Abstract
Objective
Recently, Christianson syndrome (CS) has been determined to be caused by mutations in the X-linked Na+/H+ Exchanger 6 (NHE6). We aimed to determine the diagnostic criteria and mutational spectrum for CS.
Methods
Twelve independent pedigrees (14 boys, ages 4 to 19) with mutations in NHE6 were administered standardized research assessments and mutations were characterized.
Results
The mutational spectrum was composed of 9 single nucleotide variants (SNVs), 2 indels and 1 CNV deletion. All mutations were protein-truncating or splicing mutations. We identified two recurrent mutations (c.1498 c>t, p.R500X; and c.1710 g>a, p.W570X). Otherwise, all mutations were unique. In our study, seven of 12 mutations (58%) were de novo, in contrast to prior literature wherein mutations were largely inherited. We also report prominent neurological, medical and behavioral symptoms. All CS participants were non-verbal and had intellectual disability, epilepsy and ataxia. Many had prior diagnoses of autism and/or Angelman syndrome. Other neurologic symptoms included eye movement abnormalities (79%), postnatal microcephaly (92%) and MRI evidence of cerebellar atrophy (33%). Regression was noted in 50%, with recurrent presentations involving loss of words and/or the ability to walk. Medical symptoms, particularly gastrointestinal symptoms, were common. Height and body mass index measures were below normal ranges in most participants. Behavioral symptoms included hyperkinetic behavior (100%) and a majority exhibited high pain threshold.
Interpretation
This is the largest cohort of independent CS pedigrees reported. We propose diagnostic criteria for CS. CS represents a novel neurogenetic disorder with general relevance to autism, intellectual disability, Angelman syndrome, epilepsy and regression.
Mutations in the X-linked endosomal Na+/H+ Exchanger 6 (NHE6, also known as SLC9A6) cause a newly-recognized, neurogenetic syndrome with variable expressivity. In a systematic, large-scale resequencing screen of X-chromosome exons in pedigrees consistent with X-linked intellectual disability (XL-ID), protein-truncating mutations occurred in NHE6 at a rate of approximately 1% (2 in nearly 200 pedigrees). This high rate of mutation placed NHE6 among the top six most recurrently mutated genes in XL-ID.1 The first reports of mutations in NHE6 were associated with an Angelman-like syndrome (AS).2 The association of NHE6 with Angelman syndrome represents an exciting new opportunity to deepen our understanding of this condition through the exploration of an alternative gene pathway that may provide novel mechanistic perspectives on the disease. However, NHE6 mutations may not be uniformly associated with Angelman syndrome3, and therefore, this relationship needs to be studied in larger groups of patients with NHE6 mutations.
A large South African pedigree previously reported by Christianson et al.4 was one of the pedigrees determined to have an NHE6 mutation by Gilfillan et al.2 The “Christianson syndrome” (CS) reported in the original publication was not noted to include Angelman syndrome; instead, the affected males in the pedigree were reported to have X-linked severe ID associated with craniofacial dysmorphology, mutism (i.e. non-verbal status) despite normal hearing, generalized tonic-clonic epilepsy, postnatal microcephaly, ataxia, ophthalmoplegia and cerebellar and brainstem atrophy. Christianson et al. also posited a limited life expectancy, although he studied only the single pedigree. In addition to the features described above, Christianson reported an association with autistic symptoms as has been reported subsequently.3
In parallel to the description of autistic symptoms associated with mutations in NHE6, Morrow et al.5 published mutations in the highly related endosomal protein NHE9 in autism with epilepsy. Further, in a recent transcriptome study in cortex from postmortem autism brain, we demonstrated that genes encoding NHE6 and NHE9 were dysregulated in brains from patients with idiopathic autism -- NHE6 gene expression was decreased and NHE9 gene expression is increased in a fashion that was highly correlated with gene expression decreases in synapse-related genes.6 These findings demonstrate that, in addition to the role of NHE6 in monogenic CS, NHE6 and NHE9 may play critical roles in the pathophysiology of complex autism, likely participating in a convergent cellular mechanism involving synapse and circuit development.
In addition, NHE proteins have broad importance in neurology, particularly, given the spontaneous mutation in Nhe1 in the slow-wave epilepsy mouse7, and the various anti-epileptic medicines that alter regulation of proton concentrations.8, 9 The structure of NHE proteins generally involves a twelve-membrane spanning motif harboring the Na+ and H+ exchange activity that is highly conserved across family members, and the proteins also contain a large, less conserved carboxyl domain that is thought to involve protein localization and regulation.10 Other studies indicate that NHE1-5 are localized to the cell membrane, while NHE6-9 are thought to be organellar. NHE6 is localized to early, recycling and late endosomes in hippocampal neurons.11, 12 Recent studies indicate that NHE6 plays a role in neuronal arborization and synapse development through modulation of neurotrophin signaling.12
To date, all studies of mutations in NHE6 in the literature have reported the associated clinical phenotype from three or less pedigrees.2–4, 13–18 The vast majority of prior publications have reported inherited mutations. Here we report results from the prospective recruitment of twelve independent pedigrees affected by CS with confirmed mutations in NHE6. In contrast to prior literature, we find a high frequency of de novo mutations (seven to date). In the current study, we quantify the clinical features of this cohort in order to address the following questions: (1) What are the core features of CS?; (2) Quantitatively, what is the range of clinical symptoms and outcomes?; and (3) Are there important differences between the inherited mutations and the de novo mutations, either at the level of the genetic mutation or at the level of neuromedical features?
Patients and Methods
Patients
Families were recruited by advertising and word-of-mouth among families. Pedigrees in which a diagnosis of Christianson syndrome was suspected or families with prior diagnoses were enrolled. Identified probands and the extended pedigree were enrolled, including all available parents, grandparents, aunts, uncles and unaffected siblings. The phenotype of the affected proband was considered to be consistent with Christianson syndrome if: 1) occurred in boys; 2) involved intellectual disability; 3) seizures; 4) ataxia; and 5) there was a plausible deleterious NHE6 mutation. Families were assessed by a standardized neuromedical history and behavioral assessment that included: Autism Diagnostic Interview – Revised (ADI-R) 19, Social Communication Questionnaire (SCQ) 20, Social Responsiveness Scale (SRS) 21, neurological examination (e.g. head circumference, examination of tone and reflexes, eye movements), Vineland II 22 and Leiter-R 23. Twelve families in total were enrolled in this study. Thirteen families were screened; however, in one family we determined that the variant found in NHE6 was unlikely to be pathogenic, and a distinct, likely causative mutation was found in a different gene (data not shown). The institutional review board (IRB) at Brown University and Lifespan Healthcare gave permission to perform this study and informed consent was obtained from all enrolled participants. Some aspects of the pedigrees are changed to assure anonymity. Information regarding phenotype of carriers is based on pedigree and family history.
