (Fig. 6B, middle panel). SVP expression in brm-1 was drastically decreased to less than half that of svp-31 heterozygous plants. In brm-1 ProBRM:BRM-GFP plants, both the flowering time and SVP expression were restored to the wild-type level (Fig. 6A and 6B), further confirming that BRM activity is responsible for the normal expression level of SVP. Our quantification of flowering time and SVP transcript levels in brm-1, when compared quantitatively to those from svp-31 mutants, suggests that 1) BRM is a major activator of SVP expression; and 2) The early flowering phenotype of the brm-1 mutant can largely be accounted for by the down regulation of SVP transcription in the mutant, although other BRM targets also have minor contributions.
Figure 6. BRM represses flowering mainly through regulating SVP transcription.
(A) Comparison of flowering phenotypes of plants with various genetic backgrounds shortly after bolting. For direct comparison, pictures of wild-type and brm-1 ProBRM:BRM-GFP, svp-31 /+ (heterozygous) and svp-31, and brm-1 and brm-1 svp-31 were taken at the same age, respectively. All plants were grown at 22°C under long-day conditions. Scale bar: 2 cm. (B) Reduction of SVP expression is associated with the early flowering of brm-1 at 22°C. Top panel: rosette leaf number at bolting of plants in different genetic backgrounds. Error bar indicates standard deviations from at least 20 plants. Lowercase letters indicate significant differences between genetic backgrounds, as determined by Post-hoc Tukey’s HSD test. Middle panel: expression analysis of SVP. Bottom panel: expression analysis of FT. The expression of SVP and FT was calculated relative to that of GAPDH. Error bars indicate standard deviations among three technical replicates from one representative experiment. (C) Overexpression of SVP rescues the early flowering phenotype of brm mutant. Top panel: flowering phenotype of brm-5, Pro35S:SVP and brm-5 Pro35S:SVP plants grown for five weeks at 22°C under long-day conditions. Scale bar: 2 cm. Bottom panel: rosette leaf number of brm-5, Pro35S:SVP and brm-5 Pro35S:SVP plants at bolting. Lowercase letters indicate significant differences between genetic backgrounds, as determined by Post-hoc Tukey’s HSD test.
To provide additional evidence to strengthen our conclusion, we tested whether restoration of SVP in brm mutant background could overcome its early flowering phenotype by expressing SVP from a promoter that is not controlled by BRM (Pro35S:SVP) [51] in brm-5, a chemically induced mutant that has a single nucleotide change in the region encoding the ATPase domain [33]. Indeed, introduction of Pro35S:SVP into brm-5 could rescue the early flowering of the brm-5 mutant (Fig. 6C). We also generated a brm-1 svp-31 double mutant to test the genetic interaction between BRM and SVP in flowering time control. The brm-1 svp-31 double mutant flowered only slightly earlier than either single mutant (Fig. 6B), suggesting that BRM and SVP act largely in a common pathway in determining flowering time, and only minor contributions from other BRM targets. It is worth mentioning that three other flowering time genes also displayed increased H3K27me3 levels in the brm mutant (S2 Dataset and S8 Fig.). When we checked the expression of these genes, we only saw a clear decrease of AGAMOUS-LIKE24 (AGL24) expression but not the other two in brm-1 (S8 Fig.). AGL24 is a MADS-box protein involved in flowering time control. agl24 mutants show delayed flowering while agl24 svp double mutants are early flowering as svp single mutants [52]. The data thus suggest that the early flowering phenotype of brm mutants is unlikely caused by these flowering time genes. In addition, we also examined the expression of FT, a well-established SVP target, in the various genetic backgrounds (Fig. 6B, bottom panel). As expected, FT transcript levels correlated negatively with those of SVP and positively with flowering time in the corresponding genetic backgrounds. In summary, our observations strongly suggest that BRM represses flowering mainly through activating SVP.
Discussion
In both animals and plants, a group of proteins that counteract PcG function have been described and referred to as TrxG proteins [9,13]. Several putative TrxG proteins have been proposed in Arabidopsis, including the H3K4 methyltransferase ATX1 [53], the SAND-domain DNA binding protein ULTRAPETALA1 (ULT1) [54], the chromatin remodeling ATPase PICKLE (PKL) [55], the H3K27me3 demethylase REF6 [19], and the SWI2/SNF2 ATPases SPLAYED (SYD) and BRM [34]. A challenge for the field is to understand the specific roles of the putative TrxG proteins and the functional relationship among them in antagonizing PcG.
