Abstract
Simian hemorrhagic fever virus is an arterivirus that naturally infects species of African nonhuman primates causing acute or persistent asymptomatic infections. Although it was previously estimated that 1% of baboons are SHFV-positive, more than 10% of wild-caught and captive-bred baboons tested were SHFV positive and the infections persisted for more than 10 years with detectable virus in the blood (100–1000 genomes/ml). The sequences of two baboon SHFV isolates that were amplified by a single passage in primary macaque macrophages had a high degree of identity to each other as well as to the genome of SHFV-LVR, a laboratory strain isolated in the 1960s. Infection of Japanese macaques with 100 PFU of a baboon isolate consistently produced high level viremia, pro-inflammatory cytokines, elevated tissue factor levels and clinical signs indicating coagulation defects. The baboon virus isolate provides a reliable BSL2 model of viral hemorrhagic fever disease in macaques.
Keywords: Simian hemorrhagic fever virus, Persistent infection, Baboon, Japanese macaque, Hemorrhagic fever disease, Coagulopathy
Highlights
-
•
More than 10% of wild-caught and captive-bred baboons tested are SHFV positive.
-
•
Baboons remain persistently infected for longer than 10 years.
-
•
The sequences of baboon SHFV isolates were similar to each other and to SHFV LVR.
-
•
A baboon SHFV (100 PFU) consistently induced hemorrhagic fever disease in macaques.
-
•
A reliable BSL2 macaque model of viral hemorrhagic fever disease was developed.
Introduction
Simian hemorrhagic fever virus (SHFV) was first isolated in 1964 and shown to be the causative agent of fatal hemorrhagic fever outbreaks in macaque colonies in the United States and the USSR (Allen et al., 1968, Palmer et al., 1968, Tauraso et al., 1968, Shevtsova, 1969, Lapin and Shevtsova, 1971). Mortality approached 100% by 2 weeks after infection (Tauraso et al., 1968, Gravell et al., 1980). The clinical signs of an SHFV infection in macaques closely resemble those of other hemorrhagic fever viruses such as Ebola, Marburg and Lassa viruses (Mahanty and Bray, 2004, Bray, 2005). Viral hemorrhagic fever disease is characterized by the release of pro-inflammatory cytokines from infected macrophages (MΦs) and dendritic cells (DCs) that induces tissue factor production and subsequent disseminated intravascular coagulopathy (Geisbert et al., 2003b, Bray, 2005, Levi et al., 2006). Previous SHFV outbreaks in primate facilities are thought to have been initiated by inadvertent blood-to-blood transmission from a persistently infected African nonhuman primate (NHP) to a macaque (London, 1977). In the case of an outbreak at NIH, virus was likely transmitted by the use of the same needle for tattooing or tuberculosis testing of multiple NHPs of African and Asian origin (Allen et al., 1968, Palmer et al., 1968, Tauraso et al., 1968). SHFV is typically transmitted between African NHPs during fighting but can spread efficiently among macaques by both direct and indirect contact (London, 1977, Renquist, 1990).
SHFV is a member of the Family Arteriviridae, which also includes equine arteritis virus (EAV), porcine reproductive and respiratory syndrome virus (PRRSV), and lactate dehydrogenase-elevating virus (LDV), in the order Nidovirales (Snijder and Kikkert, 2013). A related virus, wobbly possum virus, was recently identified (Dunowska et al., 2012, Snijder and Kikkert, 2013). Arteriviruses typically have restricted cell tropisms and host ranges; MΦs and DCs are infected by EAV in horses and donkeys, by PRRSV in pigs, by LDV in mice and by SHFV in several species of African NHPs and macaques but not in chimpanzees or humans (Snijder and Meulenberg, 1998). EAV and PRRSV infections can cause diseases in susceptible host species characterized by fever, anorexia, tissue necrosis, inflammation of the respiratory tract and reproductive failure, such as spontaneous abortions or delivery of weak offspring (Snijder and Kikkert, 2013). In mice, LDV typically causes lifelong, asymptomatic, persistent infections that are characterized by increased serum levels of lactate dehydrogenase (Brinton and Plagemann, 1983, Snijder and Kikkert, 2013). Due to the significant agricultural impact of diseases caused by EAV and PRRSV, the majority of research on arteriviruses has been focused on these two viruses.
Only a single SHFV isolate, LVR v42-0/M6941, obtained from a stump-tailed macaque that died of SHF during the Bethesda 1964 SHFV epizootic (Tauraso et al., 1968), survived from earlier studies of SHFV and was available from the American Type Culture Collection (ATCC). Although the origin of this virus is not known for certain, patas monkeys (Erythrocebus patas) housed in the same facility were later found to be persistently infected with SHFV and to be the source of the SHFV infecting new rhesus macaques in subsequent epizootics in the same facility (London, 1977). Although SHFV LVR efficiently induced SHF disease in macaques in the 1960s (Tauraso et al., 1968), in a recent study, no correlation was observed between virus dose (ranging from 50 to 500,000 PFU/ml) and efficiency of induction of fatal hemorrhagic fever disease in macaques (Johnson et al., 2011). Bacterial sepsis was detected in 75% (12 of 16) of the SHFV-infected animals that did not survive suggesting that this played a major role in mortality. Genome sequences for a number of divergent strains of SHFV have recently been reported (Lauck et al., 2011, Lauck et al., 2013). However, viruses containing these genomes have not yet been biologically characterized.
In the present study, a survey for SHFV in wild caught and captive baboons was done using two RT-PCR assays targeting conserved regions of the SHFV genome. The data indicated that more than 10% of both populations were SHFV-infected compared to the previous estimate of 1% (London, 1977). Data from serial archival samples used for testing indicated that animals remained infected for at least 10 years. Persistently infected baboons had very low levels of viremia. SHFV isolates from two persistently infected baboons (>10 years) were amplified once in primary macaque MΦs and subsequently sequenced. The genome sequences of these isolates were very similar both to each other and to that of SHFV LVR, an isolate from the 1960s from a macaque that may have been infected with virus originating from a patas monkey. Intravenous injection of Japanese macaques (Macaca fuscata) with 100 PFU of one of the baboon SHFV isolates resulted in consistent induction of severe hemorrhagic fever disease, characterized by high level viremia, the production of pro-inflammatory cytokines, elevation of tissue factor and coagulopathy, in each of four infected animals. Cells co-expressing viral nonstructural proteins and the MΦ marker CD68 were detected in liver and spleen. Sepsis was not identified in any of the macaques. SHFV infection in macaques has been proposed as a BSL2 model for viral hemorrhagic fever disease (Johnson et al., 2011). Using the new baboon isolate of SHFV and Japanese macaques, we demonstrated consistent disease induction in the absence of bacterial infection with a low dose virus inoculum.
Results
Survey of wild-caught and current baboons at the Southwest National Primate Research Center
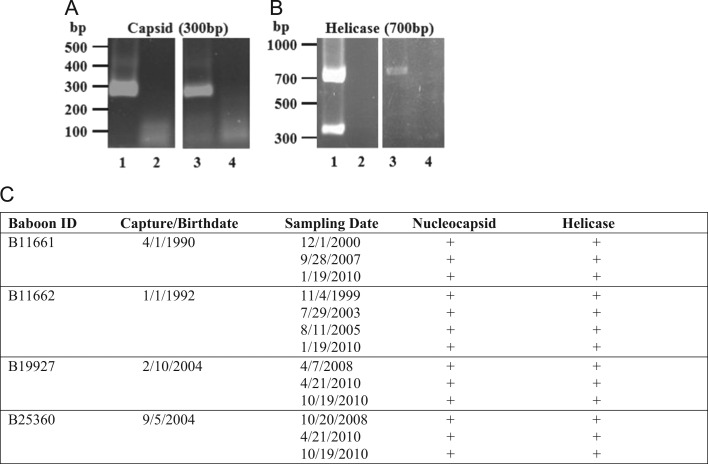
Because we required PBMCs from SHFV-negative baboons for studies comparing MØ and DC responses to experimental SHFV infections (Vatter and Brinton, 2014) and it had previously been reported that 1% of baboons were SHFV positive (London, 1977), two RT-PCR assays were developed to stringently detect SHFV in serum from positive animals. Primer sets were designed to target sequences in the SHFV-LVR nucleocapsid and helicase genes that are well conserved among different arteriviruses. The primers were first tested using RNA extracted from an SHFV-LVR pool made from a three times plaque purified virus stock. A single 300 bp PCR product was detected with the nucleocapsid primers ( Fig. 1A, lane 1). With the helicase primer set, the expected 700 bp PCR product and also a second unexpected 350 bp product were observed indicating the presence of defective genomes with an internal deletion in the helicase region in the LVR pool (Fig. 1B, lane 1). No PCR products were detected in the RNA-free water lanes (lane 2, Fig. 1A and B). The ability of the two RT-PCR assays to detect SHFV RNA in NHP sera was next tested using a single archived serum sample from each of 120 different wild-caught baboons introduced into the colony at the Southwest National Primate Research Center between 1981 and 1997. RNA from each serum sample was isolated and separately amplified by RT-PCR with each of the two primer sets. PCR bands of the expected sizes were detected for 17 of the 120 samples in both the nucleocapsid and helicase assays (lane 3, Fig. 1A and B, data not shown). However, the additional 350 bp product detected with SHFV LVR RNA in the helicase assay was not detected in helicase assays done on sera from the 17 positive baboons (lane 4, Fig. 1A and B, data not shown). No PCR bands were detected for the remaining 103 baboon serum samples with either assay (data not shown). All 17 of the positive wild caught animals were Olive baboons (Papio hamadryas anubis). Two of the 26 male samples and 15 of the 94 female samples were positive. The data indicate that a total of 14.2% of the 120 wild-caught baboons that entered the colony during this 16 year period were SHFV positive.