Sequencing and Variant Calling
All coding exons (exons 1 through 16) in the NHE6 gene, and exon/intron junctions including greater than fifty base pairs into the intron, were PCR-amplified and sequenced using Sanger methods in the proband of each family. The first cDNA position was defined as the first adenine base pair in the SLC9A6 start codon (exon 1). This corresponded to Ensembl cDNA ENST00000370695. PCR primers for each exon are shown in Supplementary Table 3. Variants were identified by verifying the chromatogram using Chromas Lite software. All exons were screened in the proband; however, only the putative mutation was tested in relatives in the extended pedigree in order to discern the pattern of inheritance. All references to genomic coordinates are based on human genome build hg19 (www.genome.ucsc.edu).
Results
Phenotypic presentation in male probands
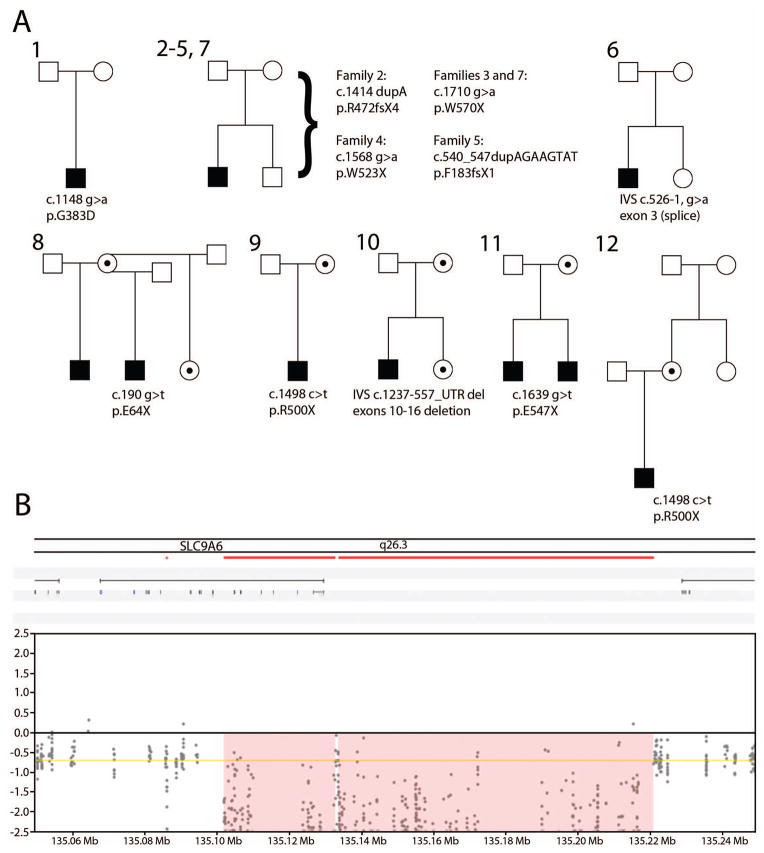
All CS participants were males between ages 4 and 19 at the time of assessment. See Figure 1 for pedigree structure and mutations and Supplementary Table 1 for a summary of clinical features.
Figure 1. Christianson syndrome pedigrees.
(A) Pedigrees for each CS family (#1–12) are shown, with male members (square) and female members (circles). Affected male probands are shaded and heterozygous female carriers are represented by dots inside circles. The cDNA and protein coordinates for each mutation (derived from Ensembl transcript ENST00000370695 and Ensembl protein sequence ENSP00000359729, respectively) are also shown beneath each pedigree. Note that the probands from families 3 and 7 share a recurrent de novo mutation with no known biological relationship; in addition, the probands from families 9 and 12 have a recurrent mutation which was maternally inherited in each respective family, with no known biological relationship between families.
(B) Genomic deletion in NHE6 (SLC9A6) identified in patient 10. The deletion spanned 120.7 kb (ChrX:135098247-135218928; GRCh37/hg19).
Developmental progress and regression
All CS participants demonstrated profound developmental delays in most or all domains, including gross and fine motor, social, language and cognitive domains. All of the boys were non-verbal or had minimal words at the time of presentation to the study. A majority of CS participants had a history of hypotonia (79%). Walking was delayed in many but not all cases, with onset ranging from 1 to 3 years (mean age 20.2 months). All boys exhibited some degree of truncal ataxia with unsteady gait.
We administered the Leiter-R to two probands (patients 2 and 3, ages 7 and 9) as a measure of non-verbal cognitive functioning. Both boys had IQs in the deficient range (<1%) with a Brief IQ of 36 (lowest score obtainable). We also administered the Vineland II to three patients (patients 1, 2 and 3), who ranged in age from 7 through 17. For all 3 patients, composite adaptive functioning was in the low range (<1%), which in combination with IQ testing is indicative of severe to profound ID. Receptive language age equivalents were between 1 month and 8 months. Expressive language age equivalents were between <1 month and 5 months. Daily living skills (e.g. personal, domestic and community) age equivalents ranged from <1 month to 1 year 8 months. Socialization was in the low range (<1 percentile) for all 3 patients and subdomain (e.g. interpersonal relationships, play and leisure time and coping skills) age equivalents ranged from <1 month to 1 year 2 months. Motor skills, measured in two patients, were both in the low range (<1 percentile) and subdomain (e.g. gross and fine motor skills) age equivalents ranged from 7 months to 1 year 6 months.
All parents reported that their child had been diagnosed with ID, and 50% of parents reported regression in walking (57%), eating (14%), loss of a few words/sounds (57%), eye contact/facial expressions (14%) and other fine/gross motor skills (14%). In these patients, the onset of regression was reported to follow a medical illness and/or seizure cluster. The age at regression was reported as 15 months, 2–3 years, 4 years, 5 years (2 cases), 9 years and 16 years (Supplementary Table 1).
Postnatal brain growth
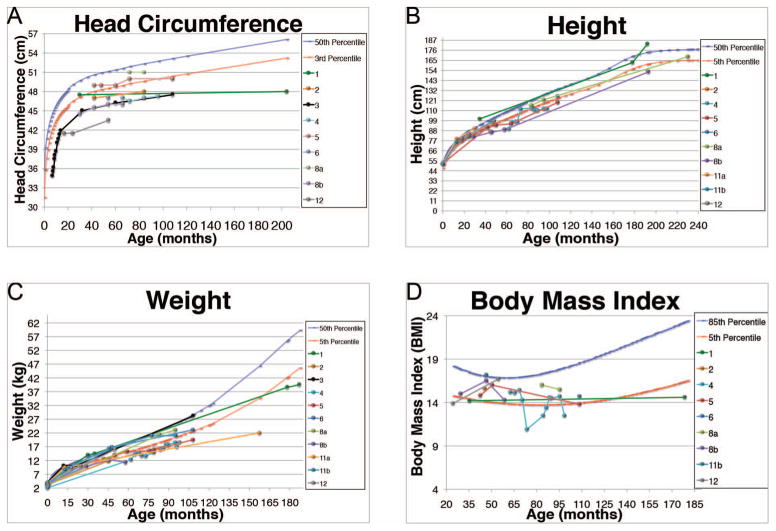
A majority (92%) of CS participants had histories of microcephaly with delayed trajectories of postnatal brain growth (Figure 2A). One participant provided a history of premature closure of cranial sutures.
