The type IV restriction endonuclease ScoMcrA, which cleaves both Dcm-methylated DNA and phosphorothioated DNA, was crystallized and the crystals diffracted to 3.35 Å resolution. The space group and the unit-cell parameters were determined.
Keywords: ScoMcrA, DNA phosphorothioation, Dcm methylation
Abstract
ScoMcrA is a type IV modification-dependent restriction endonuclease found in the model strain Streptomyces coelicolor. Unlike type I, II and III restriction endonucleases, which cleave unmodified DNA, type IV restriction endonucleases cleave modified DNA, including methylated, hydroxymethylated, glucosyl-hydroxymethylated and phosphorothioated DNA. ScoMcrA targets both Dcm-methylated DNA and phosphorothioated DNA, and makes double-strand breaks 16–28 nt away from the modified nucleotides or the phosphorothioate links. However, the mechanism by which ScoMcrA recognizes these two entirely different types of modification remains unclear. In this study, the ScoMcrA protein was overexpressed, purified and crystallized. The crystals diffracted to 3.35 Å resolution and belonged to space group P212121. The unit-cell parameters were determined to be a = 130.19, b = 139.36, c = 281.01 Å, α = β = γ = 90°. These results will facilitate the detailed structural analysis of ScoMcrA and further elucidation of its biochemical mechanism.
1. Introduction
The dnd gene cluster, which consists of five genes (dndA–dndE), in Streptomyces lividans is known to make a sulfur modification to the genomic DNA (Zhou et al., 2005 ▶). This natural modification was later found to be DNA phosphorothioation, in which a nonbridging O atom of the phosphodiester bond is substituted by a sulfur (Wang et al., 2007 ▶). Homologues of dnd genes have been found in more than 50 bacterial and archaeal genomes (Ou et al., 2009 ▶), and most of them are located in mobile elements (He et al., 2007 ▶). The phosphorothioation governed by Dnd proteins from different organisms varies in DNA-sequence specificity (Liang et al., 2007 ▶; Wang et al., 2011 ▶; Cao, Chen et al., 2014 ▶) and has recently been implicated in DNA restriction-modification systems (Liu et al., 2010 ▶; Xu et al., 2010 ▶; Cao, Cheng et al., 2014 ▶) and a bacterial antioxidation function (Xie et al., 2012 ▶).
The activity of many restriction endonucleases is inhibited if a phosphorothioate modification is found at the cleavage site (Verma & Eckstein, 1998 ▶). In contrast, the type IV restriction endonuclease ScoMcrA from S. coelicolor specifically targets DNA bearing phosphorothioation at the sequence 5′-CG*GCCG-3′ (where * represents the phosphorothioate link) and Dcm-methylated DNA with the sequence 5′-CCWGG-3′ (a methyl group was added to the inner cytosine), and cleaves 16–28 nucleotides away from the modification sites in the presence of Mn2+ (Liu et al., 2010 ▶).
A BLAST search with ScoMcrA revealed seven close homologues with at least 75% amino-acid identity in GenBank. ScoMcrA and its homologues all contain an HNH motif, which is usually present in restriction endonucleases and homing endonucleases and plays a role in hydrolysis of the DNA phosphodiester bond (Yang, 2011 ▶).
To understand the way that ScoMcrA specifically recognizes phosphorothioated DNA and Dcm-methylated DNA, we crystallized apo ScoMcrA, collected diffraction data and processed the data to 3.35 Å resolution. The results presented in this report will provide a basis for the ultimate structure elucidation of ScoMcrA, which should shed light on the recognition mechanism of this unique DNA modification.
2. Materials and methods
2.1. Macromolecule production
ScoMcrA was expressed on pJTU1655 (Liu et al., 2010 ▶) as an N-terminally His-tagged protein in Escherichia coli strain BL21(DE3). When the OD600 reached 0.6–0.8, the E. coli cell culture was induced with 0.2 mM IPTG, grown for a further 8 h at 298 K (or overnight at 289 K) and harvested by centrifugation at 2500g for 20 min.