Fig. 1.
Detection of SHFV RNA by RT-PCR in baboon sera. RNA isolated from 100 μl of baboon serum was analyzed by one-step RT-PCR. Pairs of primers were designed to amplify (A) nucleocapsid or (B) helicase regions of the SHFV genome. RNA extracted from an aliquot of a stock pool of SHFV-LVR containing 107 PFU/ml of virus was used as the positive control and nuclease-free water was used as the negative control. PCR products were separated on 1% agarose gels. Lane 1, SHFV-LVR RNA; lane 2, water; lane 3, SHFV-positive baboon serum; and lane 4, SHFV-negative baboon serum. The results shown are representative of the results obtained for sera from a total of 33 SHFV-positive and 166 SHFV-negative baboons. (C) Assay of SHFV RNA in multiple archived samples from the same animal. RNA extracted from 100 μl of an archived serum sample was tested for SHFV using both RT-PCR assays.
The RT-PCR assays were then used to test a plasma sample from each baboon from the Southwest National Primate Research Center colony used to collect whole blood samples for differentiation of MΦs and mDCs (Vatter and Brinton, 2014). The animals sampled were ones that were in the clinic for their yearly check-up or for treatment of lacerations due to fighting between animals housed together and therefore did not represent a random sampling of the entire colony. Of the 85 colony-born baboons tested (18 females and 67 males), 16 (19%) were positive by both the nucleocapsid and helicase assays (data not shown). Two of the positive animals were females and 14 were males. The helicase assays detected only the 700 bp band. Eight of the colony-born, SHFV-positive animals were Olive baboons (P. hamadryas anubis), 6 were Olive/Yellow hybrids (P. hamadryas anubis/Papio hamadryas cynocephalus), one was a Hamadryas (Sacred) baboon (Papio hamadryas) and one was a Chacma/Olive hybrid (Papio ursinus/P. hamadryas anubis). The ages of the positive animals ranged from 8 to 23 years. The data indicated that SHFV is being transmitted between baboons within the colony.
The availability of archived samples for four of the living animals found to be virus positive allowed analysis of the length of time baboons could remain SHFV-positive. Serial archived samples were available for two of the wild caught animals found to be SHFV positive in the initial survey. Each of two archived samples from B11661 and each of three samples from B11662, as well as samples collected in 2010, were positive for SHFV RNA (Fig. 1C). Both B11661 (born in 1990) and B11662 (born in 1992) were from Kenya and 11661 is known to have come from the Kajiado region of south central Kenya. These two animals were housed together at the US Naval Research Unit in Cairo from 1992 to 1994 and also from January 1994 through April 1996 at the Southwest Foundation for Biomedical Research (which became the Southwest National Primate Research Center in 1999). The sequential identification numbers were given to these two animals upon entry into the US colony in 1994. Serial archived serum samples were also available for two colony-bred animals found to be positive in 2010 during opportunistic sampling. All of the archived samples from both B19927 and B25360 were positive for SHFV RNA (Fig. 1C).
Sequence analysis of baboon SHFV isolate genomes
To semi-quantify the SHFV RNA present in the baboon serum samples, RNA isolated from 100 µl of B15701, B14815, B11661 or B11662 sera was serially diluted and analyzed with the nucleocapsid region RT-PCR assay. The B15701 and B14815 sera contained ~102 genome copies/ml while the B11661 and B11662 sera contained ~103 genome copies/ml (data not shown). These data indicated that the virus levels in the blood of persistently infected baboons were low. To analyze the viral genomic sequence, serum samples (200 μl) were first filtered to remove bacteria and then virus particles were concentrated by ultracentrifugation. The pellet was treated with RNase and DNase and then nucleic acid was extracted and subjected to 454 sequencing as described in Materials and methods. The reads were aligned with the SHFV LVR sequence (accession number AF180391.2). The genome coverage obtained was very low (15% or less). The two samples with higher copy numbers (B11661 and B11662) gave the highest levels of coverage. Sequences for primate viruses, including Torque teno virus and several primate herpesviruses as well as bacteriophages, such as Bacillus phage SPO1 and Prochlorococcus phage P-SSM2, were detected in high abundance. It was expected that the genomes of these other infectious agents outcompeted the SHFV genome for the random 454 primer sets.
To increase the amount of SHFV RNA available for sequencing and reduce the levels of other infectious agents, virus from persistently infected baboon sera was amplified by a single passage in primary macaque MΦs. Serum (200 µl) from 4 baboons (B11661, B11662, B19927 and B25360) was separately used to infect rhesus macaque MΦ cultures. Cells from a different macaque were used for each baboon sample. Culture fluids were collected at 24 h intervals and tested by RT-PCR. By 120 h after infection, both the nucleocapsid and helicase RT-PCR assays detected the presence of SHFV RNA in all of the culture fluids (data not shown). The culture fluids were also analyzed for infectivity by plaque assay on MA104 cells. Virus titers for the B11661 and B11662 120 h culture fluids were 104 PFU/ml and for B19927 and B25360 120 h culture fluids were 103 PFU/ml.
Rhesus macaque MΦ-amplified B11662 and B11661 culture fluids (1 ml) were then processed as described above and analyzed by 454 sequencing. These two isolates were chosen for sequencing because they were from animals infected for more than 10 years and contained 10 fold higher infectivity titers than the other two isolate samples. Nearly complete sequences for both (B11662 94.8% and B11661 99.7% coverage of LVR) and a low background of other sequences were obtained. Compared to the SHFV-LVR sequence (accession number AF180391.2), the sequence of the B11661 isolate varied at 7 nt positions while the genome of the B11662 isolate varied at 10 nt positions ( Table 1). Most of the changes in B11662 differed from those in B11661. The partial B11661 and B11662 genomic sequences have been deposited in GenBank under accession numbers KM655765 and KM655766, respectively. The very high sequence conservation observed between the genomes of two SHFV isolates obtained from baboons that were each persistently infected for more than 10 years and also between the genomes of these two baboon viruses and SHFV-LVR which was isolated in the 1970s was unexpected.
Table 1.
Variation of two baboon isolates compared to the SHFV LVR sequence.
| Position | B11661 | B11662 | SHFV-LVR | Amino acid change | Gene |
|---|---|---|---|---|---|
| 26 | G | C | C | Non-coding | 5′UTR |
| 151 | A | G | A | Non-coding | 5′UTR |
| 618 | G | A | G | Glu to Lys | nsp1α |
| 619 | A | G | A | Glu to Gly | nsp1α |
| 4928 | G | A | G | Silent | nsp4 |
| 7965 | T | C | T | Leu to Pro | nsp9 |
| 9910 | G | T | G | Silent | nsp11 |
| 10750 | G | A | G | Silent | nsp12 |
| 12408 | T | C | T | Silent | GP4′ |
| 13597 | A | T | T | Val to Glu | GP3 |
| Silent | GP4 | ||||
| 13604 | A | G | G | Silent | GP3 |
| Glu to Lys | GP4 | ||||
| 13605 | C | A | A | Asp to His | GP3 |
| Glu to Ala | GP4 | ||||
| 13606 | G | A | A | Asp to Ser | GP3 |
| Silent | GP4 | ||||
| 14032 | T | T | C | Pro to Ser | GP5 |
| Ser to Phe | GP5a | ||||
| 14736 | A | G | A | Silent | GP5 |
| 15278 | C | T | T | Silent | GP6 |
Infection of Japanese macaques with a baboon SHFV isolate
In studies conducted in the 1960s, SHFV LVR efficiently induced fatal hemorrhagic fever disease in rhesus macaques (Tauraso et al., 1968). However, in a recent study in rhesus macaques, inefficient disease induction was observed with a current stock of SHFV LVR (Johnson et al., 2011). Because of the high sequence similarity between the B11661 and B11662 baboon isolates and LVR, we postulated that the baboon isolates would have a virulence phenotype similar to LVR in macaques. The baboon SHFV isolates had undergone only a single tissue culture passage in primary macaque MΦ while LVR had been passaged multiple times in culture (Tauraso et al., 1968, Johnson et al., 2011) and the results obtained with the helicase RT-PCR assay (Fig. 1) indicated that defective genomes, which would be expected to reduce virulence, were still present in our LVR stock after three serial plaque purifications.
In the 2011 macaque infection study performed with SHFV LVR, bacterial sepsis was observed in 75% of the fatal cases (Johnson et al., 2011). The authors hypothesized that virus-induced immunosuppression led to the development of sepsis in animals that had existing or opportune bacterial infections. The presence of bacterial sepsis in many of the animals made it difficult to discriminate the relative contributions of the SHFV infection and of the bacterial infection to the host response parameters measured. To try to avoid bacterial co-infection, healthy animals from a Japanese macaque (JM) colony at the Oregon National Primate Research Center were used to test the ability of the B11661 isolate to induce hemorrhagic disease. The B11661 isolate was chosen because a complete coding region sequence was obtained for this isolate. Although data on SHFV infections in several macaque species have been previously reported (rhesus, cynomolgus and stump-tailed) (Palmer et al., 1968, London, 1977, Johnson et al., 2011), no data on SHFV infections in JMs has been reported. Four animals were infected in two experimental cohorts, each comprised of two animals. Each animal was injected intravenously with 100 PFU of macaque MΦ-amplified SHFV B11661 diluted in PBS. The first two animals (JM-23333 and JM-24054) developed fever and lethargy by 3 days post-infection (dpi). By 7 dpi, one macaque (JM-23333) was febrile, lethargic and anorexic and was euthanized. Because of the possibility that the non-specific clinical signs noted in JM-23333 could have been associated with bacterial sepsis in addition to SHFV infection, the second macaque (JM-24054) was treated with an antibiotic on day 9. At 13 dpi, JM-24054 displayed severe hemorrhagic fever disease signs that reached a clinical score of 10 and was euthanized ( Table 2).
