Abstract
The Catharanthus roseus Receptor-Like Kinase 1-like (CrRLK1L) family of 17 receptor-like kinases (RLKs) has been implicated in a variety of signaling pathways in Arabidopsis, ranging from pollen tube (PT) reception and tip growth to hormonal responses. The extracellular domains of these RLKs have malectin-like domains predicted to bind carbohydrate moieties. Domain swap analysis showed that the extracellular domains of the three members analyzed (FER, ANX1, HERK1) are not interchangeable, suggesting distinct upstream components, such as ligands and/or co-factors. In contrast, their intercellular domains are functionally equivalent for PT reception, indicating that they have common downstream targets in their signaling pathways. The kinase domain is necessary for FER function, but kinase activity itself is not, indicating that other kinases may be involved in signal transduction during PT reception.
Keywords: Arabidopsis, FERONIA, pollen tube reception, receptor-like kinases, signaling
Introduction
To accomplish double fertilization in flowering plants (angiosperms), the pollen (male gametophyte) hydrates on the stigma and germinates to form a pollen tube (PT), which transports two non-motile sperm through the stigma, the style, and the transmitting tract toward the ovule that harbors the embryo sac (female gametophyte). The PT's growth direction is influenced by long- and short-range attractants, ultimately guiding the PT to the female gametophyte, which consists of two female gametes—egg and central cell—and five accessory cells, including two synergids at the micropylar pole (reviewed in 1,2). The synergids mediate the last step of PT guidance toward the site of PT reception by secreting LUREs, small cysteine-rich peptides 3,4. PT reception occurs at the filiform apparatus, a membrane-rich region at the micropylar end of the synergids, the first point of contact between male and female gametophyte. The PT interacts with the synergid, ruptures, and releases the sperm to effect double fertilization, initiating seed development.
The first evidence that PT reception requires an active signaling process came from the identification of the Arabidopsis feronia (fer) mutant, whose embryo sacs develop normally but remain unfertilized because the PT continues to grow inside the female gametophyte and does not rupture to release the sperm (Supplementary Fig S1; 5,6). FER encodes a receptor-like kinase (RLK) of the CrRLK1L subfamily 7, which consists of 17 members (Fig1, reviewed in 8). FER is expressed throughout the plant except for mature pollen and localizes to the membrane-rich filiform apparatus of the synergids 7.
Figure 1. FER is a member of the CrRLK1L family of receptor-like kinases.
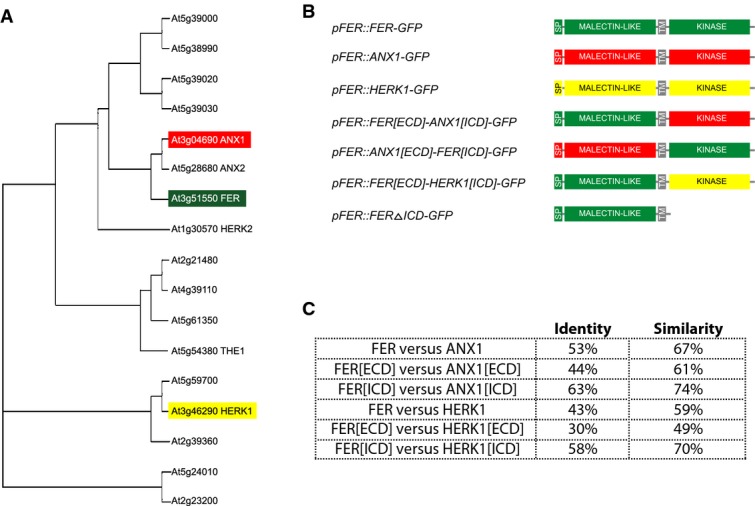
- Phylogenetic tree showing the relationships between CrRLK1L proteins (the members highlighted in color were investigated by domain swap experiments).
- Chimeric and truncated proteins assessed in this study. SP refers to the predicted signal peptides.
- Comparisons of amino acid similarity and identity between FER, ANX1, and HERK1 (determined by pairwise blastp alignments at http://blast.ncbi.nlm.nih.gov).
CrRLK1L proteins have a predicted intracellular S/T kinase domain (ICD) with relatively high conservation, a transmembrane (TM) domain, and a variable extracellular domain (ECD). Boisson-Dernier and colleagues 9 discovered two malectin-like domains in the ECDs of some CrRLK1L proteins. More recent domain searches, however, predict a malectin-like domain in all CrRLK1L members (pfam.sanger.ac.uk). This malectin-like domain has limited homology to the ER-localized, carbohydrate-binding malectin protein of Xenopus laevis (reviewed in 9), suggesting that CrRLK1L ligands may be glycosylated.
The functions of only six of the 17 CrRLK1L family members have been identified: FER, ANXUR1 (ANX1), ANX2, THESEUS1 (THE1), HERCULES1 (HERK1), and HERK2 (reviewed in 8,9). The two closest homologues of FER, ANX1 and ANX2, are only expressed in pollen, where they localize to the plasma membrane (PM) of the growing PT tip. Whereas single anx1 and anx2 mutants show no phenotype, anx1;anx2 double mutant PTs burst immediately after germination 10,11. ANX1/2 modulate the level of NADPH-oxidase-dependent reactive oxygen species (ROS) and the tip-focused Ca2+ gradient to sustain secretion of membrane and cell wall material to the PT tip 12. ANX1/2 function in tip-growing PTs seems to have been adopted by FER in polarly growing root hairs where FER acts upstream of several guanine exchange factors (ROPGEFs), activating Rho-like GTPases (RAC/ROPs) and leading to ROS-mediated root hair development 13–15. Additionally, FER-RopGEF-RAC/ROP modules seem to negatively regulate abscisic acid responses and positively regulate auxin-promoted root hair initiation and growth 15.