The nature of the antagonism between SWI/SNF-type chromatin remodeling ATPases and PcG proteins has been investigated in several studies in animals; and several models of counteraction have been proposed [23,25,56–58]. Interestingly, a very recent report in Arabidopsis showed that BRM overcomes the repression of AG and AP3 by the PcG pathway during the initiation of floral development [34], however, how it does so and to what extent BRM is required for antagonizing PcG function in plants remains unclear. Our genome-wide study shows that BRM deficiency led to an increase in H3K27me3 levels at several hundred genes across the genome during vegetative development in Arabidopsis. We further observed increased occupancy of CLF/SWN-containing PcG complex(es) at these genes when BRM is not located there (Fig. 3; S1 Table). Considering that there are low but significant levels of H3K27me3 (Fig. 2B–2D) and CLF/SWN occupancy (Fig. 3) at SVP in wild-type plants, we favour a model of antagonism between BRM and PcG, in which BRM might function to prevent high levels H3K27me3 and CLF/SWN occupancy instead of excluding them (Fig. 7). It is also possible that BRM could function to keep PcG in an inactive state. In addition to chromatin remodelers, plants might employ transcription factors to counteract PcG. A recent study showed that the binding of transcription factor AG to the promoter of zinc finger repressor KNUCKLES (KNU) causes the eviction of the PcG proteins from the locus, leading to the induction of KNU [59]. Thus, both transcription factors and chromatin remodeling proteins could be involved in counteracting PcG. It will be interesting to determine whether and how these two machineries work together in antagonizing PcG function.
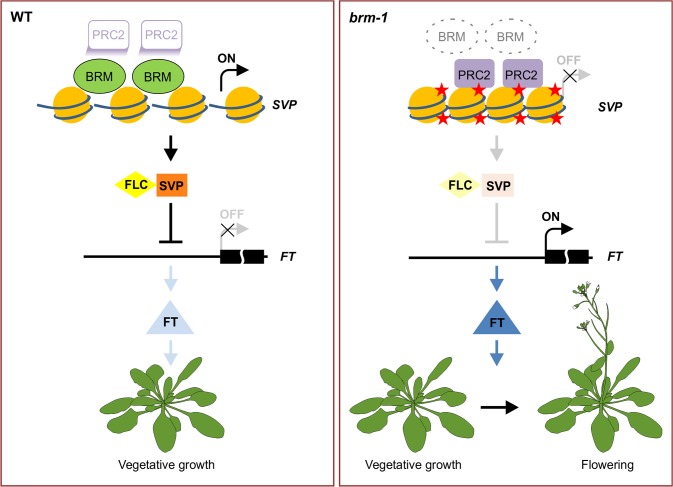
Figure 7. A model for BRM in preventing inappropriate PcG activities at SVP to promote vegetative growth.
In wild-type plants, BRM is physically present at the target chromatin sites, and suppresses the inappropriate silencing of target genes by PcG, thus maintaining the expression of the target loci where such expression is needed at the specific developmental phase. Without BRM, PcG is allowed access to some inappropriate genomic sites, resulting in increased H3K37me3 levels and consequently down-regulation of the expression of the genes. For example, SVP is highly expressed in wild-type seedlings and its downstream target FT is repressed, therefore, vegetative growth is promoted. Conversely, SVP expression is repressed by the mistargeting of PRC2 in brm mutants, and FT is de-repressed as a result to lead to early flowering. Red star: the H3K27me3 mark.
Our genome-wide analysis of H3K27me3 occupancy in brm mutant indicates that BRM does not only antagonize PcG function during plant development, but also cooperates with PcG at some loci (Fig. 1D and 1E). For example, the H3K27me3 level at WRKY23 is decreased and the expression of the gene is up-regulated in both brm and fie mutant (FIE is a PcG subunit) seedlings [17] (this study), suggesting that both BRM and PcG are required for the proper expression of WRKY23. Further, we show that the decreased H3K27me3 observed at WRKY23 loci in brm mutant could be because of, at least partly, the decreased CLF binding. Therefore, this observation suggests that BRM may work with the PcG proteins at some of the common loci and thus repress the targets expression. WRKY23 was recently found to be needed for proper root development and the over-expression of WRKY23 results in the reduction of root length [60]. It will be interesting to test whether the increased transcription of WRKY23 could explain the short root phenotype of brm [28]. The synergistic relationship between BRM and PcG reported here was also observed by a study in human embryonic stem cell showing that an embryonic stem cell specific SWI/SNF complex acts synergistically with PRC2 at all four Hox loci [25]. The mechanism by which BRM cooperates with PcG is currently unknown. One possibility would be that BRM directly interacts with PcG and facilitates the targeting of PcG to genes. Indeed, we found that BRM co-localizes with H3K27me3 at the WRKY23 locus in wild-type seedlings (Fig. 4), suggesting that BRM might interact with PcG proteins. However, no study so far has demonstrated a direct physical interaction between BRM and PcG proteins. It is possible that these two complexes might interact transiently or indirectly. Nevertheless, the synergistic relationship between BRM and PcG found in both animals and plants might suggest its biological relevance and warrants further studies.