Figure 2. Patient growth trajectories.
(A) Postnatal head growth of current Christianson syndrome patients. The 50th (blue) and 3rd (red) head circumference percentiles are shown as a reference. Head circumference < 3rd percentile is indicative of microcephaly. Head circumference percentiles are reported according to Rollins et al.37
(B) Height trajectories of current Christianson syndrome patients. Height percentiles were plotted according to CDC recommendations38, whereby the 2006 WHO child growth standards39 were used for measurements taken ≤ 24 months of age and the 2000 CDC growth charts40 for measurements taken > 24 months.
(C) Weight trajectories of current Christianson syndrome patients. Weight percentiles were plotted according to CDC recommendations38, whereby the 2006 WHO child growth standards39 were used for measurements taken ≤ 24 months of age and the 2000 CDC growth charts40 for measurements taken > 24 months. Birth weight and percentile for probands are as follows: patient 1 (3.6 kg, 50–75%), patient 2 (3.2 kg, 25–50%), patient 3 (4.0 kg, 90–95%), patient 4 (2.1 kg, <2%), patient 5 (3.5 kg, 50–75%), patient 6 (3.1 kg, 25–50%), patient 8a (3.2 kg, 25–50%), patient 8b (3.8 kg, 75–90%), patient 11a (3.7 kg, 75–90%), patient 11b (3.5 kg, 50–75%), patient 12 (3.0 kg, 10–25%).
(D) Body Mass Index (BMI) scores of current Christianson syndrome patients. BMI percentile values were plotted according to the 2000 CDC growth charts.40 The 85th (blue) and 5th (red) percentiles were chosen as reference percentiles since they designate overweight and underweight categories, respectively.
Epilepsy
All 14 boys with confirmed NHE6 mutations had epilepsy. All participants had epilepsy onset between the ages of 4 months and 3 years (mean onset was 16.4 +/− 7.86 months). Seizure types included infantile spasms (one case), tonic seizures, tonic-clonic seizures, myoclonic seizures, drop seizures (unknown whether tonic or atonic) and episodes described as staring spells in most cases. While most patients’ seizure semiologies suggested generalized epilepsy, one patient had seizures described with left facial grimacing and another had focal eye deviation, suggesting that some cases also had focal onset seizures.
We were able to review EEGs for four cases (Patients 4, 6, 11a and 11b) and EEG findings from formal reports or clinical notes from four additional cases (Patients 1, 2, 3 and 5). While two patients reportedly had normal EEG results, the majority of EEGs were abnormal with both background abnormalities and epileptiform abnormalities. Two EEGs normalized after treatment, and one child had an initially normal EEG prior to losing the normal background. Background abnormalities included generalized slowing and absence of normal sleep features in at least one case. EEG reports mention epileptiform features including frequent generalized spike-wave complexes, irregular generalized spike-wave pattern and multifocal independent and sometimes synchronous spikes. One patient had hypsarrhythmia in infancy and later focal spikes (Patient 11a). EEG findings are summarized in Supplementary Table 1.
At least four patients had clinical and EEG features consistent with epileptic encephalopathy, both Lennox-Gastaut Syndrome (Patients 1, 6 and 11b) and infantile spasms (Patient 11a), which are known to be often genetically mediated.24 Three patients had clinical symptoms and EEG findings suggestive of Lennox-Gastaut Syndrome according to the information available (Figure 3A). Patient 1 had tonic, tonic-clonic, myoclonic and clinical absence seizures and an EEG reporting 2–4 Hz slow spike-wave. Patient 6 had generalized tonic-clonic seizures and drop seizures as well as focal motor features with some seizures; the EEG showed bilateral frequent semi-rhythmic slow spike-and-wave discharges. Patient 11b had generalized seizures, in the form of tonic seizures and drop attacks, as well as complex partial seizures; interictal EEG showed frequent slow spike-wave complexes with absent normal background. Patient 11a, mentioned above, had infantile spasms and hypsarrythmia. Further review of CS patients’ EEG will allow more detailed syndrome classification. While most patients had generalized seizure types, the other patients’ clinical and EEG features did not fall into clear epilepsy syndromes.
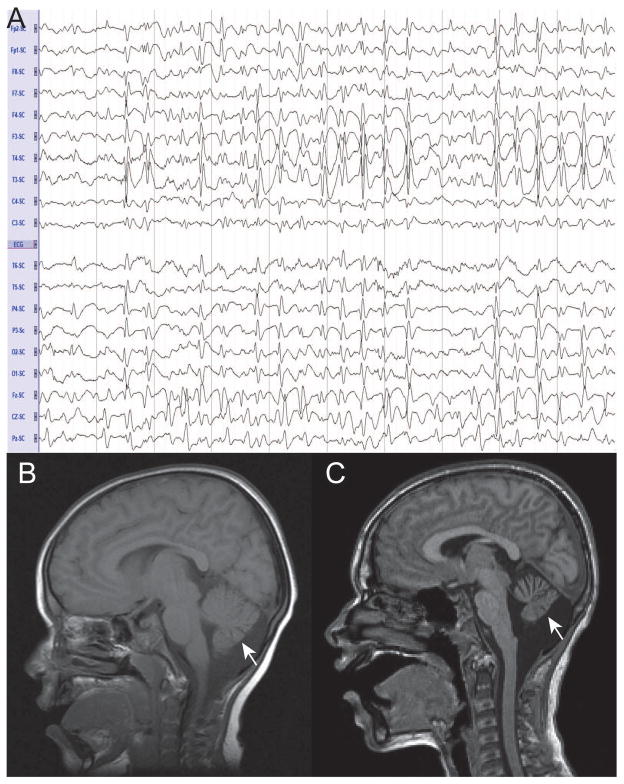
Figure 3. Neurological investigations in Christianson syndrome patients.
(A) EEG referential montage of CS patient (11a) showing diffuse slow spike-and-wave complexes with lack of normal background features, characteristic electrographic findings of Lennox-Gastaut Syndrome.