CrRLK1L proteins have also been implicated in cell elongation during vegetative growth. Expression of FER and two other CrRLK1L members, HERK1 and THE1, are upregulated in vegetative tissue after brassinosteroid treatment 16. All three genes are strongly expressed in elongating cells during vegetative growth, where they localize to the plasma membrane (PM). Both fer and the1;herk1 double mutant plants show a vegetative dwarf phenotype, indicating that FER, HERK1, and THE1 might act in the same pathway to regulate cell elongation.
In this study, we used the fer PT reception phenotype to investigate the functional similarities and differences between three CrRLK1L members (FER, ANX1, and HERK1) in a domain swap analysis. FER proteins with altered kinase domains were used to define the role of kinase activity for FER function. All constructs were expressed under the pFER promoter in fer-1/FER mutant plants, which normally show ∼50% unfertilized ovules due to defects in PT reception 5–7. Complementation analysis revealed common downstream signaling events but distinct ligand activation. In addition, we show that FER kinase activity is not necessary for FER function in PT reception, indicating that additional kinases may play a role in the FER signal transduction cascade.
Results
Domain swap analysis of FER and related proteins
Like most Receptor-like kinases (RLKs), the main determinant of specificity in the CrRLK1L family is predicted to be receptor–ligand interactions determined by specific amino acids in the ECD. This is supported by the fact that the CrRLK1L family members have highly divergent ECDs (Fig1). The ICDs of CrRLK1L proteins have highly conserved S/T kinase domains, but divergent C-terminal tails that could play a role in downstream specificity. A third determinant of specificity could be the distinct expression patterns of the various family members in combination with other upstream and downstream regulators. In order to determine the factors controlling CrRLK1L specificity, we tested whether closely related (ANX1) and more distantly related (HERK1) proteins (Fig1 and Supplementary Fig S2), and/or replacements of the ECD and ICD of FER with the respective domains of ANX1 and HERK1, are able to complement the PT reception phenotype in fer-1/FER mutants, when expressed under control of the pFER promoter.
We previously showed that a 1.2-kb pFER promoter fragment drives the expression of a FER-GFP fusion protein that localizes to the region of the filiform apparatus in synergids and complements the fer PT reception phenotype (Fig2A; 7). Primary transformants are hemizygous for pFER::FER-GFP such that only half of the fer embryo sacs carry the complementing construct (Fig3). We used the same promoter fragment to drive expression of full-length ANX1-GFP and HERK1-GFP fusion proteins in the fer-1/FER background. The fusion proteins were detected in the filiform apparatus (Fig2B, 2C), and in the PM of transiently transformed onion epidermal cells (Supplementary Fig S3). All primary transformants were either FER/FER or fer-1/FER and did not show significantly different numbers of unfertilized ovules compared to untransformed control plants (Fig3 and Supplementary Table S1). These results indicate that full-length ANX1-GFP and HERK1-GFP constructs cannot complement the fer-1 phenotype when expressed in synergids and that no dominant-negative effects on PT reception are conferred by expressing ANX1-GFP and HERK1-GFP.
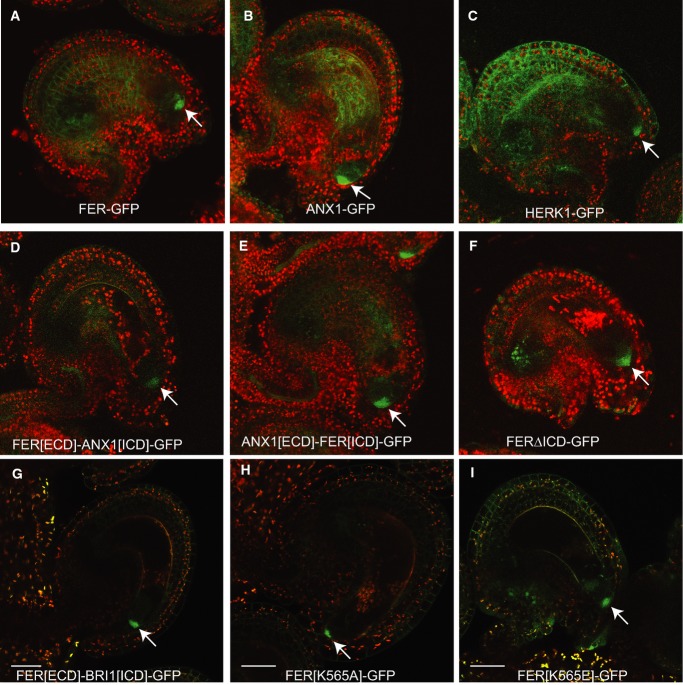
Figure 2. Representative confocal images illustrating that ANX1, HERK1, and domain swap constructs exhibit a FER-like subcellular localization in the filiform apparatus of synergids.
