A protease in the common commercial enzyme cleaner Zymit used for the cleaning of crystallization robots cannot be completely inactivated by EDTA.
Keywords: crystallization robotics, protease, protease inhibitor, Zymit, cleaning protocol
Abstract
The protease in the commonly used commercial low-foam enzyme cleaner Zymit cannot be completely blocked by EDTA, a widely used inhibitor of metalloproteases, at concentrations of up to 5 mM. Severe protein degradation was observed in crystallization drops after EDTA-containing wash steps unless residual Zymit protease was removed with NaOH at a concentration of at least 0.1 M. Wash steps with 0.1% SDS were also ineffective in completely removing the remaining Zymit activity. Protocols including wash steps with at least 0.1 M NaOH, as for example specified in the original ZENM protocol, are recommended to completely deactivate Zymit protease activity.
1. Introduction
Automated robotic setup of crystallization experiments has become widespread in structural biology laboratories (Shaw Stewart & Mueller-Dieckmann, 2014 ▶). The major benefit of automation is miniaturization and reproducibility, allowing the consistent setup of crystallization drops in the low-nanolitre range. Crystallization robots can generally be classified into two groups: the first category includes devices that exclusively use disposables for any liquid transfer or single-use devices such as capillaries (Ng et al., 2003 ▶), fluid plugs (Gerdts et al., 2008 ▶) or microfluidic systems and chips (Hansen et al., 2002 ▶). For these systems, thorough cleaning after each use is generally not required and cross-contamination does not occur. In contrast, robots which use nondisposable tips, syringes or needles for liquid dispensing (Shaw Stewart & Mueller-Dieckmann, 2014 ▶) may need frequent or daily cleaning to prevent blockage and contamination. This is particularly true for parts of the system that are in contact with the protein samples, because proteins tend to adsorb onto metal and plastic surfaces, especially if the protein sample is denatured. A common step in the removal of protein contamination is to include a cleaning step utilizing a protease cocktail. Subsequent deactivation (neutralization) of the proteases, which tend to adhere to the surfaces of the protein-sample dispensing device, is required. The cleaning routines originally evolved to relieve clogging problems that various laboratories experienced, and such procedures have become a standard way to prophylactically clean the nozzles. We caution that careful removal of any proteases is necessary to prevent degradation and proteolysis of the sample protein, and we show that cleaning protocols must be carefully examined for effective deactivation of proteases to prevent partial proteolysis of sensitive protein samples.
2. Preventing proteolysis using adequate Zymit protease removal
A commonly used, inexpensive, commercial protease-containing ‘low-foam enzyme cleaner’ is Zymit, a proprietary cocktail distributed by International Products Corporation, Burlington, New Jersey, USA. According to its MSDS, Zymit contains an unspecified ‘protease enzyme’ as a trade secret (http://www.ipcol.com/pdfs/Zymit_msds.pdf). Zymit finds use in a number of applications to decontaminate labware and diagnostic or surgical instruments from residual proteins, and to prevent clogging of precision devices. It is therefore specified in a recommended cleaning protocol for the crystallization robots manufactured by Art Robbins Instruments (ARI), Sunnyvale, California, USA. The Phoenix or Gryphon robot consists of a 96- or 384-channel syringe dispenser for crystallization-cocktail transfer and one to four Nano-nozzle noncontact dispensers (Lee-valves) for the protein sample. An optional contact syringe dispenser arm for lipidic cubic phase (LCP) setup is also available for the Gryphon model. The prototype of these robots was originally conceived in an academic laboratory (Krupka et al., 2002 ▶), but no specifications for a cleaning protocol were published.
To prevent clogging of the nano-dispenser with residual proteins, an effective ‘end of the day’ or ‘long wash’ protocol (ZENM) was developed. It consists of aspirating a 2% Zymit solution followed by a neutralizing wash step with 1 mM solutions of the metal chelator ethylenediaminetetraacetate (EDTA; Science Stuff Inc., Austin; Texas, USA) in 20 mM tris(hydroxymethyl)aminomethane (Tris) buffer (Sigma–Aldrich, St Louis, Missouri, USA) pH 8.0, intended to inactive the protease of the Zymit solution, and a third wash cycle using 0.1 M sodium hydroxide (NaOH; Merck, Darmstadt, Germany) followed by a rinse of the system with distilled water. A fourth wash cycle with 2–10% Micro90 (International Products Corporation, Burlington, New Jersey, USA) and finally a rinse and checking of the system with distilled water concludes the original ZENM protocol.