The proper transition from vegetative growth to flowering is critical for the reproductive success of angiosperm plants and must be controlled precisely. BRM has been proposed as a repressor of flowering as suggested by the early flowering phenotype and the elevated FT expression of brm mutants [39,40]. However, it was not clear whether BRM acts directly or indirectly to repress FT. SVP has been demonstrated to be a direct repressor of FT [5,6], and thus serves as a key repressor of floral transition. The precise regulation of SVP is obviously of critical importance for our understanding of flowering control. Thus far, however, no direct upstream activator of SVP has been identified. In this work, we provide evidence demonstrating that BRM represses FT by directly maintaining a high level of SVP expression (Fig. 7). First, loss of BRM activity results in decreased expression of SVP (Fig. 5A–5E), which is associated with increased H3K27me3 levels (Fig. 1D and 1E) and increased occupancy of CLF and SWN (Fig. 3C and 3D). Second, BRM directly binds to the SVP locus in vegetative tissues where SVP is highly expressed (Fig. 4B and 4C). Together, these observations suggest that BRM represses the floral transition through directly activating SVP. This is consistent with the genetic evidence showing that the brm-1 svp-31 double mutant displays almost the same early flowering phenotype as brm-1 and svp-31 single mutants (Fig. 6A and 6B). Although our data support a scenario that BRM represses flowering mainly through SVP, some evidence suggests that BRM may also repress flowering through other pathways. For example, the expression of CONSTANS (CO), an activator of FT in the photoperiod pathway, was increased in brm mutants [40]. In addition, elevated expression of both FLC and FT in brm mutants was also reported previously [39,40]. Since FLC is a repressor of FT expression [61], it seems hard to understand why the expression levels of both FLC and FT were increased in brm mutants. Our results presented here provide an explanation for this apparent discrepancy: mutation of BRM results in reduced expression of SVP and consequently lower abundance of the SVP-FLC repressor complex, ultimately leading to activation of FT, regardless of the increased expression of FLC.
It is also relevant to note that down-regulation of BAF60/SWP73B was recently reported to cause increased FLC expression and delayed floral transition [37]. The apparently opposing flowering time phenotype of brm mutants and the BAF60 knockdown line is puzzling. It is unknown whether and how BAF60 regulates SVP expression. It might be possible that the presence of BAF60 in a SWI/SNF complex inhibits the activity of BRM, thus reduction of BAF60 could allow BRM to activate SVP expression, which, in turn, leads to delayed floral transition. Alternatively, it might also be possible that BRM and BAF60 are present in distinct complexes that differ in their regulatory activities and target genes, e.g., BRM activates SVP, while BAF60 represses FLC.
Our genome-wide H3K27me3 profiling data also reveal that BRM is involved in the regulation of a number of other important developmental genes including, most noticeably, members of the miR166 and miR156 families (S2 Dataset). It is well established that the miR166 family miRNAs target the transcripts of the HD-ZIPIII genes, controlling the level and domain of their expression to allow their proper functions in plant development [62–64]. More recently, we uncovered a new role for miR166 in repressing the seed maturation program during vegetative development [65]. An earlier study demonstrated the involvement of BRM in repression of the seed maturation genes in leaves [33] – a brm mutation was isolated in a reporter-assisted genetic screen for Arabidopsis mutants exhibiting ectopic expression of seed storage protein genes [33,65,66]. Our new data presented here thus provide a potential link between the two early studies [33,65]: it strongly suggests that BRM promotes the accumulation of miR166, which in turn represses seed maturation genes in developing seedlings. In conclusion, our work demonstrates that BRM promotes vegetative development by harnessing PcG proteins (mainly by preventing their activities) at key developmental genes.
Materials and Methods
Plant Materials and Growth Condition
Arabidopsis seeds were stratified at 4°C for 3 days in dark condition. Then the seeds were sown on soil or on agar plates containing 4.3 g/L Murashige and Skoog nutrient mix (Sigma-Aldrich), 1.5% sucrose, 0.5 g/L MES (pH 5.8), and 0.8% agar. Plants were grown in growth rooms with 16-h-light/8-h-dark cycles (Long-day, LD) at 22°C or 16°C. T-DNA insertion mutants were obtained from the ABRC, unless otherwise indicated. The brm-1 (SALK_030046), brm-5, clf-29 (SALK_021003), tfl2-1 (CS3796), svp-31 (SALK_026551) and SWN-4 (SALK_109121) mutants are all in the Col background and have been described previously [6,28,33,46,67,68]. Homozygous T-DNA insertion mutants were identified by PCR-based genotyping. Transgenic plants ProBRM:BRM-GFP, ProSVP:GUS, Pro35S:SVP, Pro35S:GFP-CLF and Pro35S:GFP have been described [5,43,47,69].