- A FER-GFP is localized to the filiform apparatus of synergids (arrow) and can also be detected at the periphery of sporophytic cells of the ovule.
- B-I All of the GFP fusion constructs show filiform apparatus localization (arrow). While synergid expression and localization to the filiform apparatus was consistent in all lines examined, sporophytic expression in the ovules varied depending on the transgene insertion site. Scale bars are 30 micrometers.
Data information: For all overlayed images, GFP fusion protein signal is shown in green and chlorophyll autofluorescence is shown in red.
Figure 3. Complementation of the fer-1 mutation with related CrRLK1L proteins and domains.
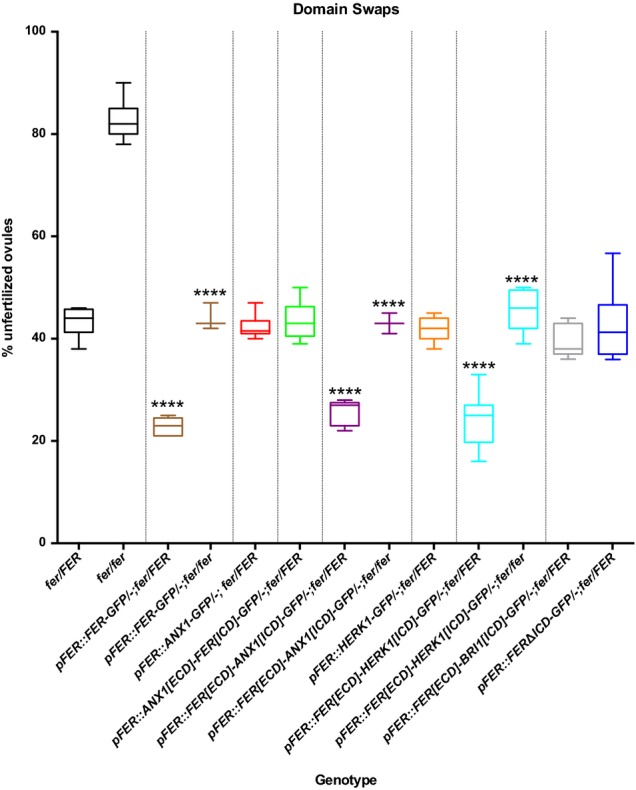
Box plots from complementation assays showing the percentage of unfertilized ovules for control plants and T1 transformants for each of the constructs. **** indicates lines with % unfertilized ovules significantly different from untransformed controls of the same genotype (P-values < 0.0001 in t-tests). At least 3 independent T1 plants (> 300 ovules counted for each) were analyzed for complementing and at least 8 T1 plants for non-complementing lines. See Supplementary Table S1 for raw seed count data.
The failure of ANX1 and HERK1 to complement the fer phenotype could be due to a lack of ligand recognition, an inability to perform downstream functions, or a combination of both. In order to distinguish between these possibilities, domain swaps between ANX1 and FER were constructed and driven under the pFER promoter as GFP fusion proteins (Fig1B). Primary transformants expressing the ANX1 ECD fused to the FER ICD showed expression of the fusion protein in the filiform apparatus of synergids in multiple transformants (Fig2E), and in the PM of onion epidermal cells (Supplementary Fig S3). However, only FER/FER and fer-1/FER plants were recovered. The percentages of unfertilized ovules in the primary transformants were not significantly different from controls (Fig3), indicating that the ANX1[ECD]-FER[ICD]-GFP protein is not able to complement fer-1 and that no dominant-negative effect was conferred by the ANX1 ECD. Thus, the FER ECD is indispensible for PT reception in synergids.
We also exchanged the FER ICD with the ICDs of ANX1 or HERK1 and expressed GFP fusion proteins under control of the pFER promoter (Fig1B). Both constructs were able to complement the fer-1 phenotype (FER[ECD]-ANX1[ICD]-GFP localization is shown in Fig2D). The percentage of unfertilized ovules was reduced by 50% in fer-1/FER and fer-1/fer-1 primary transformants that were hemizygous for the complementation constructs (Fig3). These results indicate that either the ICDs of ANX1 or HERK1 proteins are interchangeable with the FER ICD, that the ICD is dispensable for FER function, or that any kinase domain is sufficient. In order to distinguish between these possibilities, we deleted the ICD of FER in a GFP fusion construct driven by the pFER promoter. The FERΔICD deletion construct was expressed in synergids and localized to the filiform apparatus (Fig2F) but did not complement fer-1 (Fig3), indicating that the ICD is necessary for FER function in PT reception. We also exchanged the FER ICD with an ICD from an unrelated RLK, BRASSINOSTEROID INSENSITIVE1 (BRI1). BRI1 is a member of the leucine-rich repeat (LRR) subfamily and is the receptor for brassinosteroids 17,18. The pFER::FER[ECD]-BRI1[ICD]-GFP construct localized to the filiform apparatus but was unable to complement fer-1 (Figs2G and 3), indicating that a CrRLK1L ICD is necessary for transmitting the signal perceived by the FER ECD in synergids.