The cell pellets were suspended in buffer A (20 mM Tris–HCl pH 8.0, 300 mM NaCl, 20 mM imidazole), which was supplemented with 1 mg ml−1 aprotinin, 1 mg ml−1 leupeptin, 30 mg ml−1 lysozyme and 0.05 mM phenylmethylsulfonyl fluoride (PMSF), and lysed using a homogenizer at 277 K. The cell lysate was centrifuged at 15 000g for 30 min at 277 K, and the supernatant was applied onto a HisTrap HP column (GE Healthcare) and resolved using an ÄKTA FPLC (GE Healthcare) by eluting with an linear gradient of imidazole (20–500 mM). The collected protein sample was further purified sequentially by anion-exchange chromatography using a Source 15Q column (GE Healthcare) and gel-filtration chromatography using a Superdex 200 10/300 GL column (GE Healthcare) pre-equilibrated with 10 mM Tris–HCl pH 8.0, 100 mM NaCl, 2 mM dithiothreitol (DTT). The purity of the protein was verified by SDS–PAGE with Coomassie Brilliant Blue staining (Fig. 1 ▶). The purified protein from Superdex 200 gel-filtration chromatography was concentrated to 10 mg ml−1, cooled in liquid nitrogen and stored at 193 K until use. Macromolecule-production information is summarized in Table 1 ▶.
Figure 1.
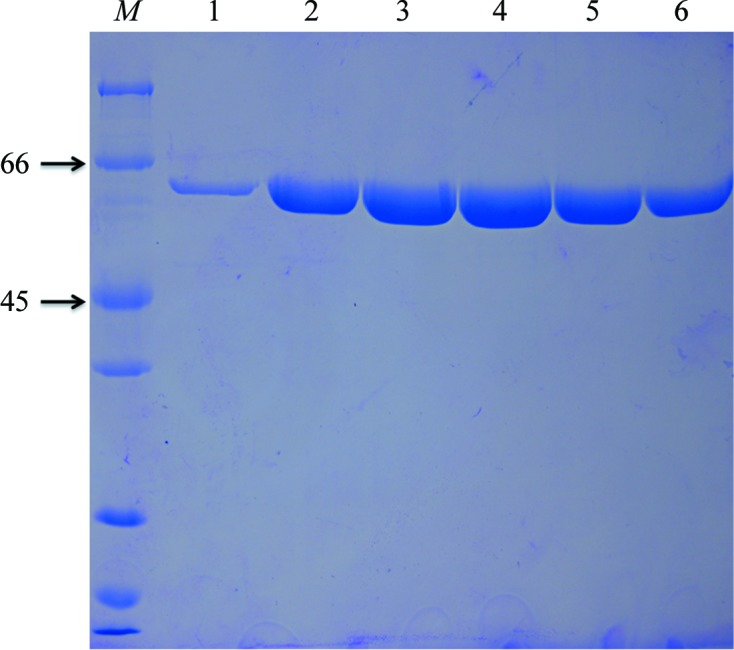
Purified ScoMcrA was analyzed using 15% SDS–PAGE followed by Coomassie Brilliant Blue staining. Lane M, protein molecular-weight marker (labelled in kDa). Lanes 1–6 contain fractions from tubes 26–31 (the fraction volume in each tube is 0.5 ml) from gel-filtration chromatography using a Superdex 200 column, and the elution peak volume was 14.08 ml. Samples of lanes 3 and 4 (corresponding to tubes 28 and 29 or an elution volume of 13.5–14.5 ml) were collected and concentrated to 10 mg ml−1 for crystal screening.
Table 1. Macromolecule-production information.