(B and C) MRI results of CS patient showing moderate to severe atrophy of the cerebellar hemispheres and vermis (arrow) in one patient in the current study at two distinct ages. (B) Sagittal T1-weighted MRI of patient at 4 years (1.5T). (C) Sagittal T1-weighted MRI at 10 years (1.5T).
Seizure frequency and control varied substantially with the most mildly affected having a period of seizure freedom for over a year, to some participants experiencing seizure clusters daily. A variety of anti-epileptic medications have been used (Supplemental Table 1). Non-medication treatment included ketogenic diet and vagus nerve stimulator.
Symptoms and prior diagnoses of Angelman syndrome and/or Autistic Disorder
Over a third of the patients (43%) were originally diagnosed with Angelman syndrome by clinicians. In this prospective study, clinical diagnostic criteria by Tan et al. 25 were utilized. Upon direct assessment, the majority of CS patients (93%) exhibit phenotypes similar to Angelman syndrome. For major criteria, all patients had ID and limited speech. Approximately 100% of patients had ataxia/unsteady gait (1 patient is currently non-ambulatory). For behavioral features, a majority were reported to have a happy disposition (100%) and unprovoked laughter (64%). For minor AS criteria, all had seizures and drooled, 92% were microcephalic, 64% had sleep problems and 29% reported a fascination with water. Many children also sat with arms in flexed posture.
The majority of patients displayed autistic symptoms. Six out of 14 (43%) patients were previously diagnosed with autism clinically. Three patients (1, 2 and 3) were assessed with the ADI-R and all met criteria for autism. Evident symptoms included absence of social play or interest in sharing, lack of a variety of facial expressions to communicate and poor eye contact. Patient 1 did not point to express interest, nod or shake his head to communicate. Patient 2 had the least severe autistic symptoms of the patients directly assessed; yet he did not engage in play with other children. He regularly uses other’s hands as a tool (e.g. to turn a doorknob). Patient 3 had unusual preoccupations and sensory interests involving laundry baskets, metal objects and windows. We assessed nine patients with the SCQ. Patient scores ranged from 14 to 28, with an average score of 23.2 (autism cutoff score is 15). Eight out of nine participants tested (89%) met autism criteria. Parent responses consistently reported poor eye contact, use of a caregiver’s body parts as tools, unusual sensory interests and a lack of reciprocal play and/or social interest.
Eye movement abnormalities and vision
The majority of patients (79%) had abnormal eye movements. Almost a third of patients (29%) required surgery, usually to treat strabismus-related issues. The majority of participants (54%) had known visual acuity problems. Ophthalmic symptoms found in individual patients include cortical visual impairment, refractive error and right V1 nerve palsy. None of the patients in our study had a history consistent with known retinal degeneration as presented previously in a young man with CS at age 22.16
Sleep
A majority of families (64%) reported sleep problems in the proband. Two families noted that the affected males appear to have no sleep pattern and rarely sleep through the night. Parents generally reported that affected males have difficulty sleeping as they will be active during the night time.
Gastointestinal-related symptoms
GI-related symptoms were prominent in the CS patients assessed. Half (50%) had gastroesophageal reflux disease (GERD), for which two patients (14% of total patients) required Nissen fundoplication. Many parents (71% of families) reported current and past feeding difficulties such as difficulty chewing and times where the patient refuses to eat. Feeding difficulties were also notable during the neonatal period with latching problems. Some parents also reported that their boys had history of swallowing difficulties (29%) including dysphagia, choking and aspiration.
Height and weight growth
The majority of CS patients (91%) were born at average or above-average weight. However, patients were notable for small stature and attenuated weight gain. As infants, some were diagnosed with failure to thrive (FTT). Parents reported that CS patients were unable to gain weight despite typical, and even high, caloric intake. Many of these height and weight measurements were in the <3–4 percentile range (Figure 2B and 2C). Many CS boys exhibit a body-mass index well below average that may get progressively worse with age (Figure 2D).
Other medical, neurologic and behavioral features
All participants (100%) were hyperkinetic. Also, in a majority of families, parents reported that the proband had an unusually high pain threshold. Medical conditions which were found in a minority of participants included failure to thrive (21%), cyanosis (21%) and eczema (21%). Three patients required extended hospitalization due to infectious disease problems, including two with suspected encephalitis. Infectious disease hospitalizations included: pneumonia; status epilepticus and possible viral encephalitis; and influenza B infection with myositis and possible encephalitis. A range of other medical problems were observed in only a single child and are listed in Supplementary Table 1.
MRI Findings
There were 3 participants (33%) with documented cerebellar atrophy. MRI studies at sequential time points for one of these patients with moderate to severe atrophy of the cerebellar hemispheres and vermis, associated with regression – loss of the ability to walk, is shown in Figure 3B and C (arrow). Another patient had bilateral lesions in the inferior cerebellum with minimal volume loss. There were also notable findings regarding increases in ventricles as well as changes in white matter (Supplementary Table 1).
Phenotypic presentations in female carriers
Female carriers presented with diverse clinical presentations that included neurotypical functioning, mild to moderate ID and psychiatric illness. One female carrier (confirmed by our lab) had a myriad of diagnoses including moderate ID, speech and language delay, selective mutism, sensory integration disorder, separation anxiety disorder, oppositional defiant disorder, reactive attachment disorder and attention deficit/hyperactivity disorder (ADHD). Providers have also proposed childhood onset schizophrenia and autism spectrum disorder in this patient. Her neuropsychological testing at 10 years 10 months revealed overall cognitive functioning in the impaired range (WISC-IV Full Scale IQ=44, <1st percentile) with impaired academic abilities (WIAT-II reading and math, <1st percentile) and adaptive functioning in the extremely low range (ABAS-II, <1st percentile), consistent with her diagnosis of moderate intellectual disability. Another female carrier has been characterized as having mild ID and ADHD. She is also microcephalic with a head circumference of 50.9 (−3 SD).
