Abstract
Hepatocellular carcinoma (HCC) accounts for most cases of liver cancer worldwide; contraction of hepatitis C (HCV) is considered a major risk factor for liver cancer even when individuals have not developed formal cirrhosis. Global, untargeted metabolic profiling methods were applied to serum samples from patients with either HCV alone or HCC (with underlying HCV). The main objective of the study was to identify metabolite based biomarkers associated with cancer risk, with the long term goal of ultimately improving early detection and prognosis. Serum global metabolite profiles from patients with HCC (n=37) and HCV (n=21) were obtained using high performance liquid chromatography-mass spectrometry (HPLC-MS) methods. The selection of statistically significant metabolites for partial least-squares discriminant analysis (PLS-DA) model creation based on biological and statistical significance was contrasted to that of a traditional approach utilizing p-values alone. A PLS-DA model created using the former approach resulted in a model with 92% sensitivity, 95% specificity, and an AUROC of 0.93. A series of PLS-DA models iteratively utilizing three to seven metabolites that were altered significantly (p<0.05) and sufficiently (FC≤0.7 or FC≥1.3) showed the best performance using p-values alone, the PLS-DA model was capable of generating 73% sensitivity, 95% specificity, and an AUROC of 0.92. Metabolic profiles derived from LC-MS readily distinguish patients with HCC and HCV from those with HCV only. Differences in the metabolic profiles between highrisk individuals and HCC indicate the possibility of identifying the early development of liver cancer in at risk patients. The use of biological significance as a selection process prior to PLSDA modeling may offer improved probabilities for translation of newly discovered biomarkers to clinical application.
Keywords: metabolomics, metabolic profiling, hepatocellular carcinoma, hepatitis C, liver cancer, HPLC-MS, LC-MS, early cancer detection, biomarker
Introduction
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, and is responsible for an estimated 660,000 deaths worldwide. [1] It is believed that HCC develops as a consequence of significant damage to the cellular machinery in the liver after sustained viral infections or cirrhosis. [2–5] Hepatitis C (HCV) is of particular interest since an estimated 130–170 million people are infected with the virus worldwide, and HCV causes approximately 25% of all reported cases of HCC. [6–8] HCV infections can be currently diagnosed with HCV antibody enzyme immunoassays but testing cannot distinguish acute and chronic infections or stratify HCV patients by cancer risk. [9–13] The main objective of this study was to investigate the ability to differentiate patients with HCV who did not develop cancer from those HCV patients who developed HCC. This differentiation is currently difficult due to the profound changes associated with the inflammatory response in both groups. Any improvement in the ability to identify the subset of patients at highest risk for HCC would have a substantial impact on the current treatment paradigm as it would provide earlier interventions, including liver transplant.
The field of metabolomics provides a powerful approach to identify small molecule biomarkers associated with cancer and other diseases. [14–19] By focusing on the concentrations and fluxes of low molecular weight metabolites (< ~1000 m/z) in biofluids, detailed information on biological systems and their concordant correlations across related disease states can be obtained. [20, 21] A significant potential exists to clinically translate a subset of promising biomarker candidates with good diagnostic or prognostic capacity, and therefore metabolomics has become a popular approach in recent years. [22–25] Metabolomics studies based on liquid chromatography mass spectrometry (LC-MS) have been reported for detecting both aberrant lipid metabolism and characteristic metabolites of interest for individuals with HCC. [26] Recent metabolomics studies have focused on serum biomarkers for HCC [28–32] while others have focused on the metabonomic response. [33] However, far fewer studies have focused exclusively on biomarkers that could differentiate at risk individuals in the HCV population from those who have developed HCC. [27] The concept of targeted pathway screening has previously been presented as a viable alternative to the current paradigm focused on statistically driven selection on the subset of LC-MS features which are currently identifiable in non-targeted metabolomics databases. [34] The contextually supervised approach we present is similar to the concept of targeted pathway screening but our selection is driven by historical clinical precedent.
In this study we applied global LC-MS metabolite profiling methods to investigate blood serum metabolites sensitive to patients diagnosed with HCC. We first built a supervised model derived from five putative metabolite markers known to be related to liver diseases and evaluated this model in terms of its ability to differentiate patients with HCV-HCC from those with HCV alone. A second supervised model was then created by adding two high performing metabolite markers from the global analysis to the five and then evaluated for improvements in sensitivity or specificity. The results indicate that an approach involving the monitoring of the seven metabolites in individuals diagnosed with HCV may provide information that would enable health care providers to administer earlier treatments for HCC.