Table 2.
Summary of experimental SHFV infection studies.
| Animal ID# | Virus infection: 100 plaque forming units (PFU) intravenously | Prothrombin time (PT) seconds (pre-infection) | Prothrombin time (PT) seconds (euthanasia) | Activated partial thromboplastin time (PTT) seconds (pre-infection) | Activated partial thromboplastin time (PTT) seconds (euthanasia) | Survival (days) post-SHFV infection | Clinical signs |
|---|---|---|---|---|---|---|---|
| Cohort 1 | |||||||
| JM-23333 | SHFV B11661 | NA | NA | NA | NA | 7 | Fever, lethargy, anorexic, increased respiratory rate |
| JM-24054 | SHFV B11661 | NA | NA | NA | NA | 13 | Fever, diarrhea, scrotal edema, petechiae, epistaxis, |
| Cohort 2 | |||||||
| JM-22015 | SHFV B11661 | 10.5 | 23.0 | 29.4 | 48.8 | 9 | Fever, petechiae and periocular and scrotal edema |
| JM-23328 | SHFV B11661 | 11.5 | 19.5 | 32.4 | 54.8 | 9 | Fever, petechiae and periocular edema |
The two animals in the second cohort (JM-22015 and JM-23328) were prophylactically treated with antibiotic prior to infection with the SHFV B11661. Both of these animals developed fever by 3 dpi and displayed clinical signs of hemorrhagic fever disease, such as fever, epistaxis, petechiae and subcutaneous edema by 8 dpi and were humanely euthanized on day 9 due to clinical scores of 10 or greater (Table 2). At necropsy, no evidence of bacterial sepsis was detected in any of the four SHFV B11661 infected animals.
Host responses to SHFV infection
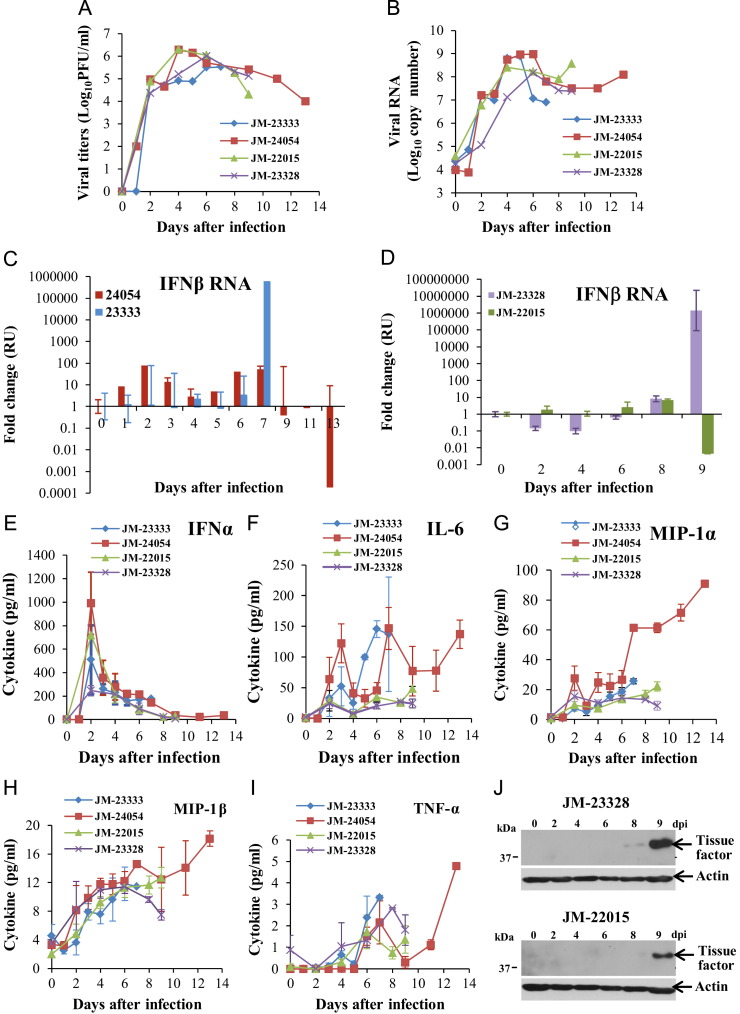
Plasma and PBMC samples were collected daily from the first set of animals and every other day from the second set. The every other day collection schedule allowed larger samples to be obtained. High levels of infectious virus were detected in plasma from all four animals by plaque assay on MA104 cells by 2 dpi, and the levels remained high ( Fig. 2A), which was in stark contrast to the lack of virus detection in plasma at later times after infection in a previous study (Johnson et al., 2011). Intracellular viral RNA levels in PBMCs collected at different times after infection were analyzed by qRT-PCR using primers targeting the nucleocapsid region. The nucleocapsid region primers detect both genomic and subgenomic intracellular viral RNAs. Peak viral RNA levels were detected between 4 and 6 days after infection and remained elevated (Fig. 2B).
Fig. 2.
Analysis of SHFV replication kinetics, cytokine production and tissue factor expression in Japanese macaques. (A) Virus infectivity titers in plasma. Plasma samples collected at the indicated times after infection were titrated for infectivity by plaque assay in MA104 cells. Each data point is the average of duplicate titrations. (B) Viral RNA copy numbers in PBMCs. Total cell RNA was extracted from PBMCs collected at the indicated times after infection and plus-sense, genome and subgenomic viral RNA was amplified by real-time RT-PCR and quantified using a standard curve generated with a known concentration of viral RNA. (C, D) Relative quantification (RQ) of intracellular IFNβ mRNA in PBMCs by real-time quantitative RT-PCR. PBMC RNA was used to measure IFNβ mRNA levels. IFNβ mRNA levels are expressed as the fold change in the levels of IFNβ mRNA in PBMCs from an infected animal versus the level in the same animal at day zero. The amount of IFNβ mRNA in each sample was normalized to the level of 18S rRNA in the same sample. Values shown are the averages of assays done in triplicate. Error bars represent standard error (E–I) IFNα and pro-inflammatory cytokine levels in the plasma were quantified by multiplexed ELISA. Values shown are the averages of assays done in triplicate. Error bars represent standard deviation. (J) Tissue factor expression in PBMCs collected on different days after infection was analyzed by Western blotting using anti-tissue factor antibody. Actin was used as a loading control. Representative gels are shown.
As one approach to monitoring the host antiviral response, IFNβ mRNA levels in PBMCs were measured by qRT-PCR. At early times after infection, IFNβ mRNA expression levels increased by only low amounts in three of the four animals (JM-24054, JM-23333, and JM-23328) and decreased in the fourth (JM-22015). However, at the time that severe disease signs were observed, PBMC IFNβ mRNA levels were greatly increased for two animals but greatly decreased for the other two (Fig. 2C and D). IFNα protein levels in plasma were quantified by ELISA. Peak plasma levels of IFNα protein were observed at 2 dpi for all four animals but the levels rapidly declined thereafter (Fig. 2E). Pro-inflammatory cytokine levels in plasma were analyzed with a multiplexed ELISA as described in Materials and methods. Increased levels of IL-6, MIP-1α, MIP-1β and TNFα were detected starting at 2 dpi in the plasma of all four of the infected animals ( Fig. 3F–I). No increase in IL-1β, IL-10 or IL-12/23 (p40) levels was detected in any of the plasma samples (data not shown). The data support the hypothesis that the production of pro-inflammatory cytokines is a major contributor to SHFV-induced pathology.
Fig. 3.
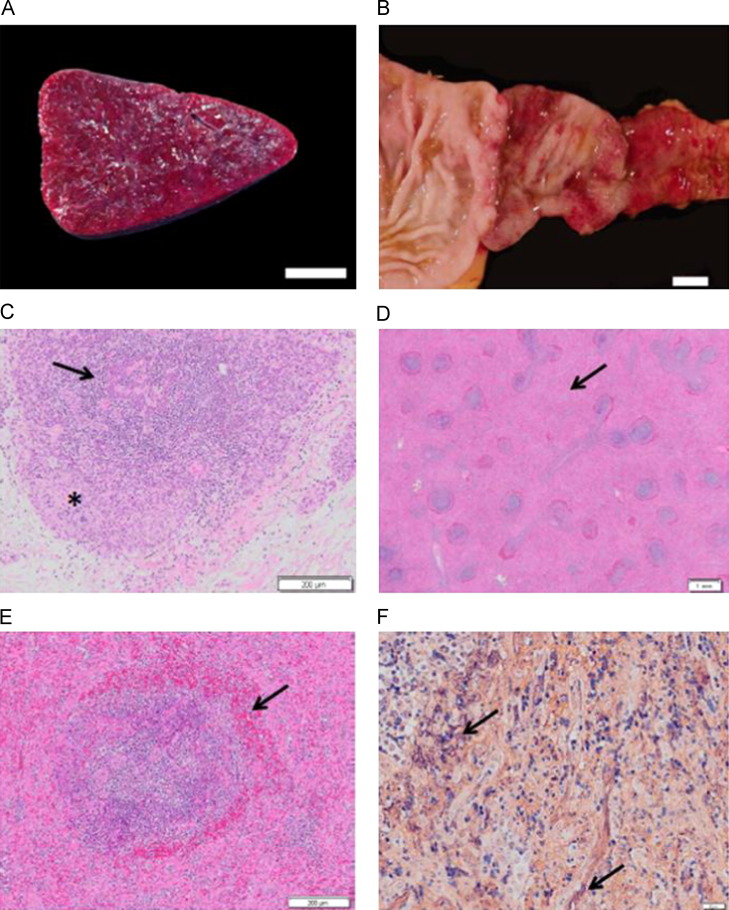
Gross and microscopic lesions of SHF. (A) Spleen, cut surface (JM-24054). White pulp is markedly diminished within diffusely expanded red pulp, bar=1 cm. (B) Duodenum (JM-22015). The mucosa exhibits ecchymotic to diffuse hemorrhage that extends to the pyloric valve, bar=1 cm. (C) Thymus stained with H&E (JM-24054). There is diffuse lymphocyte necrosis of cortical lymphocytes (asterisk) and depletion of medullary lymphocytes (arrow), bar=200 μm. (D) Spleen stained with H&E (JM-24054). Depleted and necrotic follicles are rimmed with a zone of hemorrhage. Red pulp is markedly expanded (arrow), bar=1 mm. (E) Spleen stained with H&E (JM-24054). A splenic follicle exhibiting lymphocyte necrosis within the central portion of the germinal center, the mantle and marginal zones with an adjacent rim of hemorrhage (arrow), bar=200 μm. (F) Spleen (JM- 24054). Splenic red pulp is filled with abundant fibrin strands stained purple by phosphotungstic acid hematoxylin (arrows), bar=20 μm.