The NdeI restriction site and the amino acids added to the wild-type ScoMcrA are underlined.
| Source organism | S. coelicolor |
| DNA source | Genomic DNA |
| Forward primer | CATATGGCACCTTCGGAGAT |
| Reverse primer | GGGCTCAGGCAGCGTAAT |
| Cloning vector | pBluescript |
| Expression vector | pET-28a |
| Expression host | E. coli BL21(DE3) |
| Complete amino-acid sequence of the construct produced | MGSSHHHHHHSSGLVRGSHMASEITRAGILQAIAEHDRIGEAFRATYGFHAATSYFLEHEGRLYDSKAIAGVAHMYDFGVALKSSGLSGGLKHAVAWLRREGFTIREAKTFHRRVGDVRARRAMGALHRVLLLWAIGQAVARARLQWSTTRDAVALMEKYGQVEDGVDGVRYFWALVRDDLWCVEQAEELTLTSRGRRTLESLNAVDSAGLREDDYNLLRSQEAAASAAAGLIARYFHLLAGLLEDFGLHELLAGRWDALRLLGETFKDRDAIWRAYGGQKMAGIGCLADGILSAFSDDKGYADGRIDTTWIAYVGDGLSGDQKLTDGNELMAEHQAVGRALRYWHKFQGQWSFETWAVIVQRRLRWGLGEDKLRREFLWVLAVSERETWEVLEALEADTGELHDDTGDYRSDLALTGADGTESDDEAYRRLAQKAEANAERRGQLKKTVADKYVRDSARGAVLKRCQKRCENECAGHTELTKAGLILQVDHVNDLAKGGDVWNMIALCNCHALKTYGANKVRLQRLLAATARRLHEEKLQ |
2.2. Crystallization
Crystallization trials for ScoMcrA were performed at 287 K using the hanging-drop vapour-diffusion method in 24-well plates. Typically, 1 µl reservoir solution was mixed with 1 µl protein solution and equilibrated against 160 µl reservoir solution. Initial crystallization screening trials were performed using Crystal Screen, Crystal Screen 2, Crystal Screen Lite, Index, PEG/Ion, PEGRx and SaltRx kits from Hampton Research. After 3 d, small crystals of ScoMcrA were obtained from condition No. 17 of the Crystal Screen Lite kit, which consists of 0.2 M lithium sulfate monohydrate, 0.1 M Tris–HCl pH 8.5, 15%(w/v) polyethylene glycol (PEG) 4000. In our subsequent optimization efforts, various concentrations of PEG 4000 from 10 to 14%(w/v) with a stepwise increase of 0.5% were screened. It was found that changing the concentration of PEG 4000 from 10 to 13% resulted in a substantial enlargement of the crystal size. After streak-seeding, diffracting crystals were obtained using the hanging-drop vapour-diffusion method in 24-well plates at 287 K after one week. To obtain better crystals, we screened conditions including changing the buffer to MES (pH 5.2–7.1), sodium citrate (pH 4.2–6.5), sodium cacodylate (pH 5.1–7.4), bis-tris (pH 5.5–7.4), Tris–HCl (pH 7.0–9.0) and HEPES (pH 6.8–8.2), changing the lithium sulfate monohydrate to 47 salts from the StockOptions Salt kit (Hampton Research), changing the PEG 4000 to PEG 200, PEG 1000, PEG 3350, PEG 6000, PEG 10 000, PEG 20 000, polyethylene glycol monomethyl ether (PME) 550, PME 2000 and PME 5000 with concentrations from 10 to 25%, and changing the temperature to 277 and 297 K. A total of more than 40 000 droplets were set up during the course of this crystallization campaign, but there was still no significant improvement in the crystal quality. Crystallization information is summarized in Table 2 ▶.
Table 2. Crystallization.