Functional analysis of the FER intracellular domain
The ICDs of CrRLK1 proteins contain a typical S/T kinase motif with a K at the active site (Fig4A and Supplementary Fig S4). In an in vitro kinase assay, the kinase domain of FER was capable of autophosphorylation and a point mutation changing K at the active site to R abolished in vitro kinase activity 7. In order to determine whether kinase activity is necessary for FER function in PT reception, a pFER::FER[K-R]-GFP construct, carrying the K-R change in the active site (K565), was transformed into fer-1/FER plants to check for complementation. Surprisingly, this dead kinase version of FER was able to complement the fer-1 phenotype (Fig4B and Supplementary Table S2). Two additional non-conserved substitutions, K-A and K-E, were properly localized to the filiform apparatus (Fig 2H and I) and also complemented fer-1 (Fig4B), indicating that kinase activity is not necessary for FER function in PT reception.
Figure 4. Site-directed mutagenesis of FER kinase domain.
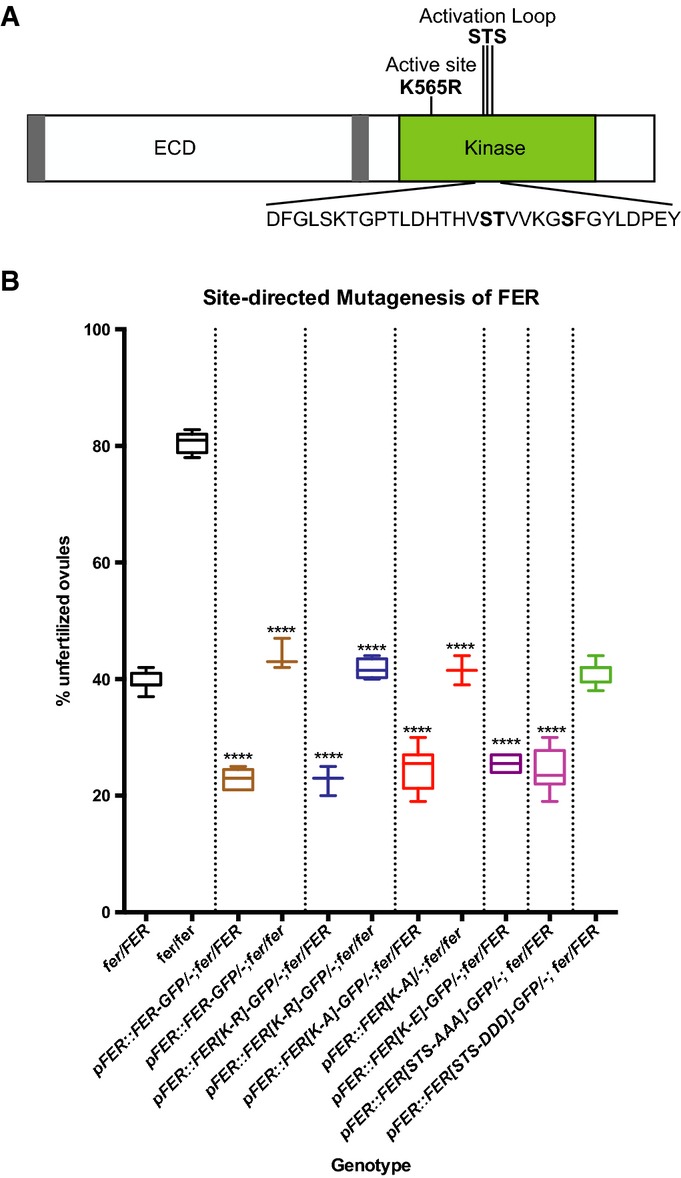
- FER protein domains and positions of the mutations introduced for complementation assays. The bold S, T, and S indicate targets for mutagenesis in the activation segment.
- Box plots from complementation assays showing the percentage of unfertilized ovules for control plants and T1 transformants for each of the site-directed mutagenesis constructs. **** indicates lines with % unfertilized ovules significantly different from untransformed controls of the same genotype (P-values < 0.0001 in t-tests). See Supplementary Table S2 for raw seed count data.
Structures based on X-ray crystallography show that kinases contain an activation segment C-terminal to the catalytic domain 19. In many kinases, phosphorylation of residues in the activation loop of this segment determines the conformation of the loop, which modulates kinase function. While most structure-function studies have been done with animal kinases, site-directed mutagenesis experiments have shown that activation loop phosphorylation is important for the function of some plant RLKs 20–22. Each of the CrRLK1L proteins has a predicted activation loop with S and T residues that are targets for phosphorylation (Fig4A). S695 and T696 of FER were shown to be phosphorylated in a membrane phosphoproteomics study on Arabidopsis seedlings, and the corresponding S residues were also found to be phosphorylated in HERK1 23. In seedling and mature pollen phosphoproteomic studies 23,24, an activation loop peptide with a phosphorylated S (S701 in FER) was identified; however, this peptide is highly conserved in CrRLK1 family members and could not be assigned to a specific protein. FER function could be modulated through phosphorylation changes at these residues. In order to investigate this possibility, site-directed mutagenesis was used to convert these residues (S695, T696, and S701) to A and D in order to mimic constitutively dephosphorylated and phosphorylated states, respectively. Single and double changes of these residues had no effect on the ability of the construct to complement fer-1 (Supplementary Table S2). A GFP fusion protein with all three amino acids changed to A (STS-AAA) was able to complement fer-1 (Fig4B), indicating that phosphorylation of the activation loop is not necessary for FER function. In contrast, the corresponding changes of these residues to D led to a failure to complement fer-1 (Fig4B). However, we could not recover any transformants that expressed the pFER::FER[STS-DDD]-GFP construct in synergids (12 independent hygromycin-resistant primary transformants were analyzed), indicating that the STS to DDD change in the FER activation loop likely leads to protein instability. Whether this change in protein stability is functionally significant in planta, that is whether it mimics what happens to a phosphorylated RLK, remains to be determined.