Disseminated intravascular coagulopathy is one of the hallmarks of viral hemorrhagic fever disease. Data from attempts to measure coagulation parameters on samples from the first cohort using commercially available human-specific kits were inconclusive likely due to the small volumes of the daily blood samples. However, data was obtained with the larger samples collected from the second cohort. Specifically, JM-22015 and JM-23328 exhibited marked defects in coagulation with prothrombin time (PT) starting at 10.5 s and 11.5 s, respectively, prior to infection and reaching 23.0 s and 19.5 s, respectively, at the time of euthanasia (Table 2). A similar pattern was observed for activated partial thromboplastin times (APTT), with pre-infection sample values of 29.4 s and 32.4 s, respectively, and values at the time of euthanasia reaching 48.8 s and 54.8 s, respectively (Table 2).
Disseminated intravascular coagulopathy in Ebola virus infected macaques is characterized by coagulation defects initiated by increased expression of tissue factor on monocytic cell surfaces induced by pro-inflammatory cytokine production (Bray, 2005). Lysates of PBMCs obtained from the infected animals were analyzed by Western blotting using an anti-human tissue factor antibody. Tissue factor was detected by 8–9 dpi (Fig. 2J). The detection of tissue factor in PBMCs at the time that clinical signs of coagulopathy (petechiae and epistaxis) were also observed was consistent with the development of disseminated intravascular coagulopathy (Table 2).
Pathological findings associated with SHFV infection
Gross and histologic lesions were generally similar to those described previously (Allen et al., 1968; Abildgaard et al., 1975; Renquist, 1990; Zack, 1993; Johnson et al., 2011). At necropsy, consistent findings in all four animals included splenomegaly, as well as petechial and ecchymotic hemorrhages in varied and multiple tissues. The spleens were pale and markedly to severely enlarged with tense, smooth capsules and rounded margins. On the sectioned surface, there was expansion of the red pulp (Fig. 3A) and two animals (JM-23333 and JM-24054) exhibited attenuation of the white pulp. Hemorrhages were observed in the skin, at venipuncture sites, and also in the subcutis, sclera, base of the tongue, spleen, liver, kidney, lung, myocardium, pancreas, cervical spinal cord, trachea, different lymph nodes, mucosa of the stomach, duodenum, jejunum and the muscular tunic of the cecum. Mucosal hemorrhage involving the cranial-most segment of the duodenum was noted in two animals (JM-24054 and JM-22015) (Fig. 3B). The key histologic feature in all animals was lymphoid necrosis involving the spleen, lymph nodes, gut-associated lymphoid tissue (GALT) and thymus (Fig. 3C). Splenic lesions included lymphoid necrosis of splenic follicles, mantle and marginal zones, and periarteriolar lymphoid sheaths (Fig. 3D). Hemorrhages were also present within and surrounding the marginal zone (perifollicular hemorrhage) and expansion of the red pulp with an eosinophilic coagulum and fibrin was observed (Fig. 3D–F). Fibrin strands are stained purple by the phosphotungstic acid hematoxylin histochemical procedure (Fig. 3F). Moderate to severe lymphoid necrosis was observed in the cortex of the thymus. The thymic medulla was less affected. All lymph nodes examined demonstrated mild to moderate lymphoid necrosis. Generally, the follicular germinal centers were more affected than the paracortical regions or medullary cords. A minimal to mild degree of lymphoid necrosis affecting the GALT was noted. Sections of tonsil were available for evaluation in three of the four animals and all exhibited lymphoid necrosis.
Other changes included mild hepatic lipidosis in two animals (JM-22015 and JM-23333) and mild hepatocellular degeneration and necrosis in three animals (JM-22015, JM-23328, and JM-24054). One animal (JM-24054) demonstrated minimal lymphohistiocytic and neutrophilic hepatitis. Another animal (JM-23328) had random, multifocal mild to moderate hepatic infiltrates composed of lymphocytes, histiocytes, plasma cells and neutrophils. Marked hepatic amyloid deposits in one animal (JM-23333) hampered detection of subtle microscopic changes. Renal tubular degeneration and necrosis were present in all animals and varied in degree from minimal to moderate. Mild renal tubular protein was present in one animal (JM-24054). A lymphohistiocytic interstitial pneumonia was also present in all of the animals. One animal (JM-24054) had marked interstitial pneumonia and also exhibited vasculitis of the pulmonary vasculature. All animals had minimal lymphocytic or lymphohistiocytic infiltrates in the myocardium. Three animals showed minimal lymphocytic or lymphohistiocytic inflammation in various regions of the central nervous system. One animal (JM-22015) exhibited minimal perivascular lymphohistiocytic inflammatory infiltrates in the meninges, choroid plexus, cerebral gray matter and spinal cord. One animal (JM-23228) had minimal perivascular lymphohistiocytic inflammatory infiltrates in the brainstem and gray matter of the cervical spinal cord. One animal (JM-24054) showed minimal perivascular lymphocytic inflammatory infiltrates limited to the choroid plexus. In addition, necrotizing epididymitis was present in three animals (JM-22015, JM-23333, and JM-24054) and lymphohistiocytic orchitis was observed in one of these (JM-24054). No histologic evidence of concurrent bacteremia or septicemia was observed in any of the animals.
Detection of SHFV-infected cells in tissues of infected animals
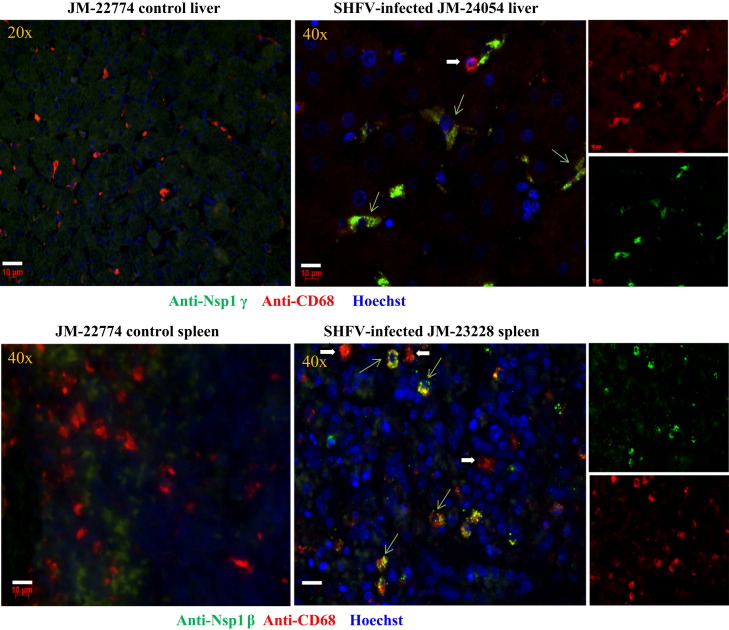
Evidence of virus replication in PBMCs was detected throughout the course of the infection (Fig. 2B). To determine whether virus replication was also occurring in other tissues when severe disease signs were observed, liver and spleen tissues were obtained from each animal at necropsy and 5 μm thick sections were prepared and processed for antigen detection as described in Materials and methods. Infected cells were detected with rabbit antibodies generated against peptides from either SHFV nonstructural protein nsp1β or nsp1γ (Vatter et al., 2014). Anti-CD68 antibody was used to detect macrophage/monocytes. Some CD68-positive cells but no viral antibody positive cells were detected in tissue sections from a control animal. Cells that were positive for both CD68 and viral antigen were detected in liver and spleen tissue sections from infected animals ( Fig. 4). Viral antigen was located in the cytoplasm. Not all of the CD68 cells detected were SHFV-positive. The results indicate that tissue associated, SHFV-infected MΦs are still present after animals develop severe hemorrhagic fever disease.
Fig. 4.
Detection of SHFV-infected cells in tissues from SHFV-infected and control Japanese macaques by immunofluorescence. Post-mortem tissue was obtained, formalin-fixed and embedded in paraffin. Sections (5 μm thick) were cut, processed as described in Materials and methods and incubated with a rabbit antibody to SHFV nsp1β or nsp1γ, then biotinylated goat anti-rabbit antibody and then streptavidin Alexa Fluor 488. Next, the sections were incubated with a mouse anti-CD68 antibody, then a biotinylated goat anti-mouse secondary antibody and then streptavidin Alexa Fluor 594. Nuclei were visualized with Hoechst 33258 staining. Cells were visualized using a Zeiss Axioscope 2 plus microscope equipped with a digital camera. Thin green arrows indicate cells detected by both anti-SHFV nonstructural protein and anti-CD68 antibodies. Thick white arrows indicate cells detected only by anti-CD68 antibody. The objective used to capture the image is indicated in the upper left corner, bars=10 μm.