Deviating from the original recommendation, the version of the Phoenix manual provided upon setup of the system specified 1 mM NaOH instead of the 0.1 M NaOH concentration of the original ZENM protocol for the second wash step following the EDTA treatment (1 mM EDTA pH 8.0). The EDTA solution used for the experiments in §3 was effective in chelating Ni2+ as tested in stripping procedures that remove Ni2+ from immobilized metal-affinity chromatography (IMAC) resins.
3. Sodium hydroxide, not EDTA, is critical in Zymit protease neutralization
After unsuccessful crystallization trials of different proteins or protein complexes submitted to our platform, SDS–PAGE analysis of the crystallization drops from the crystallization plates revealed a disturbing degree of degradation for each protein compared with the stock solution (Fig. 1 ▶). Proteins 2 and 4 almost completely degraded within seven and six months, respectively. In contrast, proteins 1 and 3 seemed to be more stable: protein 3 was found to be only partially degraded after two months and protein 3 was the protein that showed the highest stability over five months. Given that almost every protein was affected, regardless whether expressed in bacterial, insect or mammalian cell-culture systems, and in different laboratories, we concluded that the degradation was unlikely to be the result of protease contamination introduced during the expression/purification process.
Figure 1.
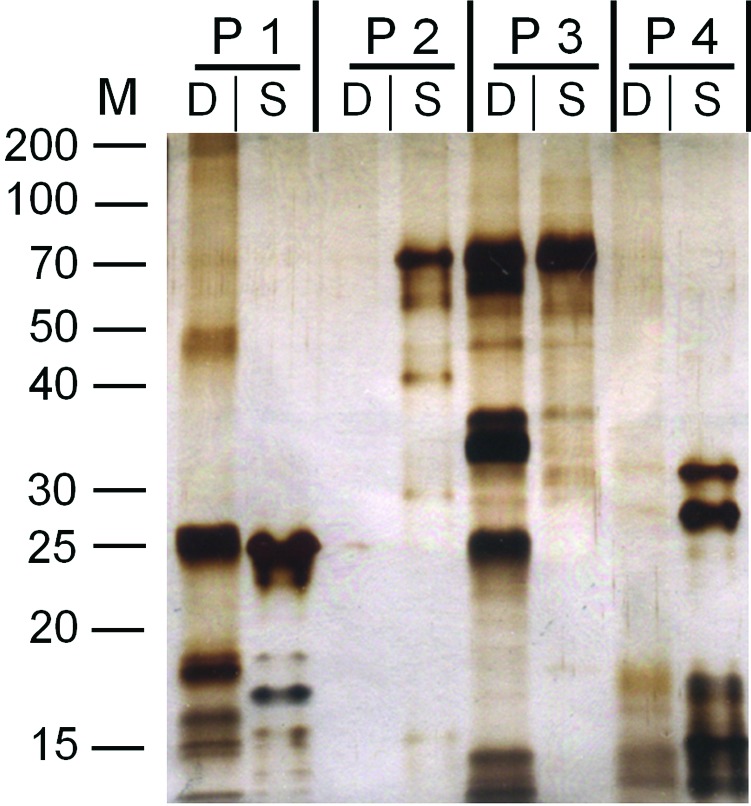
Analysis of protein stability during crystallization experiments. Samples from crystallization experiments involving/targeting different proteins (P1–P4) were recovered from drops initially set up for crystallization for two to seven months (lane D) and analyzed by SDS–PAGE (15% acrylamide). Corresponding protein stock solutions stored at −80°C were also loaded for comparison (lane S). Bands were visualized by silver staining. The proteins from the drops were aspirated through a nano-needle, which was washed with 5 mM EDTA and 1 mM NaOH following Zymit cleaning treatment. Most of the protein samples from the drops show significant degradation. The positions of molecular-weight markers are indicated on the left and labelled in kDa.