Discussion
Arabidopsis has over 400 predicted RLKs, but functions are known only for relatively few of these 25. The majority of plant RLKs are predicted S/T kinases, the largest family being LRR kinases with variable numbers of LRRs in their ECDs that are predicted to bind molecules such as hormones, elicitors, or peptide ligands 26. The common theme in mutant phenotypes associated with CrRLK1L genes is cell wall sensing 8,9,27,28, indicating that these RLKs probably bind similar types of ligands. The presence of malectin-like domains in CrRLK1L RLKs indicates that the ligands could be carbohydrates from the cell wall or peptide ligands with specific glycosylation patterns 8. It was recently shown that, in roots, FER binds to the secreted RALF peptide 29. However, RALF itself is not expressed in pollen, such that the ligand binding to FER during PT reception is likely another member of the large RALF-like family. RALF and 9 of the 34 RALF-like peptides have potential glycosylation sites (Supplementary Fig S5), including the pollen-expressed RALFL4 and RALFL26, indicating that glycosylation may be important for recognition by the malectin-like domains in the CrRLK1L ECDs. However, these glycosylation sites have not been experimentally verified and, thus, their role in CrRLK1L signaling pathways remains to be determined.
ANX1 and HERK1 are not functionally interchangeable with FER
FER and ANX1/2 perform seemingly opposite functions during pollination, with the ANX1/2 proteins maintaining PT growth and integrity 10,11, while FER signaling leads to cessation of PT tip growth, PT rupture, and release of the sperm to achieve double fertilization 5–7. One hypothesis for the function of the FER and ANX proteins is that they compete for the same ligand, with FER. This would lead the ligand at higher affinity upon PT arrival at the synergids. This would lead to inactivation of the ANX pathway and bursting of the PT, similar to that seen in anx1;anx2 double mutant PTs upon germination 10,11. If this hypothesis is true, then expressing ANX1 in synergids should complement the fer-1 phenotype, since the ANX1 ECD in the filiform apparatus should also compete for the ligand at the PT tip. However, neither the pFER::ANX1-GFP nor the pFER::ANX[ECD]-FER[ICD]-GFP construct could complement the fer-1 phenotype, indicating that ANX1 and FER probably bind different ligands and some other mechanism is responsible for inhibiting ANX signaling upon PT reception.
As expected, the more distantly related HERK1 RLK was also not able to complement fer-1 when expressed in synergids. However, both the HERK1-GFP and ANX1-GFP fusion proteins showed a subcellular localization very similar to FER-GFP in synergids. FER is highly enriched in the filiform apparatus, a membrane-rich region where PTs first contact the micropylar end of the synergid. This localization pattern is not seen with all membrane-localized proteins in synergids: the ROP6C protein, for instance, is evenly expressed in synergids 7; however, the molecular basis for this localization pattern remains to be determined.
The filiform apparatus consists of highly invaginated membranes. FER accumulates in this region of the synergid and the kinase domain is predicted to reside inside the cell 7. Even though ANX1 and HERK1 localize to the filiform apparatus when expressed in synergids, we cannot exclude the possibility that they are not integral membrane proteins or their topology differs from that of FER. In this case, the failure to complement fer-1 could reflect a distinct topology rather than a difference in ligand specificity. We consider this unlikely, however, because FER and HERK1 have been detected in membrane phosphoproteomics studies 23,30, and ANX1-GFP is localized to the PM at the tip of growing PTs 10,12 and onion epidermal cells (Supplementary Fig S3), indicating that these three CrRLK1Ls are indeed membrane proteins. Furthermore, GFP is pH sensitive 31 and no signal could be detected in onion epidermal cells when GFP was extracellular in the context of a fusion to a GPI-anchored protein 32. In contrast, the same protein fused to Citrine, which is less pH sensitive 33, was readily detectable in the filiform apparatus 34. Given that we can detect a signal in the filiform apparatus from all GFP fusion proteins, it is likely that their ICDs are indeed intracellular and that the proteins have a topology similar to that of FER.
The intracellular domains of CrRLK1L proteins are interchangeable
The CrRLK1L RLKs are predicted to perceive distinct ligands and transmit the signal through signal transduction cascades. Recent publications have shown that FER function in root hair elongation involves binding of ROPGEFs, which transduce a phosphorylation signal to a RAC/ROP, leading to ROS production 13,14. In yeast-two-hybrid assays, the FER kinase domain can bind with at least 4 different ROPGEFs 14. This FER/GEF/ROP pathway has also been implicated in suppression of abscisic acid signaling in roots by activating the ABSCISIC ACID INSENSITIVE2 phosphatase 15. Furthermore, ANX1/2 regulate NADPH-oxidase-dependent ROS production during PT elongation, indicating that the same signaling pathway may be shared in tip-growing root hairs and PTs 12. FER was also shown to control the production of ROS in the filiform apparatus of the synergids 35. While the exact role of ROPGEF/RAC/ROP signal transduction in FER-mediated PT reception and in other CrRLK1L-mediated processes has yet to be determined, our results, showing that the ANX1 and HERK1 ICDs can complement the fer-1 PT reception phenotype when linked to the FER ECD, clearly indicate that various CrRLK1L family members share common downstream signaling components.