Allelic series and genotype-phenotype correlations
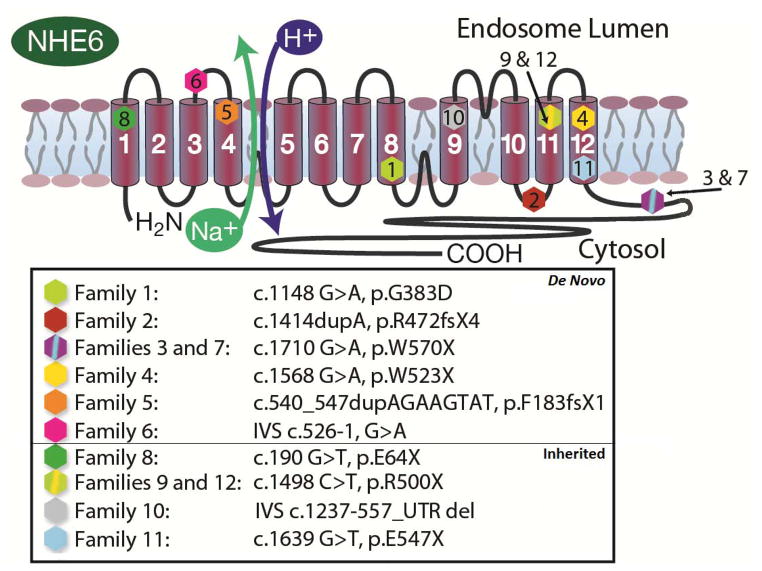
Mutations from twelve families are represented on the NHE6 protein in Figure 4 and pedigrees are shown in Figure 1. Interestingly, in contrast to prior literature regarding NHE6 mutations wherein mutations have been largely inherited (Supplementary Table 2), a majority of mutations in our study (58%) were de novo. Families 1 through 7 had confirmed de novo mutations (ie mutations were not found in the mother).
Figure 4. Patient mutations in NHE6 protein.
Mutations from all families in the present report (n = 12) are shown in the NHE6 protein. The NHE6 protein is a twelve-membrane spanning motif with the Na+/H+ exchange occurring between transmembrane segments S4 and S5. While most mutations are located in the transmembrane domain, two mutations (3 and 7) are located in the carboxyl domain. cDNA positions are based on Ensembl transcript ENST00000370695 and peptide positions are based on ENSP00000359729. Probands from families #1–7 exhibit de novo mutations, whereas probands from families #8–12 exhibit inherited mutations.
Another notable finding was two separate recurrent mutations. The first was found at c.1498 c>t, p.R500X. This mutation was observed in two families not known to be related, and in one family this mutation was determined to occur de novo in a maternal carrier (Family 12, Figure 1). The mutation was carried in the mother of the proband with CS but not in the maternal grandparents. The same mutation was inherited in another proband (Family 9). The two families share a small haplotype around the point mutations that is approximately 171kb long (Supplementary Table 4). We also observed a second recurrent mutation, c.1710 g>a, p.W570X, in Families 3 and 7. Here, the mutations were determined to be de novo in both families, although the families again share a haplotype around the point mutation spanning 260 kb.
In total, we observed 9 SNVs, 2 indels and 1 CNV deletion. The majority of mutations were protein truncations (10/12, 83%) and occurred in exons 1, 3, 11, 12, 13 and 14. With the possible exceptions of Families 3 and 7 all protein truncating mutations occurred prior to the end of the predicted transmembrane domains of the protein (Figure 4). Two mutations (17%) were splice mutations. Family 1 represented a g>a change in the first base of exon 9 which did result in a 20% reduction in the protein levels (data not shown) and also a missense mutation (glycine to aspartate) in the region of the exchanger domain that was predicted to be highly deleterious by Polyphen-2.27 A second likely splice mutation was found in Family 6, a G to A transition in the last base of the intronic sequence upstream of the splice acceptor for exon 3. There was one genomic deletion discovered in the proband in Family 10 spanning from ChrX:135098247-135218938 (c.1237-557). This CNV deletion is predicted to delete exons 10 through 16 and the 3′-untranslated region (UTR).
Our data support a diversity of genetic findings as well as phenotypic findings in this cohort. Upon examination of the genetic mutations -- which may be characterized as early truncating versus late truncating, or alternatively, de novo versus inherited – there are no statistically strong genotype-phenotype correlations that emerge at present.
Discussion
Core and secondary symptoms to guide Christianson syndrome diagnosis
In this study, we prospectively ascertained 12 independent pedigrees, 14 affected boys in total, with mutations in NHE6. Interestingly, in contrast to prior literature, a majority of the pedigrees enrolled exhibited de novo mutations. Through quantitative study of clinical features in probands across these independent pedigrees (Supplementary Table 1) and through review of prior literature (Supplementary Table 2), we propose core diagnostic features of NHE6 mutations and Christianson syndrome that are present in greater than 85% of the affected males. These symptoms appear to be consistent regardless whether the mutation is inherited or de novo. These core diagnostic symptoms of CS (Table 1), presenting in boys in childhood, include: non-verbal status; intellectual disability (moderate to severe range); epilepsy; truncal ataxia; postnatal microcephaly and/or attenuation in postnatal growth of head circumference; and hyperkinetic behavior. Secondary symptoms that are often present (greater than 35%) include: symptoms of autism; symptoms of Angelman syndrome (AS); eye movement problems such as strabismus; regressions (particularly loss of the ability to walk after age 10); low weight for age, and cerebellar vermis atrophy by MRI (particularly after age 10). Low weight and cerebellar atrophy are key features, particularly with age, that distinguish CS from AS due to 15q11 locus anomalies. Another possible distinguishing feature is that epilepsy in AS may not occur in 100% of patients25, 28 whereas in CS, as the data currently stand, epilepsy occurs in all patients. Additional medical and behavioral symptoms are notable, including GERD and high pain threshold.
Table 1.
Diagnostic criteria for Christianson syndrome
|
Core Diagnostic Symptoms (>85%)
|
| Non-verbal |
| Intellectual disability (moderate to severe range) |
| Epilepsy# |
| Ataxia (truncal) |
| Postnatal microcephaly or attenuation of growth in head circumference |
| Hyperkinesis
|
|
Secondary Symptoms (>35%)
|
| Symptoms of autism |
| Symptoms of Angelman syndrome symptoms (particularly in first 5 years) |
| Eye movement problems (e.g. strabismus) |
| Hypotonia |
| Gastroesophageal reflux disease (GERD) |
| Regressions (especially after 1st decade of life) |
| Low height and/or weight for age group (progressing with age)* |
| Cerebellar vermal atrophy (particularly after first decade)* |
CS may be among the most common X-linked developmental brain disorders. In a large-scale sequencing project in 208 pedigrees with suspected X-linked intellectual disability, two protein-truncating mutations in NHE6 were found.1 These data place NHE6 among the top six genes that were found to have recurrent protein-truncating mutations, and suggest that CS may constitute approximately 1–2% of X-linked developmental brain disorders. Similarly in a whole-exome sequencing project of multiplex, non-consanguineous pedigrees with ID, one in nineteen families exhibited a protein truncating mutation in NHE6.29 If we assume that between 1–3% of the world’s population is diagnosed with an intellectual disability, and approximately 10–20% of the causes are due to X-linked genes, then we can estimate that CS may affect between 1 in 16,000 to 100,000 people. By comparison, this represents approximately 10–50% the prevalence of Fragile X syndrome, the most commonly inherited form of intellectual disability.