Chromatin Immunoprecipitation (ChIP) Assays
ChIP was carried out as described [70,71] with minor modifications. Briefly, two grams of 14-day-old seedlings grown on MS agar were cross-linked with 1% formaldehyde and then ground into fine power with liquid nitrogen. Chromatin was isolated and sheared into 200–800 base pair fragments by sonication. The sonicated chromatin was immunoprecipitated with 5 μL of anti-H3K27me3 (07–449, Millipore), anti-GFP (ab290, Abcam) or anti-H3 (Ab1791, Abcam) antibodies. The precipitated DNA was then recovered with the MiniElute PCR Purification Kit (Cat#28004, Qiagen) according to the manufacturer’s instructions. ChIP-qPCR was performed with three technical replicates, and results were calculated as percentage of input DNA according to the Champion ChIP-qPCR user manual (SABioscience). If fold enrichment was used, the calculated percentage input of the wild-type control plant at the regions tested was set to 1. The fold enrichment represents the fold change relative to the wild-type. Independent ChIP experiments were performed at least two more times and similar results were obtained. Primer sequences used for ChIP-qPCR were listed in S2 Table.
ChIP-seq Analysis
Ten ng of ChIP DNA immuoprecipitated by the anti-H3K27me3 antibody was used for ChIP-seq library construction. End repair, adapter ligation and amplification were carried out using the Illumina Genomic DNA Sample Prep Kit according to the manufacturer’s protocol. Illumina Genome Analyser IIx or HiSeq 2500 was used for high-throughput sequencing of the ChIP-seq library. The raw sequence data were processed using the Illumina sequence data analysis pipeline GAPipeline1.3.2. Then Bowtie [72] was employed to map the reads to the Arabidopsis genome (TAIR10) [73]. Only perfectly and uniquely mapped reads were retained for further analysis. Then the data were analyzed as described [19]. Briefly, the alignments were first converted to WIG files using MACS [74]. Then the data were imported to Integrated Genome Browser (IGB) [75] for visualization. Secondly, the program SICER [76] was used to identify ChIP-enriched domains (peaks) in histone modification signals. Thirdly, quantitative comparisons between wild-type Col and mutants were performed using the ChIPDiff program [77]. Regions with more than twofold changes were kept for further analysis. Finally, the identified regions were annotated according to the Arabidopsis annotation gff file (TAIR10, www.arabidopsis.org) using a customized Perl script. Two independent biological replicates were used for sequencing, and only the regions of H3K27me3 found in both replicates were included in the analyses.
Gene Expression Analysis
Total RNA was isolated from ∼100 mg of plant tissues using the RNeasy Plant Mini kit (Qiagen). One μg RNA was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription kit (ABI). Random primers from the kit were used as primers. Real-time quantitative PCR was conducted using the SsoFast EvaGreen Supermix kit with the Bio-Rad CFX96 real-time PCR detection system (Bio-Rad Laboratories, Inc.). The data shown in the figures are the average of three technical replicates. Results were repeated with two additional independent RNA samples (biological replicates). GAPDH served as the internal reference. PCR primers used in real-time PCR are listed in S2 Table.
For genome-wide expression analysis, three biological replicates of Col, brm-1, clf-29 and brm-1 clf-29 samples were analyzed on Affymetrix ATH1 arrays. Genes showing 1.5 fold changes were considered to be differentially expressed.
Construction of YFP-SWN Transgenic Line
The SWN gene without the stop codon was amplified by PCR and cloned into the pDONR221 vector (Invitrogen) by BP reaction according to the manufacturer’s instructions. The resulting transgene in the entry vector was sequenced to make sure that no mutation was introduced during PCR. The transgene was then transferred into the pEarlyGate 104 Gateway-compatible destination vector [78] by LR reaction, according to the manufacturer’s instructions, to make Pro35S:YFP-SWN. The construct was introduced into Agrobacterium tumefaciens GV3101, which was then used to transform swn-4 mutant plants using the floral dip method [79]. Transgenic plants were selected in MS agar media containing 50 μg/ml of Hygromycin B and allowed to grow in soil to maturity to yield seeds. PCR primers used in making the construct are listed in S2 Table.