Analysis of immune cell populations
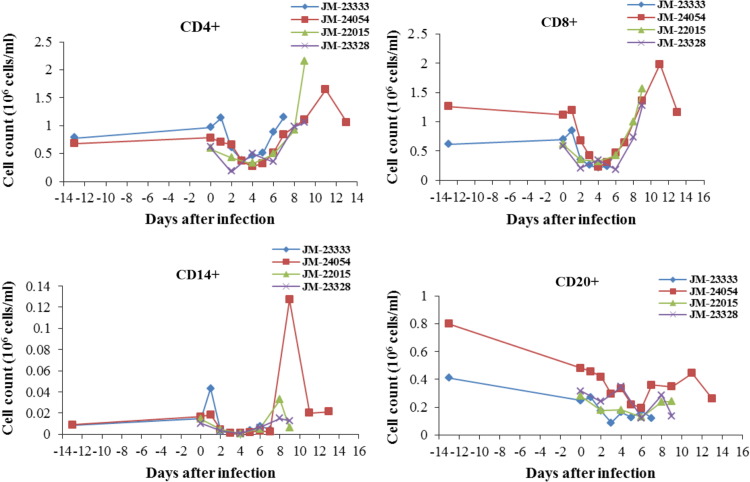
Immunosuppression as indicated by decreases in various lymphocyte cell populations in the blood between 2 and 9 days after infection was previously reported in SHFV LVR-infected macaques (Johnson et al., 2011). Lymphocyte subpopulations in PBMCs collected from each of the SHFV B11661-infected JMs at different times post-infection were quantified by flow cytometry. In all four animals, the PBMC CD4+, CD8+, CD14+ and CD20+ cell populations decreased between 2 and 6 dpi compared to the levels present in each animal prior to infection ( Fig. 5). Thereafter, the number of CD4+ and CD8+ cells increased in all four animals. Only a transient increase in the CD14+ and CD20+ populations was observed. The observed decrease in each of the PBMC cell populations starting at 2 dpi in all four animals is consistent with the development of an immunosuppressive state.
Fig. 5.
-PBMCs collected prior to or on the day of infection and either every day or every other day after SHFV infection were stained with the indicated cell surface marker and the number of positive cells was quantified by flow cytometry.
Discussion
Viruses in multiple RNA virus families, such as Filoviridae, Arenaviridae, Bunyaviridae, and Flaviviridae, can induce viral hemorrhagic fever disease in humans (Johnson et al., 2011). Due to the high human morbidity caused by these viruses, including the Filoviruses, Ebola and Marburg, experiments to elucidate how these viruses cause disease must be performed under high containment conditions in suitable animal models. Both macaque and mouse models have been developed for Ebola and Marburg, (Geisbert et al., 2003a, Mahanty and Bray, 2004, Bradfute et al., 2012). In the cynomolgus macaque-Zaire Ebola virus model disease kinetics are accelerated and infections are uniformly fatal, compared to those of Zaire Ebola infections in humans, which can incubate for three weeks and are not fatal in all infected individuals (Mahanty and Bray, 2004). Disease in both NHPs and humans is associated with viral induced suppression of the hosts׳ innate and adaptive immune response and increased survival in humans is correlated with the hosts׳ ability to minimize the negative effects of the virus infection on these responses.
Our data from SHFV-infected macaques provides multiple similarities to Ebola virus induced hemorrhagic disease. Infection of macaques with small doses of Zaire Ebola typically induces fever by 3–4 days, hemorrhagic fever disease signs by 5–6 days, and morbidity by 7–8 days. Similar disease induction kinetics and a similar progression of disease parameters were observed in the SHFV-infected macaques. The primary target cells of both Ebola virus and SHFV in macaques are MΦs and DCs (Geisbert et al., 2003c, Vatter and Brinton, 2014). At late times of infection, Ebola virus also infects parenchymal cells, hepatocytes, adrenal cortical cells and fibroblasts (Bray and Mahanty, 2003, Mahanty and Bray, 2004). Although SHFV-infected MΦs were present in the livers and spleens of moribund animals, adjacent cells in these organs were not positive for viral antigen. The similar kinetics of severe disease development observed in Ebola virus and SHFV-infected macaques indicates that infection of additional types of cells is not required for induction of morbidity.
Several Ebola viral proteins suppress Type I IFN production (Zampieri et al., 2007, Chang et al., 2009). Both the transient peak of INFα detected in the plasma at 2 days after SHFV infection and the delayed upregulation of IFNβ mRNA in PBMCs are consistent with recent data showing that all three of the SHFV nonstructural protein 1s have Type 1 IFN suppressive activity (Han et al., 2014). Ebola virus infected MΦs and DCs produce proinflammatory cytokines, chemokines and tissue factor that induce vasodilation, increase vascular permeability and disseminated intravascular coagulation (Geisbert et al., 2003d, Bray and Geisbert, 2005). Pro-inflammatory cytokines also cause bystander lymphocyte apoptosis in Ebola virus infected macaques (Geisbert et al., 2003b, Ruf, 2004, Bray, 2005). SHFV-infection of cultured macaque MΦs and myloid DCs induces proinflammatory cytokines (Vatter and Brinton, 2014) and proinflammatory cytokine production, decreased PBMC lymphocyte populations and tissue lymphocyte necrosis were detected in all four of the JMs infected with SHFV. Neither Ebola virus nor SHFV can infect lymphocytes, implying that the two viruses induce lymphocytosis by related indirect mechanisms. Although CD4+ and CD8+ cell populations were decreased in all four SHFV- infected animals between days 2 and 6 after infection, cell numbers increased thereafter suggesting the activation of an adaptive immune response. Humans who mount a rapid and robust immune response can survive an Ebola virus infection (Mahanty and Bray, 2004) and therapeutic recombinant antibodies as well as transfusions from Ebola survivors have enhanced patient survival. However, if an immune response was initiated in the SHFV infected macaques, it did not alter the progression of hemorrhagic fever disease.
Although it was previously estimated that 1% of baboons, 1% of African green monkeys and 10% of patas monkeys are SHFV-positive (London, 1977), the numbers and histories of the animals tested to obtain these estimates and which available biological test (macaque infection, cytopathic infection of macaque MΦ cultures or cytopathic infection in MA104 cells) was used were not reported. In the present study, a total of 205 baboons from a colony in the USA were tested for SHFV using RT-PCR assays targeting two conserved regions of the SHFV genome. These assays provided a sensitive and consistent method for detecting persistently infected animals. The data obtained from archived samples from wild-caught Olive baboons introduced into the colony over a 16 year period indicated that 14.2% not 1% of these animals were infected with SHFV. However, because blood samples obtained at the time of capture were not available for any of the wild-caught animals, it could not be determined whether these animals were infected in the wild or after capture. The percentage of opportunistically sampled animals living in the colony that were SHFV positive was 19% which was also much higher than 1%. However, the majority of the colony animals that were sampled were males that were in the clinic for treatment of lacerations obtained during fighting and this percentage is therefore expected to be higher than that for the entire colony. Multiple baboon species as well as hybrid animals in the colony were found to be SHFV positive. The data obtained on the colony-bred animals clearly indicated that SHFV continues to be transmitted between animals after introduction into a captive baboon colony. Additional species of African NHPs, red colobus (Procolobus rufomitratus tephrosceles) and red-tailed guenons (Cercopithecus ascanius) were recently reported to be infected with SHFV in the wild (Lauck et al., 2013, Bailey et al., 2014). The sequences of these SHFV isolates are divergent from LVR and whether they can infect macaques and cause hemorrhagic fever disease is not known.
A single previous study reported that patas monkeys could remain SHFV positive for up to 13 years as indicated by detection of antibody to purified SHFV LVR in an ELISA (Gravell et al., 1986). The duration of persistent SHFV infections in baboons had not previously been tested. Baboons in the present study were also found to remain SHFV-positive for at least 10 years. Data from SHFV RT-PCR assays done on serially diluted sera from persistently infected baboons indicated that the viremia levels in these animals are low (~100 to 1000 genome copies/ml) and SHFV was also reported to grow to low titers in baboon MΦs and mDCs (Vatter and Brinton, 2014). Attempts to sequence SHFV genome RNA directly from baboon plasma by 454 sequencing yielded only short contigs representing 15% or less of the genome. However, nearly complete sequences were obtained from virus that was amplified by a single passage in rhesus macaque MΦ cultures. The single passage in macaque MΦ may have selected for a virus subpopulation that can efficiently infect and replicate in macaque MΦs and therefore the full diversity of the virus populations in the persistently infected animals was not analyzed. Baboons B11661 and B11662 were housed together only in the early 1990s and it is possible that one infected the other. However, it was still surprising that the genome sequences obtained for the SHFV isolates from these two long term (>10 years) persistently infected baboons were so similar to each other and even more surprising that these sequences were also very similar to the sequence of SHFV LVR (AF180391.2) (Vatter et al., 2014), which was isolated in 1964 (Tauraso et al., 1968). Although coronavirus genomes encode proofreading enzymes, arterivirus genomes do not (Denison et al., 2011). A major contributing factor to the low sequence divergence observed for the baboon and LVR SHFV isolates may be the low level of virus replication in the target cells in these animals (Vatter and Brinton, 2014).