FER proteins with non-conserved changes in the active site complement the fer mutant
Receptor-like kinases are defined by the presence of an ECD potentially involved in ligand binding, a transmembrane domain, and an intracellular kinase domain. Most RLKs are presumed to sense a ligand and phosphorylate another protein to initiate a signal transduction cascade 25. While FER has kinase activity in vitro that can be abolished by a K-R change in the active site 7, the dead kinase version of FER was able to complement fer-1, indicating that kinase activity is not necessary for FER function in PT reception. The K565R dead kinase was also able to partially complement the reduced response to root mechanostimulation in fer mutants 36. These results indicate that either a K565R change retains partial kinase function, or that another kinase in the complex is able to substitute for FER's role in signal transduction. Confirmation of the ability of K565 changes to completely abolish kinase activity awaits the identification of endogenous FER targets.
While most plant RLKs that have been studied extensively require kinase activity for their functions (reviewed in 37), a few do not. Among these are FEI1, a LRR RLK involved in cell wall biosynthesis 38, and Arabidopsis CRINKLY4 (ACR4), a CR4-type RLK involved in epidermal development 39. In addition, ∼20% of the more than 400 RLKs in the Arabidopsis genome have amino acid substitutions in critical active sites of their kinase domains, leading to the prediction that these RLKs perform their functions without kinase activity 40. Instead of directly phosphorylating downstream components of its signal transduction cascade, FER could function as a part of a complex with another RLK that acts synergistically with FER, and is thus able to complement the lack of kinase activity in the kinase dead version. Plant RLKs often occur in heterodimeric complexes. For example, the BRASSINOSTEROID-INSENSITIVE1-ASSOCIATED KINASE1/SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE3 (BAK1/SERK3) RLK has been shown to act in heterodimeric complexes with other LRR RLKs, such as BRI1 and FLAGELLIN-SENSING2 (FLS2), to enhance their activity 41. Like BAK1, FER has been shown to be involved in diverse developmental processes 5,7,13–16,42. It is tempting to speculate that in some of these cases, FER also acts as a co-receptor to enhance the activity of another kinase that transduces the received signal.
FER could also act as a scaffolding protein to bring other components such as ROPGEFs into a complex so that signal transduction can occur. In mice, the EPIDERMAL GROWTH FACTOR RECEPTOR (EGFR) family member ErbB3 has a non-functional kinase domain and acts in a complex with other EGFR proteins with active kinase domains. Upon ligand binding, the active EGFR phosphorylates ErbB3 at specific tyrosines, which then serve as docking sites for downstream targets (reviewed in 43). FER has been shown to directly bind ROPGEF proteins that are involved in downstream signal transduction 13, and our data showing that the kinase domain is necessary for FER function—even though kinase activity is not—indicate that FER may bind ROPGEFs but not be responsible for activating them by phosphorylation. FER might act in a complex with another CrRLK1L protein, a member of a different RLK class, or even a cytoplasmic kinase to provide a docking site for a ROPGEF, which is then phosphorylated by the FER partner. Testing of this model awaits the identification of more FER-interacting proteins.
Taken together, our results suggest that the three members of the CrRLK1L subfamily FER, ANX1, and HERK1 share common downstream signaling targets, but are activated by distinct ligand interactions. Furthermore, the kinase activity of FER may not be essential to execute its function, indicating that FER might act as an important co-receptor recruiting other co-factors or downstream targets to mediate signal transduction during PT reception.
Materials and Methods
Plant material and growth conditions
fer-1 plants (allele described in 7) were used for all transformations and Landsberg erecta was used as the wild-type control for complementation. The FER-GFP line was reported in 7. The fer/fer line was derived from the fer-1 allele 44. Plant growth conditions were as described 5. For transformation, fer-1 heterozygotes were selected by plating F2 seeds on MS plates supplemented with 50 mg/l kanamycin. After Agrobacterium-mediated transformation, seeds were harvested and plated on MS plates supplemented with 20 mg/l hygromycin to select transformants.
Domain swap and site-directed mutagenesis constructs
PCR with Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland) was used to generate all constructs. The 1.2-kb pFER promoter fragment described in 7 was used to drive expression of GFP fusion constructs in synergids as well as sporophytic tissues. Primers described in Supplementary Table S3 were used in the combinations described in Supplementary Table S4 to create overlapping fragments with Gateway attB sites. These PCR products were used to create entry clones in pDONR207 (Life Technologies) and sequenced to ensure that no undesired mutations were introduced by PCR. Entry clones were used in LR reactions with pMDC111 45 to create in-frame GFP fusions in a plant binary vector. pMDC107 (identical to pMDC111 but with a different reading frame 45) was used for the ΔICD construct. The site-directed mutagenesis constructs were generated by modifying the pFER::FER-GFP/pMDC111 construct that complemented the fer-1 mutation in 7. Overlapping primers (Supplementary Table S5) were designed to introduce the desired nucleotide changes between the Sma1 and Xba1 restriction sites in the FER kinase domain. Restriction digests and ligations were used to replace the native fragments with the mutated PCR fragments, and all constructs were sequenced to verify that the desired mutations had been introduced.