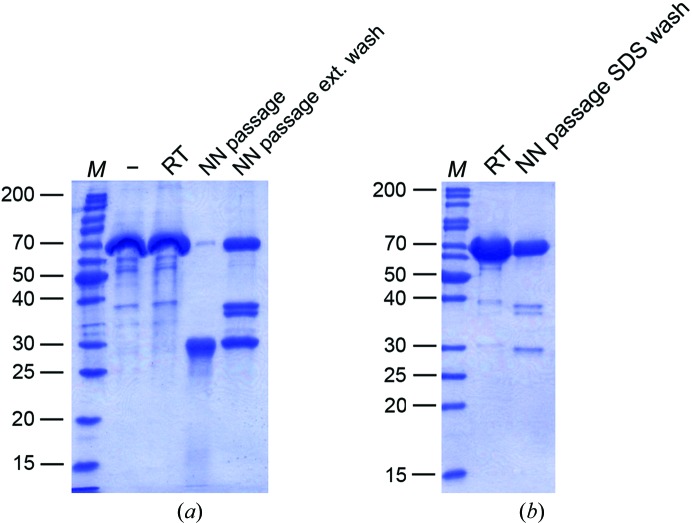
To investigate the origin of this degradation, we compared different treatment conditions of a given purified 70 kDa protein (Fig. 2 ▶ a). No degradation was observed in the original stock solution or when the protein was stored at room temperature for 2 d. However, if the protein solution was aspirated through the nano-needle cleaned by applying the low-NaOH wash protocol described above (1 mM EDTA pH 8.0 followed by 1 mM NaOH), the protein became significantly degraded within 2 d. At this point, it became doubtful that 1 mM EDTA is sufficient to completely inactivate the proteases in the Zymit solution. Cleaning the nano-needle with 500 ml 0.1%(w/v) aqueous sodium dodecyl sulfate (SDS) solution was also not sufficient to prevent protein degradation, because lower molecular weight proteins accumulated relative to the non-aspirated sample (Fig. 2 ▶ b).
Figure 2.
Analysis of protein stability after passing robotic systems washed with different cleaning solutions. (a) Samples from stock solutions of a purified protein (−), the same solution after incubation at room temperature (RT) or aspiration through the nano-needle (NN passage) or aspirated through the nano-needle after application of an extended wash protocol including 1 mM NaOH rinses (NN passage ext. wash) were analyzed by SDS–PAGE (15% acrylamide). Bands were visualized by Coomassie staining. (b) The same protein sample as in (a) after incubation at room temperature (RT) or aspirated through the nano-needle washed with SDS following Zymit cleaning (NN passage SDS wash) were analyzed as in (a). Neither the extended wash nor the SDS treatment could efficiently block protein degradation. In each case, lane M contains molecular-weight markers (labelled in kDa).
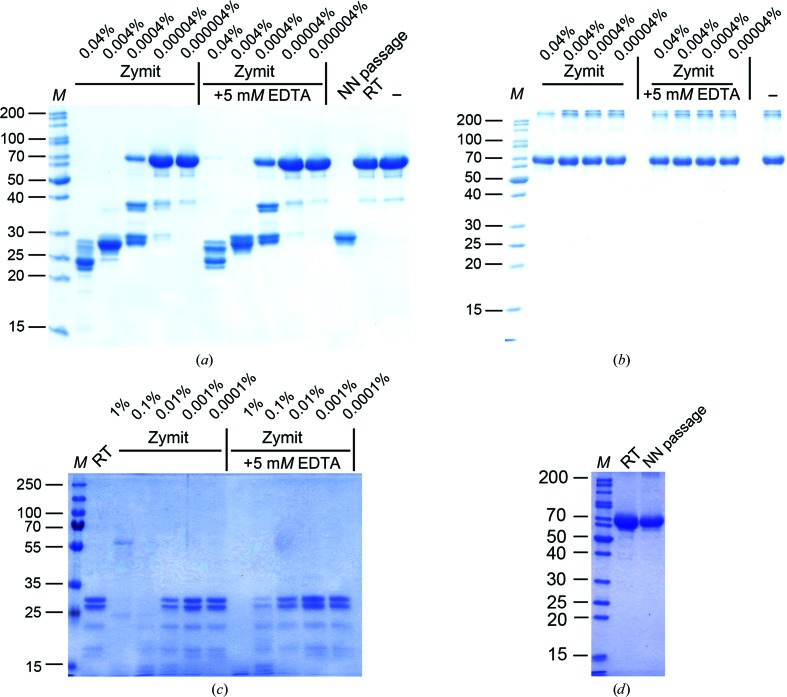
To establish the EDTA concentration required to inactivate the protease in Zymit, we incubated three different proteins in various buffers (pH 8.0) with different concentrations of Zymit in the absence and presence of 5 mM EDTA, with the latter essentially being added prior to Zymit (Figs. 3 ▶ a, 3 ▶ b and 3 ▶ c). We demonstrate that two of these three proteins were highly susceptible to degradation by Zymit even in the presence of 5 mM EDTA (Figs. 3 ▶ a and 3 ▶ c). The third protein was more stable and only minimally affected even at the highest Zymit concentration examined (0.04%). At this stage it was evident that EDTA is not the reagent of choice to block the proteolytic activity of Zymit solutions.