| Method | Vapour diffusion |
| Plate type | 24-well hanging-drop plate (Hampton Research) |
| Temperature (K) | 287 |
| Protein concentration (mgml1) | 10 |
| Buffer composition of protein solution | 10mM TrisHCl pH 8.0, 100mM NaCl, 2mM DTT |
| Composition of reservoir solution | 0.2M lithium sulfate monohydrate, 0.1M TrisHCl pH 8.5, 13%(w/v) PEG 4000 |
| Volume and ratio of drop (l) | 1.0:1.0 |
| Volume of reservoir (l) | 160 |
2.3. Data collection and processing
The ScoMcrA crystals were cryoprotected by transferring them in cryoloops into a cryoprotectant which consisted of reservoir solution supplemented with 30%(v/v) glycerol, replacing the water in the crystallant with cryoprotectant. Prior to data collection, ScoMcrA protein crystals were flash-cooled in liquid nitrogen and tested on an in-house X-ray generator at Shanghai Institute of Organic Chemistry. A complete diffraction data set was collected on beamline BL17U1 at Shanghai Synchrotron Radiation Facility (SSRF), People’s Republic of China. The data set was collected at a wavelength of 0.978918 Å and was processed to 3.35 Å resolution. Intensity data were integrated and scaled using the HKL-2000 software (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
Recombinant N-terminally His-tagged ScoMcrA was purified by Ni2+-affinity, anion-exchange and gel-filtration chromatography and was subjected to extensive crystallization screening. The gel-filtration chromatography elution peak volume was 14.08 ml, corresponding to a calculated molecular weight of about 127.2 kDa, which indicated that ScoMcrA exists as a dimer in solution.
Optimization of the crystallization conditions yielded larger irregular hexagon-shaped crystals (Fig. 2 ▶). The crystals of the ScoMcrA protein diffracted to 3.35 Å resolution (Fig. 3 ▶) and belonged to the orthorhombic space group P212121, with unit-cell parameters a = 130.19, b = 139.36, c = 281.01 Å, α = β = γ = 90.00°. Diffraction data were collected and processed (Table 3 ▶) with a final R meas value of 14.4%. The data completeness, multiplicity and the average I/σ(I) value for the collected data set were 100.0%, 7.4 and 17.2, respectively (100.0%, 7.5 and 3.7, respectively, for the highest resolution shell).
Figure 2.

Irregular hexagon-shaped crystal of ScoMcrA. The maximum dimensions of the crystal are ∼0.4 × 0.2 × 0.04 mm
Figure 3.
A typical X-ray diffraction pattern from an ScoMcrA protein crystal. The crystal diffraction data set was processed to 3.35 Å resolution.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Beamline | BL17U1, SSRF |
| Wavelength () | 0.978918 |
| Temperature (K) | 100 |
| Detector | ADSC Q315r |
| Crystal-to-detector distance (mm) | 400 |
| Rotation range per image () | 1 |
| Total rotation range () | 180 |
| Exposure time per image (s) | 1 |
| Space group | P212121 |
| Unit-cell parameters (, ) | a = 130.19, b = 139.36, c = 281.01, = = = 90.00 |
| No. of molecules per asymmetric unit | 8 |
| Matthews coefficient (3Da1) | 2.49 |
| Solvent content (%) | 50.54 |
| Mosaicity () | 0.388 |
| Resolution range () | 50.03.35 (3.473.35) |
| Total No. of reflections | 548396 |
| No. of unique reflections | 74167 |
| Completeness (%) | 100 (100) |
| Multiplicity | 7.4 (7.5) |
| I/(I) | 17.2 (3.7) |
| R meas | 0.144 (0.788) |
| R p.i.m. | 0.053 (0.287) |
| Overall B factor from Wilson plot (2) | 53.5 |
Despite the considerable effort devoted to optimization of the ScoMcrA crystals using different salts, buffers and polymers, there was no improvement in the resolution. The addition of additives (Hampton Research), change of the cryoprotectant and post-crystallization treatments such as crystal soaking, annealing and dehydration (Heras & Martin, 2005 ▶), as well as reductive alkylation (Walter et al., 2006 ▶) of ScoMcrA before crystal screening, failed to improve the diffraction quality.
Considering the Matthews coefficient, we propose that the asymmetric unit contains eight ScoMcrA molecules (four ScoMcrA dimers). In this case the Matthews coefficient is 2.49 Å3 Da−1, corresponding to a solvent content of 50.54%. Crystallization of selenomethionine-substituted ScoMcrA protein is in progress, and the crystals will be used to collect multiwavelength anomalous diffraction (MAD) data. Structure determination of ScoMcrA will provide the mechanism by which ScoMcrA recognizes phosphorothioation on the DNA backbone.