One uniform and notable feature of CS is epilepsy. Conversations with parents indicate seizure control may represent one of the leading causes of concern in the family. In the current study, we report early age of onset of epilepsy and a mixture of seizure types. Further clinical and EEG analysis of the CS group of patients will determine how many have epilepsy in the setting of epileptic encephalopathy (e.g. infantile spasms, Lennox-Gastaut Syndrome) and provide refinement of the epilepsy syndromes that these patients may experience. A variety of treatments have been tested; however, rigorous prospective studies will be required in patient cohorts with CS in order to develop most accurate treatment protocols. This appears to be among the major priorities in CS research.
Developmental and progressive symptomology in CS
Mutations in NHE6 appear to lead to clinical symptoms that reflect both developmental and progressive (perhaps degenerative) pathophysiology. Given the very early onset of CS with failures in typical development including global developmental delays, lack of language, cognitive and adaptive delays, neurodevelopmental mechanisms are likely prominent. Indeed, in recent experimental studies in mouse models, impairments in neuronal arborization and synapse development were reported.12 It seems plausible that these defects in arborization and synapse development may represent the cellular correlates for the plateau in head circumference growth demonstrated in this study in patients. This hypothesis may be confirmed by future neuropathologic studies. The paucity of white matter and leukomalacia reported on some MRI studies in the setting of developmental regression may also indicate axonal loss specifically.
In this study, as in prior studies2, 14, 15, we report a high rate of prior diagnosis of AS (43%). Additionally, patients met symptom criteria of AS (93%) upon direct examination. The likeness of CS to AS is not apparent in all participants and the rate of NHE6 mutations in AS remains to be determined and may be low.13 Of 59 patients presenting with Angelman-like phenotypes one (1.8%) had NHE6 mutations.13 Also, Gilfillan et al.2 found 4 out of 73 patients with Angelman-like phenotypes had NHE6 mutations. As indicated by Gilfillan et al.2, the likeness of CS to AS is prominent enough to have important clinical relevance, i.e. all boys with non-15q11 AS should be tested for mutations in NHE6. Also, the question of a relationship at the level of cellular mechanism remains intriguing.
CS is related to autism at both the clinical and the biologic level. A sizable proportion of boys with CS have received a previous diagnosis of autism, demonstrate autistic symptoms, and/or meet criteria for autism on standardized examinations. While we report here that a large percentage of patients who were tested using standardized assessments (ADI-R or SCQ) met criteria for autism, this result needs to be interpreted with caution given the challenges of observing a full range of autism symptoms in participants who are non-verbal and have low intellectual and adaptive functioning. However, in addition to the clinical indicators, there are important overlaps between autism and CS at the biological level, wherein in autism transcriptome studies the gene expression levels of NHE6 are reduced in postmortem cortex as compared to control.6
Despite the clear neurodevelopmental components in CS and the likenesses to related developmental disorders such as autism and AS, there appear to be progressive aspects to the CS phenotype. Based on his observations in the original pedigree, Christianson et al.4 described a potential progressive nature involving loss of cognitive and adaptive skills with aging. Further, there are clear regressions that appear to be associated with the CS. In our study, these appear to be associated with febrile illness and/or seizure, and can occur even in the first decade. In other studies, these regressions were generally described in later life past the first decade.3, 15, 16 A recurrent history involves loss of the ability to walk and loss of words or sounds. In addition to these clinical symptoms, there also may be a neurodegenerative component to CS that involves at least the cerebellum and perhaps also regions within the brain stem. We find 33% of patients with MRI evidence of cerebellar atrophy, which corresponds frequently to loss in ability to walk. This finding was also corroborated at the biological level with regard to studies in a CS mouse model that demonstrate Purkinje cell loss with age.30 Additional postmortem studies have also supported cerebellar Purkinje cell degeneration in humans.3, 4 In addition, in a study of one pedigree by Garbern et al.3 with a slightly atypical mutation, Tau deposition was prominently noted across the brain, but in particular in the subcortical regions of the brain. Finally, one single study has reported the possibility of retinal neurodegeneration, perhaps retinitis pigmentosa, in a patient over 20 with CS.16 While this finding has not been corroborated in our study, it requires identification and clinical characterization of patients after they have passed through the age window of risk. Thus far, there have been few studies identifying young or older men with CS, ie age >18 years old.2–4, 14–16, 18 Again in Christianson et al.’s4 original description of the syndrome, he reported the possibility of early mortality in affected males; however, this too requires verification through larger and longitudinal studies. Low numbers of men greater than 20 with CS could be either an issue of ascertainment, or alternatively, relate to an increased risk of mortality in CS.
Additional notable medical and behavioral findings in CS
NHE6 protein appears to be expressed in just about every cell and tissue type yet examined.31 Given this result, it is not surprising that somatic medical symptoms appear to represent a part of CS. A particularly notable symptom is gastroesophageal reflux. While GI symptoms are commonly reported in neurodevelopmental disorders such as Rett syndrome32 and 22q11.2 microdeletion syndrome33, the data that two patients in our cohort required surgery to correct extreme GERD raise the possibility that this GI condition may be related to CS. In addition, relatively low weight, particularly as the boys develop, appears to be a common somatic symptom. This finding likely warrants further clinical and pre-clinical research.
With regard to additional behavioral symptom patterns that should be emphasized, the hyperkinetic behavior in patients with CS seems fairly pervasive. Interestingly, the related endosomal NHE9 protein has been implicated previously in attention deficit/hyperactivity disorder.34–36 Further, many parents also note a high pain threshold and convey experiences involving serious injury wherein their son with CS exhibited little response to pain. This symptom has not appeared in the literature previously yet has substantial clinical relevance related to protecting males with CS from occult injuries or harm.
Prevailing genetic patterns and novel genetic findings in this study
Causative mutations in neurodevelopmental disorders are frequently de novo, X-linked recessive or autosomal recessive. In contrast to prior literature reporting largely inherited mutations in NHE6, through prospective recruitment, here we discover seven highly deleterious de novo mutations in NHE6. An eighth mutation was found, in a three-generation pedigree (Family 12), to be de novo in the mother although inherited in the proband. These de novo (and recent) mutations constitute greater than 50% of pedigrees in our cohort (Figure 1). Given a spectrum of deleterious mutations and that female carriers may exhibit intermediate neurocognitive symptoms, one might hypothesize that de novo mutations may be more highly deleterious and that inherited mutations may appear milder. However, from our data and the other mutations in the literature (Supplementary Table 2), this hypothesis does not appear to be substantiated.4 There is a statistical trend toward more regressions and earlier-onset regressions in the de novo group; however, this association will require larger studies to establish. The majority of mutations appear to be early truncating and/or splice mutations likely constituting loss of function mutations. Given our high discovery rate of de novo variants and prior publications, we anticipate that CS will constitute a spectrum of NHE6 mutations that may be either inherited or de novo. Our findings are important because they indicate that clinicians should be alert to considering mutations in NHE6 in a variety of presentations of neurodevelopmental disorders. Specifically, boys who present with new diagnoses of moderate to severe ID with seizures, language delays, and ataxia, with or without Angelman-like symptoms (AS), and with or without a pedigree consistent of X-linked ID, should be considered for NHE6 mutations.