Artificial miRNA Transgene Constructs
For generating the XVE::aMIRBRM construct, the pRS300 vector [80] was used as the backbone to first generate aMIRBRM. The primers used were designed according to WMD3 (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) and are listed in S2 Table. aMIRBRM was subcloned into the pDONR221 vector (Invitrogen), confirmed by sequencing, and then recombined into the pMDC7 Gateway-compatible destination vector [78] where the aMIRBRM transgene is controlled by a Estradiol-inducible promoter. The construct was transformed into Col wild-type plants by the floral dip method [79]. Transgenic plants were selected for Hygromycin B resistance and allowed to grow to maturity to yield seeds. Seven-day-old T2 transgenic plants were treated either by 10μmol β-Estradiol or DMSO (as the mock control) and samples were collected at different time points after the treatment.
Histochemical GUS Staining
The standard GUS staining solution (0.5 mg/mL 5-bromo-4-chloro-3-indolyl-glucuronide, 20% methanol, 0.01 M Tris-HCl, pH 7.0) was used. Seedlings immersed in GUS staining solution were placed under vacuum for 15 min, and then incubated at 37°C overnight. The staining solution was removed and samples were cleared by sequential incubation in 75% and 95% ethanol.
Flowering Time Measurement
Wild-type and mutant plants were grown side by side in soil at 22°C or 16°C with 16-h-light/8-h-dark cycles. The number of rosette leaves was counted when the length of the inflorescence stem was 1 cm. For each genotype, at least 20 plants were analyzed, and the analysis was repeated 3 times independently.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BRM (AT2G46020), SVP (AT2G22540), CLF (AT2G23380), BEL1 (AT5G41410), TCP2 (AT4G18390), WRKY23 (AT2G47260), miR156D (AT5G10945), LHP1/TFL2 (AT5G17690), EMF2 (AT5G51230), FIE (AT3G20740), SWN (AT4G02020), VRN2 (AT4G16845), AP1 (AT1G69120), TA3 (AT1G37110), ACTIN2/7 (AT5G09810), AG (AT4G18960), AP3 (AT3G54340), FLC (AT5G10140), AGL24 (AT4G24540), SMZ (AT3G54990) and FT (AT1G65480). All raw ChIP-seq dataset and ATH1 expression array dataset have been deposited in the Gene Expression Omnibus database under accession number GSE47202 and GSE53623.
Supporting Information
(A) Top panel: Rosette leaves from 14-d-old plants are shown. Scale bar: 2 mm. Bottom panel: Percentage of upwardly curled leaves in each genetic background is shown. Error bar indicates standard deviations from at least 20 plants. P values were determined by two-tailed t-test. (B) Comparison of brm-1 and brm-1 swn-4 double mutants grown in soil for 14 days. Scale: 1cm.
(PDF)
Expression data of floral homeotic genes, AG (A), AP1 (B), AP3 (C) and AGL24 (D), in different genetic backgrounds were determined by qRT-PCR with three biological replicates.
(PDF)
(PDF)
The expression levels of each gene were normalized to that of GAPDH, and the expression level in Col was set to 1. Error bars indicate standard deviation among three technical replicates from one representative experiment.
(PDF)
ChIP signals are shown as fold changes relative to that in wild-type plants. Error bars indicate standard deviation among three technical replicates from one representative experiment.
(PDF)
The expression level of AP1 gene was normalized to that of GAPDH. Error bars indicate standard deviation among three technical replicates from one representative experiment.
(PDF)
The data were extracted from Schmid et al [50] and displayed using the AtGenExpress Visualization Tool (http://jsp.weigelworld.org/expviz/expviz.jsp).
(PDF)
(A) ChIP-seq data showing an increase in H3K27me3 levels at several genes in brm-1. Data from the wild-type Col is shown in red at the top, and brm-1 is shown in orange at the bottom. (B) ChIP-qPCR validation using independent samples. Data are shown as percentage of input. Error bars indicate standard deviations among three technical replicates from one representative experiment. (C) Expression analysis of AGL24 and SMZ by qRT-PCR. The expression of each gene was normalized to that of GAPDH, and the expression level in Col was set to 1. Error bars indicate standard deviations among three technical replicates from one representative experiment. (D) Small RNA northern blot analysis of miR156 in brm-1 compared with Col. Two time points were used (10 days and 14 days after germination). The levels of small RNAs in brm-1 were compared with those in Col, which was set as 1. The numbers below the gel images indicate relative abundance. U6 served as loading control. RNA isolation and hybridization for miRNA detection was performed as described [65]. Digoxigenin-labeled miRNA probes were generated using the mirVana miRNA Probe Construction Kit (Ambion). Oligonucleotide probes used are listed in S2 Table.