In experimental SHFV infection studies done in the 1960s, inoculation of low doses of SHFV from infected macaque serum by the intramuscular route consistently induced severe hemorrhagic disease in macaques (Palmer et al., 1968, Tauraso et al., 1968). In contrast, in a 2011 study, no correlation between SHFV LVR dose (50–500,000 PFU) and hemorrhagic disease induction was observed (Johnson et al., 2011). The LVR used in the 2011 study was generated by three freeze–thaw cycles of infected MA104 cells. The large amount of cell and viral RNA present in these lysates would be expected to activate cell RNA sensors and strongly induce the host antiviral response which may have contributed to the decreased efficiency of hemorrhagic disease induction observed. It is also possible that defective SHFV genomes present in the inoculum may have reduced the efficiency of virus replication. In the present study, baboon SHFV in culture fluid from primary macaque MΦs was diluted in PBS and animals were injected with 100 PFU by the intravenous route. This inoculation route was chosen because it closely approximates natural transmission conditions and was expected to facilitate efficient infection of peripheral MΦs and DCs. Consistent hemorrhagic disease induction with similar kinetics of viremia, of proinflammatory cytokine production, and of hemorrhagic disease development was observed in all of the animals tested. Virus titers in blood and viral RNA levels in PBMCs remained high throughout the course of the infection. No evidence of bacterial sepsis was observed in any of the SHFV-infected JMs in the present study. Three of the four animals were treated with antibiotics as a precaution. The only effect of providing antibiotics and supportive care to SHFV-infected rhesus macaques observed during 1960s epizootics “was a shortening of the course of the hemorrhagic disease resulting in earlier death” (Allen et al., 1968). The consistent induction of severe hemorrhagic disease with similar kinetics achieved in macaques after inoculation of a low dose of a baboon isolate of SHFV indicates that this represents an improved BSL2 model of viral hemorrhagic fever disease for studying viral and host factors controlling disease severity. The availability of multiple animal models of viral hemorrhagic fever is expected to facilitate the development of broad based therapeutics to treat disease complications associated with hemorrhage and coagulation disorders. As has become evident during the recent Ebola outbreak in Africa, there is an urgent need for therapies targeting hemorrhagic disease complications for use in combination with virus-specific therapies to enhance the chance of patient recovery.
Materials and methods
Cells
MA104 cells obtained from O. Nainin (Centers for Disease Control and Prevention) were grown in minimum essential medium (MEM) supplemented with 10% FBS, 1% l-glutamine and 1% gentamicin at 37 °C in 5% CO2. Whole blood collected in acid citrate dextrose (ACD) BD Vacutainer® collection tubes (BD Bioscience) from rhesus macaques (Yerkes Regional Primate Research Center) was used to isolate peripheral blood mononuclear cells (PBMCs) by Ficoll® 400 (Mediatech Inc.) density gradient centrifugation according to standard protocols. Monocytes were seeded at 106 cells/well in 24-well plates and allowed to adhere for 1 h before gentle washing with Hanks buffered saline solution. MΦs were differentiated from adherent cells by incubation with RPMI-1640 culture media supplemented with 10% autologous serum or 10% heat-inactivated FBS, 50 U/ml of penicillin, 50 μg/ml of streptomycin, 5000 U/ml human recombinant MΦ colony stimulating factor (R&D Systems) for 11 days at 37 °C in 5% CO2. During differentiation, two-thirds of the culture media was replaced with fresh growth media every three days to replenish growth factors. To confirm the identity of the differentiated cells, cells were stained with fluorescently-labeled antibodies directed against the MΦ surface markers CD91 and CD163 (BD Bioscience) and then sorted and enumerated using a FACSCanto flow cytometer and FACSDiva software (BD Bioscience). Greater than 95% of cells in MΦ cultures were CD91+ and CD163+.
Virus
An aliquot of SHFV, strain LVR 42-0/M6941, was obtained from American Type Culture Collection. Analysis of virion RNA from an initial passage in MA104 cells (peak titer 105 PFU/ml) by gel electrophoresis detected the genome as well as several strong bands that were shorter than genome length (Brinton, unpublished data). The virus was serially plaque purified three times on MA104 cells and then amplified once on a MA104 cell monolayer (MOI of 0.01) to produce a stock pool. Pools of SHFV-LVR used for experiments were prepared by infecting confluent MA104 monolayers with the stock pool at an MOI of 0.2. Culture media was harvested at 32 h after infection, clarified, aliquoted and stored at −80 °C (titer ~107 PFU/ml).
The baboon SHFV isolates were amplified by a single passage on primary macaque MΦ cultures. Serum was collected in clot tubes (BD Bioscience) at Southwest National Primate Research Center. MΦ cultures prepared as described above in wells of a 24 well plate were incubated with 200 µl of baboon serum at 37 °C in 5% CO2 for 1 h. After virus adsorption, cells were washed and then culture fluid was harvested at 120 h after infection (titer ~103-4 PFU/ml).
Plaque assay
SHFV infectivity was quantified by plaque assay on confluent monolayers of MA104 cells in six-well plates. After adsorption for 1 h at room temperature, the virus inoculum was removed and the wells were overlaid with 1% SeaKem ME agarose (Bio-Whittaker Molecular Applications) mixed 1:1 with 2X MEM containing 5% FCS, and incubated at 37 °C for 72 h. After removal of the agarose plug, cells were stained with 0.05% crystal violet in 70% ethanol.
SHFV one-step RT-PCR assays
Archived serum samples collected from baboon colony animals at the Southwest National Primate Research Center were shipped on dry ice overnight to Georgia State University. To obtain plasma, blood samples from baboons were collected in ACD tubes at Southwest National Primate Research Center and shipped at room temperature overnight to Georgia State University. Plasma and serum were stored at −80 °C. RNA was isolated from 100 µl of plasma or an archived serum aliquot with TRIZOL Reagent LS (Molecular Research Center, Inc.) according to manufacturer׳s protocol and used as template for RT-PCR. Pairs of primers from either the nucleocapsid (forward primer, 5′-GGCAAACCAAAACCAAATAACAAGGG-3′ and reverse primer, 5′-ATAATCAGTGTCTGGCCTAGGTGG-3′) or the helicase (forward primer, 5′-CTCTGTCTAACTTTGTGGTGGG-3′ and reverse primer, 5′-TTTGCGAGCTGATGATGCGGG-3′) regions of the genome were designed based on the SHFV-LVR Genbank sequence (accession number AF180391.2). SHFV RNA was amplified using the SuperScript® III One-Step RT-PCR system with Platinum® Taq High Fidelity (Invitrogen). The cycling parameters were RT at 48 °C for 30 min, denaturation at 94° for 2 min, 37 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and elongation at 68 °C for 30 s. The resulting DNA was electrophoresed on a 1% agarose gel containing ethidium bromide in TBE buffer (90 mM Tris, 90 mM boric acid and 2 mM EDTA, pH 8.0) and visualized with a LAS3000 Luminescent Image Analyzer (GE Healthcare).
454 sequencing of baboon SHFV genome RNA
Diluted baboon serum or culture fluid containing a baboon SHFV was passed through a 0.45 μm filter, centrifuged at 50,000 Xg for 2 h, and the pellet was resuspended in PBS overnight at 4 °C and then treated with DNase Turbo, Benzonase, and RNase One for 2 h at 37 °C to remove any “free” RNA and DNA. A Qiagen Viral RNA kit (Qiagen, Valencia, CA) was used to extract virion RNA that was then reverse transcribed, purified, and amplified as described previously (Donaldson et al., 2010, Vatter et al., 2014). Briefly, the RNA was reverse transcribed with Superscript III (Life Technologies, Grand Island, NY) using random hexamers with a unique barcode, dsDNA was generated from the cDNA in a Klenow reaction and the dsDNA was then incubated with phosphatase and exonuclease. A 10 µl aliquot of the Klenow reaction was used as template for random amplification using a unique barcode, without the random hexamer portion, as a primer. The DNA was gel purified using a QiaQuick Gel Extraction Kit (Qiagen, Valencia, CA) and submitted to the University of North Carolina High Throughput Sequencing facility for sequencing using the Roche 454 Life Science FLX Titanium chemistry (454 Life Sciences of Roche, Branford, CT) for the first sequence run. The second sequence run used the Roche 454 Junior platform and chemistry (454 Life Sciences of Roche, Branford, CT).
The sequences obtained were binned based on the barcode sequence added, and the barcode sequences were trimmed from the reads in each unique bin, representing individual samples. Each sample bin was mapped to the SHFV reference genome (Accession no. AF180391.2) using the CLC Genomics Workbench version 5.1 default settings (CLCBio, Aarhus, Denmark). The 50% majority rule was used to determine consensus sequences for each contig. De novo assembly of each 454 sequence bin was also done to determine whether the approaches produced similar results and to detect sequences of unknown identity. The de novo assembly consensus sequences derived were then used to query the non-redundant nucleotide database using BLAST within the CLC Genomics Workbench interface. Annotation of differences between the reference genome and the genome derived by sequencing was done by a multiple sequence alignment generated with CLC Genomics Workbench.
Experimental infection and evaluation of SHFV- infected macaques
All aspects of the experimental animal infection studies were performed according to the Institutional Guidelines for Animal Care and Use at the Oregon National Primate Research Center, Beaverton, OR. Four adult (9–12 years old) male Old World Japanese macaques (M. fuscata) were inoculated intravenously with 100 PFU of macaque MΦ-amplified, baboon SHFV isolate B11661 and evaluated by physical examination on a daily basis by a clinical veterinarian. A clinical score sheet was designed that was similar to one previously described (Johnson et al., 2011) with five clinical categories to assess the disease course. Parameters included overall clinical appearance, respiratory abnormalities, activity levels and species-specific behavior, responsiveness, and core body temperature. Each category contained ratings of 0–10, with 0 as normal and 10 being severe. Animals were euthanized when their combined clinical assessment scores reached 10. Blood samples were taken daily (first cohort) or every other day (second cohort) and plasma and PBMCs were used for the assays described below. A complete necropsy was performed on each animal. Samples were collected from a standard list of tissues, fixed in 10% neutral-buffered formalin, and prepared for histopathological and immunohistochemical evaluation.