Finally, in twelve independent families we have discovered a robust allelic series of NHE6 mutations in CS. We discover nine SNVs, two splice mutations, and a 120.7 kb genomic deletion. We also report an apparently recurrent SNV (Family 9 and Family 12), c.1498 c>t transition leading to p.R500X, a protein truncating mutation in the putative eleventh transmembrane domain of the protein. In Family 12, this mutation appears likely de novo in the mother of the affected CS male (i.e. not found in the maternal grandparents). In Family 9, this mutation is inherited. These families do appear to share a haplotype around the mutation locus (Supplementary Table 4), which may be prone for recurrent mutation although further research will be necessary to clarify this. The mutation occurs at a CpG location in the genomic DNA, although not in a known CpG island or methylation hotspot; however, it remains formally possible that this C is methylated and prone to C to T transition by mechanisms involving deamination of a methylated cytosine.26 Family 9 has been reported previously in the literature as a US-based pedigree.15 In our study R500X represents approximately 16% of the mutations in CS. There are also two other reports of likely independent pedigrees with the R500X mutation.1, 2 Taking all these reports into account, conservatively R500X may represent up to 10% of mutations in the literature currently (at least three in total). Interestingly, we also discovered a second SNV (c.1710 g>a, p.W570X) which is recurrent in Families 3 and 7 and de novo in both families.
Summary
In summary, we report a robust series of NHE6 mutations in twelve independently ascertained pedigrees within which we were able to characterize quantitatively core and secondary symptom presentations. Through this study we have proposed a core set of diagnostic criteria for CS and also generated a quantitative early guide to symptom presentation in order to help establish initial clinical expectations for families and clinicians. Based on current studies, CS may be among the most common X-linked developmental brain disorders.1 CS has broad implications to common neurologic and behavioral syndromes including epilepsy, intellectual disability, autism, regression and hyperactivity.
Supplementary Material
Phenotypic features of Christianson syndrome patients in the present cohort
Phenotypic features of Christianson syndrome patients in the published literature
Sequence of PCR primers for amplification and sequencing of NHE6 exons from genomic DNA
Haplotype comparisons between two sets of patients with recurrent mutations in the NHE6 protein (patients 9 and 12, p.R500X; patients 3 and 7, p.W570X).
Acknowledgments
The authors would like to thank the families for participating in this study. The authors would also like to thank Judith Nathanson and Heather M. Thompson for their help with figures. EMM has received a Career Award in Medical Science from the Burroughs Wellcome Fund and support from NIH NIGMS P20GM103645. This work was supported by a grant from the Simons Foundation (SFARI #239834 to EMM), and also generous support to EMM from the Nancy Lurie Marks Foundation. EMM has also received generous support from the newly-formed Christianson Syndrome Association (CSA) to complete the writing of this manuscript. EMM had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
None of the authors have a financial conflict of interest.
References
- 1.Tarpey PS, Smith R, Pleasance E, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009 May;41(5):535–43. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilfillan GD, Selmer KK, Roxrud I, et al. SLC9A6 mutations cause X-linked mental retardation, microcephaly, epilepsy, and ataxia, a phenotype mimicking Angelman syndrome. American journal of human genetics. 2008 Apr;82(4):1003–10. doi: 10.1016/j.ajhg.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garbern JY, Neumann M, Trojanowski JQ, et al. A mutation affecting the sodium/proton exchanger, SLC9A6, causes mental retardation with tau deposition. Brain: a journal of neurology. 2010 May;133(Pt 5):1391–402. doi: 10.1093/brain/awq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christianson AL, Stevenson RE, van der Meyden CH, et al. X linked severe mental retardation, craniofacial dysmorphology, epilepsy, ophthalmoplegia, and cerebellar atrophy in a large South African kindred is localised to Xq24-q27. Journal of medical genetics. 1999 Oct;36(10):759–66. doi: 10.1136/jmg.36.10.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrow EM, Yoo SY, Flavell SW, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008 Jul 11;321(5886):218–23. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwede M, Garbett K, Mirnics K, Geschwind DH, Morrow EM. Genes for endosomal NHE6 and NHE9 are misregulated in autism brains. Molecular psychiatry. 2013 Mar 19; doi: 10.1038/mp.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox GA, Lutz CM, Yang CL, et al. Sodium/hydrogen exchanger gene defect in slow-wave epilepsy mutant mice. Cell. 1997 Oct 3;91(1):139–48. doi: 10.1016/s0092-8674(01)80016-7. [DOI] [PubMed] [Google Scholar]
- 8.Thiry A, Dogne JM, Supuran CT, Masereel B. Carbonic anhydrase inhibitors as anticonvulsant agents. Curr Top Med Chem. 2007;7(9):855–64. doi: 10.2174/156802607780636726. [DOI] [PubMed] [Google Scholar]
- 9.Ali A, Ahmad FJ, Dua Y, Pillai KK, Vohora D. Seizures and sodium hydrogen exchangers: potential of sodium hydrogen exchanger inhibitors as novel anticonvulsants. CNS Neurol Disord Drug Targets. 2008 Oct;7(4):343–7. doi: 10.2174/187152708786441830. [DOI] [PubMed] [Google Scholar]
- 10.Orlowski J, Grinstein S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflugers Archiv: European journal of physiology. 2004 Feb;447(5):549–65. doi: 10.1007/s00424-003-1110-3. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura N, Tanaka S, Teko Y, Mitsui K, Kanazawa H. Four Na+/H+ exchanger isoforms are distributed to Golgi and post-Golgi compartments and are involved in organelle pH regulation. The Journal of biological chemistry. 2005 Jan 14;280(2):1561–72. doi: 10.1074/jbc.M410041200. [DOI] [PubMed] [Google Scholar]
- 12.Ouyang Q, Lizarraga SB, Schmidt M, et al. Christianson syndrome protein NHE6 modulates TrkB endosomal signaling required for neuronal circuit development. Neuron. 2013 Oct 2;80(1):97–112. doi: 10.1016/j.neuron.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fichou Y, Bahi-Buisson N, Nectoux J, et al. Mutation in the SLC9A6 gene is not a frequent cause of sporadic Angelman-like syndrome. European journal of human genetics: EJHG. 2009 Nov;17(11):1378–80. doi: 10.1038/ejhg.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi Y, Hosoki K, Matsushita M, et al. A loss-of-function mutation in the SLC9A6 gene causes X-linked mental retardation resembling Angelman syndrome. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2011 Dec;156b(7):799–807. doi: 10.1002/ajmg.b.31221. [DOI] [PubMed] [Google Scholar]