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Kerstein Kaufmann for kindly providing us seeds of ProBRM:BRM-GFP; Hao Yu for seeds of ProSVP:GUS; Jeong Hwan Lee for seeds of Pro35S:SVP; Justin Goodrich for seeds of Pro35S:GFP-CLF; and ABRC for seeds of T-DNA insertion lines. We thank Steven Rothstein for critical reading of the manuscript; Vojislava Grbic for the suggestion of using the absence of AP1 expression as a marker of the vegetative developmental phase; and Alex Molnar for help with preparing the figures.
Funding Statement
This work was supported by Agriculture & Agri-Food Canada A-base, Canadian Crop Genomics Initiative, and Natural Science and Engineering Research Council of Canada (R4019A01) to YC, Natural Science Foundation of China (NSFC, 31128001) to KW and YC, and National Institutes of Health (GM061146) to XC. CC is supported by a graduate fellowship from the Chinese Scholarship Council. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. He Y (2012) Chromatin regulation of flowering. Trends Plant Sci 17: 556–562. 10.1016/j.tplants.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 2. Boss PK, Bastow RM, Mylne JS, Dean C (2004) Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16: S18–S31. 10.1105/tpc.015958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mouradov A, Cremer F, Coupland G (2002) Control of flowering time interacting pathways as a basis for diversity. Plant Cell 14: S111–S130. 10.1105/tpc.001362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639. 10.1038/nrg3291 [DOI] [PubMed] [Google Scholar]
- 5. Li D, Liu C, Shen L, Wu Y, Chen H, et al. (2008) A repressor complex governs the integration of flowering signals in Arabidopsis . Dev Cell 15: 110–120. 10.1016/j.devcel.2008.05.002 [DOI] [PubMed] [Google Scholar]
- 6. Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, et al. (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis . Genes Dev 21: 397–402. 10.1101/gad.1518407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, et al. (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis . Plant J 21: 351–360. [DOI] [PubMed] [Google Scholar]
- 8. Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwartz YB, Pirrotta V (2007) Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8: 9–22. [DOI] [PubMed] [Google Scholar]
- 10. Simon JA, Kingston RE (2013) Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49: 808–824. 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simon JA, Kingston RE (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708. 10.1038/nrm2763 [DOI] [PubMed] [Google Scholar]
- 12. Zheng B, Chen X (2011) Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr Opin Plant Biol 14: 123–129. 10.1016/j.pbi.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bemer M, Grossniklaus U (2012) Dynamic regulation of Polycomb group activity during plant development. Curr Opin Plant Biol 15: 523–529. 10.1016/j.pbi.2012.09.006 [DOI] [PubMed] [Google Scholar]
- 14. Holec S, Berger F (2012) Polycomb group complexes mediate developmental transitions in plants. Plant Physiol 158: 35–43. 10.1104/pp.111.186445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Köhler C, Hennig L (2010) Regulation of cell identity by plant Polycomb and trithorax group proteins. Curr Opin Genet Dev 20: 541–547. 10.1016/j.gde.2010.04.015 [DOI] [PubMed] [Google Scholar]
- 16. Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, et al. (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis . Development 131: 5263–5276. [DOI] [PubMed] [Google Scholar]
- 17. Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, et al. (2011) Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 7: e1002014 10.1371/journal.pgen.1002014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, et al. (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040 10.1371/journal.pgen.1002040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu F, Cui X, Zhang S, Jenuwein T, Cao X (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43: 715–719. 10.1038/ng.854 [DOI] [PubMed] [Google Scholar]
- 20. Lodha M, Marco CF, Timmermans MCP (2013) The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev 27: 596–601. 10.1101/gad.211425.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- 22. Saha A, Wittmeyer J, Cairns BR (2006) Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol 7: 437–447. [DOI] [PubMed] [Google Scholar]
- 23. Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, et al. (1992) brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68: 561 [DOI] [PubMed] [Google Scholar]
- 24. Hargreaves DC, Crabtree GR (2011) ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 21: 396–420. 10.1038/cr.2011.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, et al. (2011) esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol 13: 903–913. 10.1038/ncb2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han SK, Sang Y, Rodrigues A, F2010 B, Wu MF, et al. (2012) The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis . Plant Cell 24: 4892–4906. 10.1105/tpc.112.105114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jerzmanowski A (2007) SWI/SNF chromatin remodeling and linker histones in plants. Biochim Biophys Acta 1769: 330–345. [DOI] [PubMed] [Google Scholar]
- 28. Hurtado L, Farrona S, Reyes JC (2006) The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana . Plant Mol Biol 62: 291–304. [DOI] [PubMed] [Google Scholar]