fer-1 complementation assays
Constructs were transformed into Agrobacterium tumefaciens strain GV3101, and the resultant strain was used for transformation of Arabidopsis fer-1/FER plants using the floral dip method 46. Progeny were grown on MS plates containing 20 mg/l hygromycin to select transformants. Plants were grown to maturity and genotyped for the fer-1 mutation 7 and screened for expression of the GFP fusion proteins in synergids. Self-pollinated pistils from synergid expressing plants were collected at 2–4 days after pollination, and counts of fertilized vs. unfertilized ovules were performed to determine whether the constructs complement the fer-1 PT reception phenotype. For each construct, a minimum of 3 independent T1 transformants (average 6) of each FER genotype were used for complementation analysis and 300–500 ovules were counted for each plant. For non-complementing lines, a minimum of 8 primary transformants was analyzed for each genotype.
Confocal microscopy
In order to determine the subcellular localization of GFP fusion proteins, carpel walls were removed from mature pistils and specimens were observed by confocal microscopy. Images 2A-F were captured and processed as described 44. For images 2G-I, ovules were dissected from pistils 2 days after emasculation and mounted in DI water on standard slides. GFP and autofluorescence were imaged with a Leica TCS SP8 confocal laser scanning microscope, equipped with a 488 notch filter, using a 40× water correction objective (NA = 1.10). Fluorescence was excited using a 488 nm argon laser and detected with HyD detectors set at 489–544 nm (GFP) and 549–669 nm (autofluorescence). Single scan images were processed in ImageJ v.1.48p (NIH, Bethesda MD).
Acknowledgments
We are grateful to Michael Raissig for critical reading of the manuscript, Valeria Gagliardini, Arturo Bolaños, and Peter Kopf (University of Zürich) for general laboratory support, Christian Frey and Karl Huwiler (University of Zürich) for plant care, and Patrick Day and Nic Cejda (University of Oklahoma) for plant maintenance and seed collection. This work was funded by the University of Zürich, the University of Oklahoma, grants of Swiss National Science Foundation (SNF) to U.G., a Human Frontiers in Science Long-term Fellowship to S.A.K., and partial support of H.L. through a Research Module of the SNF ProDoc Program ‘Molecular Life Sciences’ to U.G.
Author contributions
SAK, HL, and UG conceived the experiments; SAK, HL, and DJ performed the experiments; SAK, HL, and UG analyzed the data; SAK, HL, and UG wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Information
Review Process File
References
- Dresselhaus T, Franklin-Tong N. Male-female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol Plant. 2013;6:1018–1036. doi: 10.1093/mp/sst061. [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Tsukamoto T. Pathfinding in angiosperm reproduction: pollen tube guidance by pistils ensures successful double fertilization. Wiley Interdiscip Rev Dev Biol. 2012;1:96–113. doi: 10.1002/wdev.6. [DOI] [PubMed] [Google Scholar]
- Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, et al. Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature. 2009;458:357–361. doi: 10.1038/nature07882. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Higashiyama T. A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol. 2012;10:e1001449. doi: 10.1371/journal.pbio.1001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck N, Moore JM, Federer M, Grossniklaus U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development. 2003;130:2149–2159. doi: 10.1242/dev.00458. [DOI] [PubMed] [Google Scholar]
- Rotman N, Rozier F, Boavida L, Dumas C, Berger F, Faure JE. Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr Biol. 2003;13:432–436. doi: 10.1016/s0960-9822(03)00093-9. [DOI] [PubMed] [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007;317:656–660. doi: 10.1126/science.1143562. [DOI] [PubMed] [Google Scholar]
- Lindner H, Muller LM, Boisson-Dernier A, Grossniklaus U. CrRLK1L receptor-like kinases: not just another brick in the wall. Curr Opin Plant Biol. 2012;15:659–669. doi: 10.1016/j.pbi.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Kessler SA, Grossniklaus U. The walls have ears: the role of plant CrRLK1Ls in sensing and transducing extracellular signals. J Exp Bot. 2011;62:1581–1591. doi: 10.1093/jxb/erq445. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Roy S, Kritsas K, Grobei MA, Jaciubek M, Schroeder JI, Grossniklaus U. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development. 2009;136:3279–3288. doi: 10.1242/dev.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki S, Murata T, Sakurai-Ozato N, Kubo M, Demura T, Fukuda H, Hasebe M. ANXUR1 and 2, sister genes to FERONIASIRENE, are male factors for coordinated fertilization. Curr Biol. 2009;19:1327–1331. doi: 10.1016/j.cub.2009.06.064. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Lituiev DS, Nestorova A, Franck CM, Thirugnanarajah S, Grossniklaus U. ANXUR receptor-like kinases coordinate cell wall integrity with growth at the pollen tube tip via NADPH oxidases. PLoS Biol. 2013;11:e1001719. doi: 10.1371/journal.pbio.1001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA. 2010;107:17821–17826. doi: 10.1073/pnas.1005366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GQ, Li E, Ge FR, Li S, Wang Q, Zhang CQ, Zhang Y. Arabidopsis RopGEF4 and RopGEF10 are important for FERONIA-mediated developmental but not environmental regulation of root hair growth. New Phytol. 2013;200:1089–1101. doi: 10.1111/nph.12432. [DOI] [PubMed] [Google Scholar]