- 15.Schroer RJ, Holden KR, Tarpey PS, et al. Natural history of Christianson syndrome. American journal of medical genetics Part A. 2010 Nov;152A(11):2775–83. doi: 10.1002/ajmg.a.33093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mignot C, Heron D, Bursztyn J, et al. Novel mutation in SLC9A6 gene in a patient with Christianson syndrome and retinitis pigmentosum. Brain Dev. 2012 Apr 25; doi: 10.1016/j.braindev.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Tzschach A, Ullmann R, Ahmed A, et al. Christianson syndrome in a patient with an interstitial Xq26.3 deletion. American journal of medical genetics Part A. 2011 Nov;155a(11):2771–4. doi: 10.1002/ajmg.a.34230. [DOI] [PubMed] [Google Scholar]
- 18.Riess A, Rossier E, Kruger R, et al. Novel SLC9A6 mutations in two families with Christianson syndrome. Clinical genetics. 2013 Jun;83(6):596–7. doi: 10.1111/j.1399-0004.2012.01948.x. [DOI] [PubMed] [Google Scholar]
- 19.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. Journal of autism and developmental disorders. 1994 Oct;24(5):659–85. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 20.Rutter M, Bailey A, Lord C. Social Communication Questionnaire (SCQ) Los Angeles, CA: Western Psychological Services; 2003. [Google Scholar]
- 21.Constantino J, Gruber C. The social responsiveness scale manual. Los Angeles, CA: Western Psychological Services; 2005. [Google Scholar]
- 22.Sparrow S, Cicchetti D, Balla D. Survey Forms Manual. 2. Pearson; 2005. Vineland-II: Vineland Adaptive Behavior Scales. [Google Scholar]
- 23.Roid GH, Miller LJ. Leiter International Performance Scale-Revised: Examiner’s Manual. Wood Dale, IL: Stoelting Co; 1997. [Google Scholar]
- 24.Epi KC, Allen AS, et al. Epilepsy Phenome/Genome P. De novo mutations in epileptic encephalopathies. Nature. 2013 Sep 12;501(7466):217–21. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan WH, Bacino CA, Skinner SA, et al. Angelman syndrome: Mutations influence features in early childhood. American journal of medical genetics Part A. 2011 Jan;155a(1):81–90. doi: 10.1002/ajmg.a.33775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper DN, Mort M, Stenson PD, Ball EV, Chuzhanova NA. Methylation-mediated deamination of 5-methylcytosine appears to give rise to mutations causing human inherited disease in CpNpG trinucleotides, as well as in CpG dinucleotides. Human genomics. 2010 Aug;4(6):406–10. doi: 10.1186/1479-7364-4-6-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010 Apr;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thibert RL, Conant KD, Braun EK, et al. Epilepsy in Angelman syndrome: a questionnaire-based assessment of the natural history and current treatment options. Epilepsia. 2009 Nov;50(11):2369–76. doi: 10.1111/j.1528-1167.2009.02108.x. [DOI] [PubMed] [Google Scholar]
- 29.Schuurs-Hoeijmakers JH, Vulto-van Silfhout AT, Vissers LE, et al. Identification of pathogenic gene variants in small families with intellectually disabled siblings by exome sequencing. J Med Genet. 2013 Dec;50(12):802–11. doi: 10.1136/jmedgenet-2013-101644. [DOI] [PubMed] [Google Scholar]
- 30.Stromme P, Dobrenis K, Sillitoe RV, et al. X-linked Angelman-like syndrome caused by Slc9a6 knockout in mice exhibits evidence of endosomal-lysosomal dysfunction. Brain: a journal of neurology. 2011 Nov;134(Pt 11):3369–83. doi: 10.1093/brain/awr250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Numata M, Petrecca K, Lake N, Orlowski J. Identification of a mitochondrial Na+/H+ exchanger. The Journal of biological chemistry. 1998 Mar 20;273(12):6951–9. doi: 10.1074/jbc.273.12.6951. [DOI] [PubMed] [Google Scholar]
- 32.Motil KJ, Caeg E, Barrish JO, et al. Gastrointestinal and nutritional problems occur frequently throughout life in girls and women with Rett syndrome. Journal of pediatric gastroenterology and nutrition. 2012 Sep;55(3):292–8. doi: 10.1097/MPG.0b013e31824b6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giardino G, Cirillo E, Maio F, et al. Gastrointestinal involvement in patients affected with 22q11.2 deletion syndrome. Scandinavian journal of gastroenterology. 2014 Mar;49(3):274–9. doi: 10.3109/00365521.2013.855814. [DOI] [PubMed] [Google Scholar]
- 34.Lasky-Su J, Anney RJ, Neale BM, et al. Genome-wide association scan of the time to onset of attention deficit hyperactivity disorder. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2008 Dec 5;147b(8):1355–8. doi: 10.1002/ajmg.b.30869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lasky-Su J, Neale BM, Franke B, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2008 Dec 5;147b(8):1345–54. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- 36.Markunas CA, Quinn KS, Collins AL, et al. Genetic variants in SLC9A9 are associated with measures of attention-deficit/hyperactivity disorder symptoms in families. Psychiatric genetics. 2010 Apr;20(2):73–81. doi: 10.1097/YPG.0b013e3283351209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rollins JD, Collins JS, Holden KR. United States head circumference growth reference charts: birth to 21 years. The Journal of pediatrics. 2010 Jun;156(6):907–13. 13.e1–2. doi: 10.1016/j.jpeds.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 38.Grummer-Strawn LM, Reinold C, Krebs NF. Use of World Health Organization and CDC growth charts for children aged 0–59 months in the United States. MMWR Recommendations and reports: Morbidity and mortality weekly report Recommendations and reports/Centers for Disease Control. 2010 Sep 10;59(Rr-9):1–15. [PubMed] [Google Scholar]
- 39.World Health Organization. Length/height-for-age, weight-for-age, weight-for-length, weight-for-height and body mass index-for-age. WHO; Geneva: 2006. WHO Child Growth Standards. Methods and development. [Google Scholar]
- 40.Kuczmarski RJ, Ogden CL, Grummer-Strawn LM, et al. CDC growth charts: United States. Advance data. 2000 Jun;8(314):1–27. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phenotypic features of Christianson syndrome patients in the present cohort
Phenotypic features of Christianson syndrome patients in the published literature
Sequence of PCR primers for amplification and sequencing of NHE6 exons from genomic DNA
Haplotype comparisons between two sets of patients with recurrent mutations in the NHE6 protein (patients 9 and 12, p.R500X; patients 3 and 7, p.W570X).