Figure 3.
Inhibition of Zymit-dependent protein degradation. Three different proteins (a–c) were incubated with increasing concentrations of Zymit in the absence and presence of 5 mM EDTA and analyzed by SDS–PAGE (15% acrylamide). The protein in (a) and the hetero-oligomeric sample in (c) degrade irrespective of whether or not 5 mM EDTA was added. The protein in (b) is resistant to Zymit digestion. The protein in (a) incubated with 0.04% Zymit solution displays the same degradation pattern as a sample that passed a Zymit-cleaned nano-needle (after NN passage), whereas samples kept at room temperature (RT) do not show significant degradation relative to the stock solution (−). If the original ZENM cleaning protocol with the key component of 0.1 M NaOH is applied, the susceptible protein in (a) remains stable after passing the nano-needle (d). In each case, lane M contains molecular-weight markers (labelled in kDa).
After cleaning the nano-needle with Zymit, the essential step is to apply a wash step using 0.1 M NaOH to inactivate and displace traces of remaining protease completely. Exemplarily, we demonstrate in Fig. 3 ▶(d) the stability of the same protein initially shown degraded in Figs. 2 ▶ and 3 ▶(a) after passage through a nano-needle properly rinsed with 0.1 M NaOH. The protein passed through the properly cleaned nano-needle remains as stable as the same protein not exposed to any cleaning procedure after 2 d storage at room temperature.
4. Conclusions
The protease activity of the commercial low-foam enzyme cleaner Zymit against some proteins cannot be blocked with EDTA pH 8.0, even at concentrations up to 5 mM. Wash steps using NaOH at low concentration (e.g. 1 mM NaOH pH 11) were inefficient in hydrolyzing remaining protease(s). The crucial step in any crystallization robot-cleaning protocol involving Zymit is the complete removal of the remaining protease content by applying a wash step using NaOH at a concentration of at least 0.1–0.25 M (pH 13.0–13.4, respectively), as specified in the ZENM wash protocol originally released by ARI in the USA in 2012 for decontamination of Gryphon and Phoenix robotic systems.
Acknowledgments
BR acknowledges support from the European Union under an FP7 Marie Curie People Action, grant PIIF-GA-2011-300025 (SAXCESS). AN was supported by an FWF grant (SFB021 subproject) awarded to KS. We thank Christina Mayerl (Division of Biological Chemistry) for expert technical assistance and Katherine Kantardjieff (Centre for Molecular Structure, CSU San Marcos) for critical reading of the manuscript.
References
- Gerdts, C. J., Elliott, M., Lovell, S., Mixon, M. B., Napuli, A. J., Staker, B. L., Nollert, P. & Stewart, L. (2008). Acta Cryst. D64, 1116–1122. [DOI] [PMC free article] [PubMed]
- Hansen, C. L., Skordalakes, E., Berger, J. M. & Quake, S. R. (2002). Proc. Natl Acad. Sci. USA, 99, 16531–16536. [DOI] [PMC free article] [PubMed]
- Krupka, H. I., Rupp, B., Segelke, B. W., Lekin, T., Wright, D., Wu, H.-C., Todd, P. & Azarani, A. (2002). Acta Cryst. D58, 1523–1526. [DOI] [PubMed]
- Ng, J. D., Gavira, J. A. & García-Ruíz, J. M. (2003). J. Struct. Biol. 142, 218–231. [DOI] [PubMed]
- Shaw Stewart, P. & Mueller-Dieckmann, J. (2014). Acta Cryst. F70, 686–696. [DOI] [PMC free article] [PubMed]