Acknowledgments
We thank Dr Jiahai Zhou at Shanghai Institute of Organic Chemistry for assistance in the screening for diffracting crystals. We also thank Dr Jianhua He, Dr Sheng Huang, Dr Feng Yu and the other members of the staff of beamline BL17U1 at Shanghai Synchrotron Radiation Facility for assistance during data collection. This work was supported by grants from the Ministry of Education of China (20110073130011) and the National Natural Science Foundation of China (31170083 and 31130068).
References
- Cao, B., Chen, C. et al. (2014). Nature Commun. 5, 3951. [DOI] [PMC free article] [PubMed]
- Cao, B., Cheng, Q., Gu, C., Yao, F., DeMott, M. S., Zheng, X., Deng, Z., Dedon, P. C. & You, D. (2014). Mol. Microbiol. 93, 776–785. [DOI] [PMC free article] [PubMed]
- He, X., Ou, H.-Y., Yu, Q., Zhou, X., Wu, J., Liang, J., Zhang, W., Rajakumar, K. & Deng, Z. (2007). Mol. Microbiol. 65, 1034–1048. [DOI] [PubMed]
- Heras, B. & Martin, J. L. (2005). Acta Cryst. D61, 1173–1180. [DOI] [PubMed]
- Liang, J., Wang, Z., He, X., Li, J., Zhou, X. & Deng, Z. (2007). Nucleic Acids Res. 35, 2944–2954. [DOI] [PMC free article] [PubMed]
- Liu, G., Ou, H.-Y., Wang, T., Li, L., Tan, H., Zhou, X., Rajakumar, K., Deng, Z. & He, X. (2010). PLoS Genet. 6, e1001253. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Ou, H.-Y., He, X., Shao, Y., Tai, C., Rajakumar, K. & Deng, Z. (2009). PLoS One, 4, e5132. [DOI] [PMC free article] [PubMed]
- Verma, S. & Eckstein, F. (1998). Annu. Rev. Biochem. 67, 99–134. [DOI] [PubMed]
- Walter, T. S., Meier, C., Assenberg, R., Au, K. F., Ren, J., Verma, A., Nettleship, J. E., Owens, R. J., Stuart, D. I. & Grimes, J. M. (2006). Structure, 14, 1617–1622. [DOI] [PMC free article] [PubMed]
- Wang, L., Chen, S., Vergin, K. L., Giovannoni, S. J., Chan, S. W., DeMott, M. S., Taghizadeh, K., Cordero, O. X., Cutler, M., Timberlake, S., Alm, E. J., Polz, M. F., Pinhassi, J., Deng, Z. & Dedon, P. C. (2011). Proc. Natl Acad. Sci. USA, 108, 2963–2968. [DOI] [PMC free article] [PubMed]
- Wang, L., Chen, S., Xu, T., Taghizadeh, K., Wishnok, J. S., Zhou, X., You, D., Deng, Z. & Dedon, P. C. (2007). Nature Chem. Biol. 3, 709–710. [DOI] [PubMed]
- Xie, X., Liang, J., Pu, T., Xu, F., Yao, F., Yang, Y., Zhao, Y.-L., You, D., Zhou, X., Deng, Z. & Wang, Z. (2012). Nucleic Acids Res. 40, 9115–9124. [DOI] [PMC free article] [PubMed]
- Xu, T., Yao, F., Zhou, X., Deng, Z. & You, D. (2010). Nucleic Acids Res. 38, 7133–7141. [DOI] [PMC free article] [PubMed]
- Yang, W. (2011). Q. Rev. Biophys. 44, 1–93. [DOI] [PMC free article] [PubMed]
- Zhou, X., He, X., Liang, J., Li, A., Xu, T., Kieser, T., Helmann, J. D. & Deng, Z. (2005). Mol. Microbiol. 57, 1428–1438. [DOI] [PubMed]