- Yu F, Qian L, Nibau C, Duan Q, Kita D, Levasseur K, Li X, Lu C, Li H, Hou C, et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc Natl Acad Sci USA. 2012;109:14693–14698. doi: 10.1073/pnas.1212547109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2009;106:7648–7653. doi: 10.1073/pnas.0812346106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Cano-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Klaus-Heisen D, Nurisso A, Pietraszewska-Bogiel A, Mbengue M, Camut S, Timmers T, Pichereaux C, Rossignol M, Gadella TW, Imberty A, et al. Structure-function similarities between a plant receptor-like kinase and the human interleukin-1 receptor-associated kinase-4. J Biol Chem. 2011;286:11202–11210. doi: 10.1074/jbc.M110.186171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, Vervoort J, de Vries SC. Role of threonines in the Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTORKINASE1 activation loop in phosphorylation. J Biol Chem. 2001;276:41263–41269. doi: 10.1074/jbc.M102381200. [DOI] [PubMed] [Google Scholar]
- Nuhse TS, Stensballe A, Jensen ON, Peck SC. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell. 2004;16:2394–2405. doi: 10.1105/tpc.104.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayank P, Grossman J, Wuest S, Boisson-Dernier A, Roschitzki B, Nanni P, Nuhse T, Grossniklaus U. Characterization of the phosphoproteome of mature Arabidopsis pollen. Plant J. 2012;72:89–101. doi: 10.1111/j.1365-313X.2012.05061.x. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dievart A, Clark SE. LRR-containing receptors regulating plant development and defense. Development. 2004;131:251–261. doi: 10.1242/dev.00998. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wu HM. THESEUS 1, FERONIA and relatives: a family of cell wall-sensing receptor kinases? Curr Opin Plant Biol. 2011;14:632–641. doi: 10.1016/j.pbi.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Hematy K, Hofte H. Novel receptor kinases involved in growth regulation. Curr Opin Plant Biol. 2008;11:321–328. doi: 10.1016/j.pbi.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science. 2014;343:408–411. doi: 10.1126/science.1244454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R. PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem. 2010;285:39140–39149. doi: 10.1074/jbc.M110.160531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsliger MA, Wachter RM, Hanson GT, Kallio K, Remington SJ. Structural and spectral response of green fluorescent protein variants to changes in pH. Biochemistry. 1999;38:5296–5301. doi: 10.1021/bi9902182. [DOI] [PubMed] [Google Scholar]
- Capron A, Gourgues M, Neiva LS, Faure JE, Berger F, Pagnussat G, Krishnan A, Alvarez-Mejia C, Vielle-Calzada JP, Lee YR, et al. Maternal control of male-gamete delivery in Arabidopsis involves a putative GPI-anchored protein encoded by the LORELEI gene. Plant Cell. 2008;20:3038–3049. doi: 10.1105/tpc.108.061713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greisbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. Reducing the environmental sensitivity of yellow fluorescent protein. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- Lindner H. 2014. Pollen Tube Reception in the Synergid Cells of Arabidopsis thaliana – Novel Players and Old Acquaintances. PhD Thesis, University of Zürich, Switzerland.
- Duan Q, Kita D, Johnson EA, Aggarwal M, Gates L, Wu HM, Cheung AY. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat Commun. 2014;5:3129. doi: 10.1038/ncomms4129. [DOI] [PubMed] [Google Scholar]
- Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB. The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr Biol. 2014;24:1887–1892. doi: 10.1016/j.cub.2014.06.064. [DOI] [PubMed] [Google Scholar]
- Gish LA, Clark SE. The RLK/Pelle family of kinases. Plant J. 2011;66:117–127. doi: 10.1111/j.1365-313X.2011.04518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, Kieber JJ. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell. 2008;20:3065–3079. doi: 10.1105/tpc.108.063354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Robertson FC, Soares DC, Ingram GC. ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell. 2005;17:1154–1166. doi: 10.1105/tpc.104.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells E, Casacuberta JM. Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. J Exp Bot. 2007;58:3503–3511. doi: 10.1093/jxb/erm226. [DOI] [PubMed] [Google Scholar]
- Liebrand TW, van den Burg HA, Joosten MH. Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 2014;19:123–132. doi: 10.1016/j.tplants.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Deslauriers SD, Larsen PB. FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Mol Plant. 2010;3:626–640. doi: 10.1093/mp/ssq015. [DOI] [PubMed] [Google Scholar]
- Kroiher M, Miller MA, Steele RE. Deceiving appearances: signaling by “dead” and “fractured” receptor protein-tyrosine kinases. BioEssays. 2001;23:69–76. doi: 10.1002/1521-1878(200101)23:1<69::AID-BIES1009>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U. Conserved molecular components for pollen tube reception and fungal invasion. Science. 2010;330:968–971. doi: 10.1126/science.1195211. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A. Arabidopsis thaliana floral dip transformation method. Methods Mol Biol. 2006;343:87–103. doi: 10.1385/1-59745-130-4:87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File