- 29. Bezhani S, Winter C, Hershman S, Wagner JD, Kennedy JF, et al. (2007) Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19: 403–416. 10.1105/tpc.106.048272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Archacki R, Buszewicz D, Sarnowski TJ, Sarnowska E, Rolicka AT, et al. (2013) BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in Arabidopsis . PLoS ONE 8: e58588 10.1371/journal.pone.0058588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarnowski TJ, Ríos G, Jásik J, Świeżewski S, Kaczanowski S, et al. (2005) SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell 17: 2454–2472. 10.1105/tpc.105.031203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vercruyssen L, Verkest A, Gonzalez N, Heyndrickx KS, Eeckhout D, et al. (2014) ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26: 210–229. 10.1105/tpc.113.115907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tang X, Hou A, Babu M, Nguyen V, Hurtado L, et al. (2008) The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol 147: 1143–1157. 10.1104/pp.108.121996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, et al. (2012) SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci USA 109: 3576–3581. 10.1073/pnas.1113409109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu Y, Rowley MJ, Böhmdorfer G, Wierzbicki AT (2012) A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol Cell 49: 1–12. 10.1016/j.molcel.2012.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kwon CS, Hibara K, Pfluger J, Bezhani S, Metha H, et al. (2006) A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133: 3223–3230. [DOI] [PubMed] [Google Scholar]
- 37. Jégu T, Latrasse D, Delarue M, Hirt H, Domenichini S, et al. (2014) The BAF60 subunit of the SWI/SNF chromatin-remodeling complex directly controls the formation of a gene loop at FLOWERING LOCUS C in Arabidopsis . Plant Cell 26: 538–551. 10.1105/tpc.113.114454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sarnowska EA, Rolicka AT, Bucior E, Cwiek P, Tohge T, et al. (2013) DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis . Plant Physiol 163: 305–317. 10.1104/pp.113.223933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farrona S, Hurtado L, Bowman JL, Reyes JC (2004) The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131: 4965–4975. [DOI] [PubMed] [Google Scholar]
- 40. Farrona S, Hurtado L, March-Díaz R, Schmitz RJ, Florencio FJ, et al. (2011) Brahma is required for proper expression of the floral repressor FLC in Arabidopsis . PLoS One 6: e17997 10.1371/journal.pone.0017997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- 42. Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, et al. (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis . Nature 386: 44–51. [DOI] [PubMed] [Google Scholar]
- 43. Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, et al. (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25: 4638–4649. 10.1038/sj.emboj.7601311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, et al. (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3: e86 10.1371/journal.pgen.0030086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, et al. (2007) The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol 14: 869–871. [DOI] [PubMed] [Google Scholar]
- 46. Larsson AS, Landberg K, Meeks-Wagner DR (1998) The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana . Genetics 149: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smaczniak C, Immink RGH, Muiño JM, Blanvillain R, Busscher M, et al. (2012) Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA 109: 1560–1565. 10.1073/pnas.1112871109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277. [DOI] [PubMed] [Google Scholar]
- 49. Benlloch R, Berbel A, Serrano-Mislata A, Madueño F (2007) Floral initiation and inflorescence architecture: a comparative view. Ann Bot 100: 659–676. 10.1093/aob/mcm146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, et al. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506. [DOI] [PubMed] [Google Scholar]
- 51. Mizukami Y, Ma H (1992) Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71: 119–131. [DOI] [PubMed] [Google Scholar]
- 52. Gregis V, Sessa A, Colombo L, Kater MM (2006) AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis . Plant Cell 18: 1373–1382. 10.1105/tpc.106.041798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, et al. (2003) ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 13: 627–637. [DOI] [PubMed] [Google Scholar]
- 54. Carles CC, Fletcher JC (2009) The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev 23: 2723–2728. 10.1101/gad.1812609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Aichinger E, Villar CBR, Farrona S, Reyes JC, Hennig L, et al. (2009) CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis . PLoS Genet 5: e1000605 10.1371/journal.pgen.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu C, et al. (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46. [DOI] [PubMed] [Google Scholar]
- 57. Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP (2008) SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol 28: 3457–3464. 10.1128/MCB.02019-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, et al. (2010) Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18: 316–328. 10.1016/j.ccr.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sun B, Looi L-S, Guo S, He Z, Gan E-S, et al. (2014) Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343: 1248559 10.1126/science.1248559 [DOI] [PubMed] [Google Scholar]
- 60. Grunewald W, De Smet I, Lewis DR, Löfke C, Jansen L, et al. (2012) Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci USA 109: 1554–1559. 10.1073/pnas.1121134109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high—molecular—weight protein complex. Plant J 46: 183–192. [DOI] [PubMed] [Google Scholar]