Quantification of cell IFNβ mRNA and viral mRNA by quantitative real-time RT-PCR (qRT-PCR)
Total cellular RNA from an aliquot of PBMCs was isolated using an RNeasy Mini kit (Qiagen) according to the manufacturer׳s protocol. The primer mix and TaqMan MGB probe used to detect rhesus macaque IFNβ mRNA was IFNβ Rh03648734_s1 (Applied Biosystems). qRT-PCR was performed for the target gene mRNA and for the endogenous control (eukaryotic 18S rRNA VIC/TAMRA dye labeled, Applied Biosystems) in a single-plex format with 100 ng of cellular RNA and the TaqMan one-step RT-PCR master mix reagent kit (Applied Biosystems). The cycling parameters were 48 °C for 30 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and then 60 °C for 1 min on an Applied Biosystems 7500 sequence detection system. Each sample was assayed in triplicate. The triplicate Ct values were analyzed with Microsoft Excel using the comparative Ct (ΔΔCt) method of the SDS Applied Biosystems software which also applied statistical analysis to the data (TINV test in Microsoft Excel). The values were normalized to those for 18S rRNA in the same sample and are the relative fold change compared to the mock infected 24 h calibrator sample value in relative quantification units (RQUs). Error bars represent the standard error of the mean and indicate the calculated minimum (RQmin) and maximum (RQmax) of the mRNA expression levels based on an RQmin/max of the 95% confidence level.
A primer-probe set targeting the nucleocapsid region of SHFV was designed from the SHFV-LVR Genbank consensus sequence. The primer sequences were as follows: Forward primer 5′-TCCCACCTCAGCACACATCA-3′; TaqMan probe 5′-6FAM-AACAGCTGCTGATCAGGT-MGBNFQ-3′; reverse primer 5′-CCGCCTCCGTTGTCGTAGT-3′. Total cellular RNA from SHFV-infected PBMCs was isolated with TRI Reagent (Molecular Research Center, Inc.) according to the manufacturer׳s protocol. Total cellular RNA (100 ng) was used for one-step qRT-PCR to quantify viral RNA or 18S rRNA (eukaryotic 18S rRNA VIC/TAMRA dye labeled, Applied Biosystems) with the TaqMan one-step RT-PCR master mix reagent kit (Applied Biosystems) using gene specific 20X primer mixes and TaqMan MGB probes. The cycling parameters were RT at 48 °C for 30 min, AmpliTaq activation at 95 °C for 10 min, denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min (repeated 40 times). RNA was quantified with an Applied Biosystems 7500 sequence detection system. Triplicate Ct values were analyzed by the comparative Ct (ΔΔCt) method (Applied Biosystems). The amount of target (2−ΔΔCt) was determined by normalization to an endogenous control (18S rRNA) in each sample and relative to a calibrator sample (day 0). Intracellular SHFV RNA was quantified using a standard curve generated with 10-fold serial dilutions of a known concentration of SHFV RNA in vitro transcribed with an SP6 mMessage mMachine kit (Ambion) from a SHFV cDNA template. After in vitro transcription, the DNA template was digested with Turbo DNase at 37 °C for 5 min. The RNA was purified with lithium chloride, precipitated with ethanol, washed with 70% ethanol, resuspended in RNase-free water, and quantified by UV spectrophotometry. The data obtained were used to calculate viral RNA copy number.
Quantification of secreted cytokines
Pro-inflammatory cytokines in macaque plasma were quantified using a Luminex microsphere based ELISA as previously described (Hutchinson et al., 2001, Vatter and Brinton, 2014). Briefly, polyclonal antibodies ( Table 3) were conjugated to polystyrene xMAP microspheres (Luminex Corp.) using an xMAP antibody coupling kit (Luminex Corp.). One hundred microspheres for each analyte were then incubated with 25 µl of plasma at 4 °C for 16 h. Microspheres incubated with 25 µl of each of the serially diluted human recombinant cytokines were used to construct standard curves (from 400 pg/ml to 0.128 pg/ml). Microspheres were washed thrice in PBS containing 1% BSA and then incubated with one of the analyte-specific, biotinylated antibodies at 25 °C for 1 h. Microspheres were washed thrice in 1% BSA-PBS and then incubated with 5 µg/ml streptavidin-R-phycoerythrin (Sigma-Aldrich) at 25 °C for 30 min. Microspheres were washed thrice in 1% BSA-PBS, resuspended in Luminex xMAP systems fluid and analyzed using a Luminex 100 analyzer (Qiagen).
Table 3.
Antibodies used in the Luminex ELISAs.
| Cytokine | Capture antibody | Biotinylated detection antibody |
|---|---|---|
| IFNα | MT1-3-5a | MT2/4/6a |
| TNF-α | MAb1b | MAb11b |
| IL-6 | MQ2-13A5b | MQ2-39C3b |
| MIP-1α | AF-270-NAc | BAF270c |
| MIP-1β | AF-271-NAc | BAF271c |
| IL-12/23(p40) | MT86/221a | MT618a |
| IL-10 | JES3-9D7b | JES3-12G8b |
| IL-1β | JK1B-1b | JK1B-2b |
Mabtech, Inc.
BioLegend.
R&D Systems, Inc.
Western blotting
PBMC aliquots were lysed in RIPA buffer (1X phosphate-buffered saline, 1% Nonidet P-40, 0.5% sodium deoxycholate and 0.1% SDS) containing Halt protease inhibitor cocktail (Thermo Scientific). Following separation by 10% SDS-PAGE, proteins were electrophoretically transferred to a nitrocellulose membrane. Membranes were blocked with 1X TBS containing 5% non-fat dry milk and 0.1% Tween 20 before incubation with a 1:1000 dilution of sheep anti-human tissue factor antibody (Cedarlane Labs) in the presence of blocking buffer. Actin was used as a loading control and was detected with a 1:20,000 dilution of antibody C-11 (Santa Cruz Biotechnology). Blots were washed with 1X TBS containing 0.1% Tween-20 and incubated with a 1:2000 dilution of secondary antibody (horseradish peroxidase-conjugated anti-sheep or anti-mouse; Santa Cruz). Washed blots were then processed for chemiluminescence using a Super-Signal West Pico detection kit (Pierce Scientific) according to manufacturer׳s protocol.
Detection of SHFV-infected cells in tissues of infected macaques
Samples obtained from tissue collected at necropsy from SHFV-infected Japanese macaques were incubated in 10% neutral buffered formalin for 48 h and then transferred to 70% ethanol for further processing. The samples were then embedded in paraffin and cut into 5 μm thick sections. The sections were mounted onto slides, deparaffinized and hydrated. The sections were then incubated with 5% bovine serum albumin and 10% goat serum and Avidin/Biotin Blocking Kit buffer (Vector Laboratories) to block endogenous biotin and biotin receptors and avidin binding sites present in tissues. Endogenous peroxidase was quenched by standard techniques. Tissue sections were first incubated with rabbit polyclonal anti-nsp1β or anti-nap1γ antibody diluted 1:500 in antibody diluent (DAKO) overnight at 4 °C and then with biotinylated goat anti-rabbit antibody (Vector Laboratories) for 30 m at room temperature. After washing, the sections were incubated for 30 m with peroxidase ABC (Vector Laboratories) and then with streptavidin Alexa Fluor 488 (Invitrogen) for 30 m at room temperature. After washing, the sections were blocked with 5% bovine serum albumin and 10% goat serum and then incubated with anti-MΦ antibody (mouse anti-CD68, DAKO) diluted 1:80 overnight at 4 °C in antibody diluent (DAKO). Detection of biotinylated goat anti-mouse secondary antibody was performed with ABC (Vector Laboratories) and incubated with streptavidin Alexa Fluor 594 (Invitrogen) for 30 m at room temperature. Sections were stained with Hoechst 33258 dye (Sigma-Aldrich) to visualize nuclei. Stained slides were covered with prolong gold imaging medium (Invitrogen). Images were acquired using a fluorescence microscope (Zeiss Axioscope 2 plus microscope, Carl Zeiss Microimaging Inc.) equipped with a digital camera (Zeiss Axio-Cam) and filter cubes were used to document specific Alexa 488, Alexa 594 and Hoechst fluorescence. Mouse IgG1 isotype (R&D Systems, Minneapolis, MN) was used as an isotype control antibody and did not give a positive signal on tissue from infected animals (data not shown).
Flow cytometry analysis of PBMCs
Prior to experimental SHFV inoculation, blood and lymph node biopsies were collected from each animal to serve as pre-infection control samples. Following infection, blood samples were collected daily (first cohort of two animals) or every other day (second cohort of two animals) to obtain plasma and PBMCs. PBMCs were stained for cell surface markers CD4 (eBioscience) or CD8b (Beckman Coulter) to identify CD4+ and CD8+ T cell subsets, respectively, as well as CD20 (Biolegend) to identify B cells and CD14 (Biolegend) to identify monocytes. Samples were analyzed using an LSRII instrument (BD) and the data obtained were analyzed using FlowJo software (TreeStar). Total blood cell counts were obtained to determine the number of peripheral lymphocytes of each class for each animal.
Acknowledgments
This work was supported by a Public Health Service research grant AI073824 to M.A.B from the National Institute of Allergy and Infectious Diseases, National Institutes of Health and a Pilot study award (M.A.B. and S.W.W.) from the Oregon National Primate Research Center (P51OD011092). H.A.V. was supported by a Molecular Basis of Disease Fellowship from Georgia State University. This project used biological materials from the Southwest National Primate Research Center (P51 OD011133) and also from the Yerkes National Primate Research Center Comparative AIDS Core (P51OD011132).