- 62. Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, et al. (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774. [DOI] [PubMed] [Google Scholar]
- 63. McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, et al. (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713. [DOI] [PubMed] [Google Scholar]
- 64. Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, et al. (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23: 3356–3364. 10.1038/sj.emboj.7600340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tang X, Bian S, Tang M, Lu Q, Li S, et al. (2012) MicroRNA-mediated repression of the seed maturation program during vegetative development in Arabidopsis . PLoS Genet 8: e1003091 10.1371/journal.pgen.1003091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lu Q, Tang X, Tian G, Wang F, Liu K, et al. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61: 259–270. 10.1111/j.1365-313X.2009.04048.x [DOI] [PubMed] [Google Scholar]
- 67. Schönrock N, Bouveret R, Leroy O, Borghi L, Köhler C, et al. (2006) Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev 20: 1667–1678. 10.1101/gad.377206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang D, Tyson MD, Jackson SS, Yadegari R (2006) Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis . Proc Natl Acad Sci USA 103: 13244–13249. 10.1073/pnas.0605551103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Masiero S, Li MA, Will I, Hartmann U, Saedler H, et al. (2004) INCOMPOSITA: a MADS-box gene controlling prophyll development and floral meristem identity in Antirrhinum . Development 131: 5981–5990. [DOI] [PubMed] [Google Scholar]
- 70. Gendrel AV, Lippman Z, Martienssen R, Colot V (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- 71. Li C, Wu K, Fu G, Li Y, Zhong Y, et al. (2009) Regulation of oleosin expression in developing peanut (Arachis hypogaea L.) embryos through nucleosome loss and histone modifications. J Exp Bot 60: 4371–4382. 10.1093/jxb/erp275 [DOI] [PubMed] [Google Scholar]
- 72. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210. 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE (2009) The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731. 10.1093/bioinformatics/btp472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zang C, Schones DE, Zeng C, Cui K, Zhao K, et al. (2009) A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25: 1952–1958. 10.1093/bioinformatics/btp340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xu H, Wei CL, Lin F, Sung WK (2008) An HMM approach to genome-wide identification of differential histone modification sites from ChIP-seq data. Bioinformatics 24: 2344–2349. 10.1093/bioinformatics/btn402 [DOI] [PubMed] [Google Scholar]
- 78. Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469. 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 80. Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis . Plant Cell 18: 1121–1133. 10.1105/tpc.105.039834 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Top panel: Rosette leaves from 14-d-old plants are shown. Scale bar: 2 mm. Bottom panel: Percentage of upwardly curled leaves in each genetic background is shown. Error bar indicates standard deviations from at least 20 plants. P values were determined by two-tailed t-test. (B) Comparison of brm-1 and brm-1 swn-4 double mutants grown in soil for 14 days. Scale: 1cm.
(PDF)
Expression data of floral homeotic genes, AG (A), AP1 (B), AP3 (C) and AGL24 (D), in different genetic backgrounds were determined by qRT-PCR with three biological replicates.
(PDF)
(PDF)
The expression levels of each gene were normalized to that of GAPDH, and the expression level in Col was set to 1. Error bars indicate standard deviation among three technical replicates from one representative experiment.
(PDF)
ChIP signals are shown as fold changes relative to that in wild-type plants. Error bars indicate standard deviation among three technical replicates from one representative experiment.
(PDF)
The expression level of AP1 gene was normalized to that of GAPDH. Error bars indicate standard deviation among three technical replicates from one representative experiment.
(PDF)
The data were extracted from Schmid et al [50] and displayed using the AtGenExpress Visualization Tool (http://jsp.weigelworld.org/expviz/expviz.jsp).
(PDF)
(A) ChIP-seq data showing an increase in H3K27me3 levels at several genes in brm-1. Data from the wild-type Col is shown in red at the top, and brm-1 is shown in orange at the bottom. (B) ChIP-qPCR validation using independent samples. Data are shown as percentage of input. Error bars indicate standard deviations among three technical replicates from one representative experiment. (C) Expression analysis of AGL24 and SMZ by qRT-PCR. The expression of each gene was normalized to that of GAPDH, and the expression level in Col was set to 1. Error bars indicate standard deviations among three technical replicates from one representative experiment. (D) Small RNA northern blot analysis of miR156 in brm-1 compared with Col. Two time points were used (10 days and 14 days after germination). The levels of small RNAs in brm-1 were compared with those in Col, which was set as 1. The numbers below the gel images indicate relative abundance. U6 served as loading control. RNA isolation and hybridization for miRNA detection was performed as described [65]. Digoxigenin-labeled miRNA probes were generated using the mirVana miRNA Probe Construction Kit (Ambion). Oligonucleotide probes used are listed in S2 Table.
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)