References
- Abildgaard C., Harrison J., Espana C., Spangler W., Gribble D. Simian Hemorrhagic Fever: Studies of Coagulation and Pathology. Am J Trop Med Hyg. 1975;24:537–544. doi: 10.4269/ajtmh.1975.24.537. [DOI] [PubMed] [Google Scholar]
- Allen A.M., Palmer A.E., Tauraso N.M., Shelokov A. Simian hemorrhagic fever. II. Studies in pathology. Am. J. Trop. Med. Hyg. 1968;17(3):413–421. doi: 10.4269/ajtmh.1968.17.413. [DOI] [PubMed] [Google Scholar]
- Bailey A.L., Lauck M., Weiler A., Sibley S.D., Dinis J.M., Bergman Z., Nelson C.W., Correll M., Gleicher M., Hyeroba D., Tumukunde A., Weny G., Chapman C., Kuhn J.H., Hughes A.L., Friedrich T.C., Goldberg T.L., O’Connor D.H. High genetic diversity and adaptive potential of two simian hemorrhagic fever viruses in a wild primate population. PLoS One. 2014;9(3):e90714. doi: 10.1371/journal.pone.0090714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradfute S.B., Warfield K.L., Bray M. Mouse models for filovirus infections. Viruses. 2012;4(9):1477–1508. doi: 10.3390/v4091477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M. Pathogenesis of viral hemorrhagic fever. Curr. Opin. Immunol. 2005;17(4):399–403. doi: 10.1016/j.coi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Bray M., Geisbert T.W. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int. J. Biochem. Cell Biol. 2005;37(8):1560–1566. doi: 10.1016/j.biocel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bray M., Mahanty S. Ebola hemorrhagic fever and septic shock. J. Infect. Dis. 2003;188(11):1613–1617. doi: 10.1086/379727. [DOI] [PubMed] [Google Scholar]
- Brinton M.A., Plagemann P.G. Clearance of lactate dehydrogenase by SJL/J mice infected with lactate dehydrogenase-elevating virus. J. Reticuloendothel. Soc. 1983;33(5):391–400. [PubMed] [Google Scholar]
- Chang T.H., Kubota T., Matsuoka M., Jones S., Bradfute S.B., Bray M., Ozato K. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009;5(6):e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8(2):270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Haskew A.N., Gates J.E., Huynh J., Moore C.J., Frieman M.B. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 2010;84(24):13004–13018. doi: 10.1128/JVI.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunowska M., Biggs P.J., Zheng T., Perrott M.R. Identification of a novel nidovirus associated with a neurological disease of the Australian brushtail possum (Trichosurus vulpecula) Vet. Microbiol. 2012;156(3–4):418–424. doi: 10.1016/j.vetmic.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert T.B., Young H.A., Jahrling P.B., Davis K.J., Larsen T., Kagan E., Hensley L.E. Pathogenesis of Ebola hemorrhagic fever in primate models: evidence that hemorrhage is not a direct effect of virus-induced cytolysis of endothelial cells. Am. J. Pathol. 2003;163:2371–2382. doi: 10.1016/S0002-9440(10)63592-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert T.W., Hensley L.E., Jahrling P.B., Larsen T., Geisbert J.B., Paragas J., Young H.A., Fredeking T.M., Rote W.E., Vlasuk G.P. Treatment of Ebola virus infection with a recombinant inhibitor of factor VIIa/tissue factor: a study in rhesus monkeys. Lancet. 2003;362(9400):1953–1958. doi: 10.1016/S0140-6736(03)15012-X. [DOI] [PubMed] [Google Scholar]
- Geisbert T.W., Hensley L.E., Larsen T., Young H.A., Reed D.S., Geisbert J.B., Scott D.P., Kagan E., Jahrling P.B., Davis K.J. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 2003;163(6):2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert T.W., Young H.A., Jahrling P.B., Davis K.J., Kagan E., Hensley L.E. Mechanisms underlying coagulation abnormalities in Ebola hemorrhagic fever: overexpression of tissue factor in primate monocytes/macrophages is a key event . Infect. Dis. 2003;188(11):1618–1629. doi: 10.1086/379724. [DOI] [PubMed] [Google Scholar]
- Gravell M., London W.T., Leon M.E., Palmer A.E., Hamilton R.S. Differences among isolates of simian hemorrhagic fever (SHF) virus. Proc. Soc. Exp. Biol. Med. 1986;181(1):112–119. doi: 10.3181/00379727-181-42231. [DOI] [PubMed] [Google Scholar]
- Gravell M., London W.T., Rodriguez M., Palmer A.E., Hamilton R.S. Simian haemorrhagic fever (SHF): new virus isolate from a chronically infected patas monkey. J. Gen. Virol. 1980;51(Pt 1):99–106. doi: 10.1099/0022-1317-51-1-99. [DOI] [PubMed] [Google Scholar]
- Han M., Kim C.Y., Rowland R.R., Fang Y., Kim D., Yoo D. Biogenesis of non-structural protein 1 (nsp1) and nsp1-mediated type I interferon modulation in arteriviruses. Virology. 2014;458-459:136–150. doi: 10.1016/j.virol.2014.04.028. [DOI] [PubMed] [Google Scholar]
- Hutchinson K.L., Villinger F., Miranda M.E., Ksiazek T.G., Peters C.J., Rollin P.E. Multiplex analysis of cytokines in the blood of cynomolgus macaques naturally infected with Ebola virus (Reston serotype) J. Med. Virol. 2001;65(3):561–566. [PubMed] [Google Scholar]
- Johnson R.F., Dodd L.E., Yellayi S., Gu W., Cann J.A., Jett C., Bernbaum J.G., Ragland D.R., Claire M., St, Byrum R., Paragas J., Blaney J.E., Jahrling P.B. Simian hemorrhagic fever virus infection of rhesus macaques as a model of viral hemorrhagic fever: clinical characterization and risk factors for severe disease. Virology. 2011;421(2):129–140. doi: 10.1016/j.virol.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapin B.A., Shevtsova Z.V. On the identity of two simian hemorrhagic fever virus strains (Sukhumi and NIH) Z. Vers. 1971;13(1):21–23. [PubMed] [Google Scholar]
- Lauck M., Hyeroba D., Tumukunde A., Weny G., Lank S.M., Chapman C.A., O׳Connor D.H., Friedrich T.C., Goldberg T.L. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS One. 2011;6(4):e19056. doi: 10.1371/journal.pone.0019056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M., Sibley S.D., Hyeroba D., Tumukunde A., Weny G., Chapman C.A., Ting N., Switzer W.M., Kuhn J.H., Friedrich T.C., O׳Connor D.H., Goldberg T.L. Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. J. Virol. 2013;87(1):688–691. doi: 10.1128/JVI.02433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi M., van der Poll T., ten Cate H. Tissue factor in infection and severe inflammation. Semin. Thromb. Hemost. 2006;32(1):33–39. doi: 10.1055/s-2006-933338. [DOI] [PubMed] [Google Scholar]
- London W.T. Epizootiology, transmission and approach to prevention of fatal simian haemorrhagic fever in rhesus monkeys. Nature. 1977;268(5618):344–345. doi: 10.1038/268344a0. [DOI] [PubMed] [Google Scholar]
- Mahanty S., Bray M. Pathogenesis of filoviral haemorrhagic fevers. Lancet Infect. Dis. 2004;4(8):487–498. doi: 10.1016/S1473-3099(04)01103-X. [DOI] [PubMed] [Google Scholar]
- Palmer A.E., Allen A.M., Tauraso N.M., Shelokov A. Simian hemorrhagic fever. I. Clinical and epizootiologic aspects of an outbreak among quarantined monkeys. Am. J. Trop. Med. Hyg. 1968;17(3):404–412. [PubMed] [Google Scholar]
- Renquist D. Outbreak of simian hemorrhagic fever. J. Med. Primatol. 1990;19(1):77–79. [PubMed] [Google Scholar]
- Ruf W. Emerging roles of tissue factor in viral hemorrhagic fever. Trends Immunol. 2004;25(9):461–464. doi: 10.1016/j.it.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Shevtsova Z.V. A further study of simian hemorrhagic fever virus. Vopr. Virusol. 1969;14(5):604–607. [PubMed] [Google Scholar]
- Snijder E.J., Kikkert M. Arteriviruses. In: Knipe D.M., Howley P.M., editors. Vol. 1. Lippincott Williams and Wilkins; Philadelphia, PA: 2013. pp. 859–879. (Fields Virology). [Google Scholar]
- Snijder E.J., Meulenberg J.J.M. The molecular biology of arterivirus. J. Gen. Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- Tauraso N.M., Shelokov A., Palmer A.E., Allen A.M. Simian hemorrhagic fever. 3. Isolation and characterization of a viral agent. Am. J. Trop. Med. Hyg. 1968;17(3):422–431. [PubMed] [Google Scholar]
- Vatter H.A., Brinton M.A. Differential responses of disease-resistant and disease-susceptible primate macrophages and myeloid dendritic cells to simian hemorrhagic fever virus infection. J. Virol. 2014;88(4):2095–2106. doi: 10.1128/JVI.02633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatter H.A., Di H., Donaldson E.F., Baric R.S., Brinton M.A. Each of the eight simian hemorrhagic fever virus minor structural proteins is functionally important. Virology. 2014;462–463C:351–362. doi: 10.1016/j.virol.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack P.M. Simian Hemorrhagic Fever. In: Jones T.C., Mohr U., Hunt R.D., editors. Nonhuman Primates I. Springer-Verlag; New York: 1993. pp. 118–131. [Google Scholar]
- Zampieri C.A., Sullivan N.J., Nabel G.J. Immunopathology of highly virulent pathogens: insights from Ebola virus. Nat. Immunol. 2007;8(11):1159–1164. doi: 10.1038/ni1519. [DOI] [PMC free article] [PubMed] [Google Scholar]