Materials and Methods
Chemicals
HPLC-grade methanol and acetic acid were purchased from Fisher Scientific (Pittsburgh, PA). Deionized water was obtained from an EASYpure II UV water purification system (Barnstead International, Dubuque, IA).
Serum sample collection and storage
Fasting blood samples from HCV patients with histologically proven HCC (n=37) and patients with HCV alone (n=21) were collected at the Indiana University School of Medicine (Indianapolis, IN). All patients had been diagnosed with stage 4 cirrhosis and AFP (alpha fetaprotein) values were available for approximately one third of the patients. None of the HCC patients had undergone chemotherapy prior to blood sample collection. Demographic and clinical characteristics of the patients are presented in Table 1. Each blood sample was allowed to clot for 45 min and then centrifuged at 2,000 rpm for 10 min. The sera were collected, aliquoted in separate vials, frozen, and shipped over dry ice to Purdue University (West Lafayette, IN) where they were stored at −80 °C until use. All samples were collected following the protocol approved by Indiana University School of Medicine and Purdue University Institutional Review Boards. All subjects included in the study were provided written informed consent according to institutional guidelines.
Table 1.
Summary of clinical and demographic characteristics of the patients studied.
| Clinical Diagnosisa | |||
|---|---|---|---|
| HCV | HCC | ||
| N | 21 | 37 | |
| Male | 14 | 29 | |
| Female | 7 | 8 | |
| Age | (years) | ||
| Mean | 50 | 55 | |
| Range | 37–71 | 21–72 | |
| AFPb | Mean (ng/mL) | 106 ± 17 | 90 ± 5 |
All patients were diagnosed with stage 4 cirrhosis.
p-value: 0.20
Sample preparation and data acquisition
Frozen serum samples were thawed and proteins were precipitated by adding 200 µL of cold methanol to 100 µL of serum. The mixture was then centrifuged at 14000 rpm for 10 min after which the supernatant was removed. An additional 200 µL of cold methanol was added to the recovered supernatant and the new mixture was centrifuged at 14000 rpm for 10 min. The supernatant solution obtained after this second protein removal step was then dried under vacuum and the obtained residue was reconstituted in 7.5 µL methanol and vortexed for 5 sec. An additional 7.5 µL 0.4% acetic acid solution in water was added to bring the total volume to 15 µL, which was then vortexed for 5 sec. This final 15 µL solution was again centrifuged at 14000 rpm for 10 min to remove residual particulate matter and the supernatant was transferred to an LC vial for analysis. A separate pooled sample was created by mixing together 3 µL aliquots from 15 serum samples randomly selected from all the sample vials which has previously been shown to strengthen the statistical design of applied mass spectrometry experiments. [35] This pooled sample, referred to as the quality control (QC) matrix sample, was subjected to analysis periodically between every 10 samples. QC sample data also served as technical replicates throughout the data set to assess process reproducibility.
LC-MS analysis was performed using an Agilent LC-QTOF system (Agilent Technologies, Santa Clara, CA) consisting of an Agilent 1200 liquid chromatography system coupled online with an Agilent 6520 time-of-flight mass spectrometer. A 3 µL aliquot of reconstituted sample was injected on to a 2.1×50 mm Agilent Zorbax Extend-C18 1.8 µm particle column with a 2.1×30 mm Agilent Zorbax SB-C8 3.5 µm particle guard column, both of which were heated to 60 °C. Serum metabolites were gradient-eluted at 0.6 mL/min using mobile phase A: 0.2% acetic acid in water and mobile phase B: 0.2% acetic acid in methanol (2% to 98% B in 13 min, 98% B for 6 min). An extended equilibration method was utilized to ensure consistent elution across the columns (2% to 98% B in 10 min and 98% to 2% B for 10 min (performed twice), followed by 2% to 98% B in 10 min and 98% to 2% B for 5 min) and proper reconditioning of the resistive ion capillary necessary for low intersample carryover during consecutive acquisitions. Although 98% B at 0.6 mL/min for 10 min was able to flush the column packing material, the analyte deposition on the rough inner surface of the glass capillary was not trivial at the chosen level of preconcentration and alternating strong and weak solvents was employed to ensure the consistent performance of the ion transmission element. This equilibration strategy was not required at a preconcentration level of 0.5x, 1x, or 2x but was necessary at 6.67x and had an added benefit that it extended the lifetime of the C8 column twofold. Electrospray ionization (ESI) was used in positive mode at 3.5 kV with a concurrent 35 psig emitter nebulization pressure. The interface capillary was maintained at 325 °C, with a curtain gas flow of 8 L/min. Agilent MassHunter Workstation LC-TOF and QTOF Acquisition software (B.02.01) were used for automatic peak detection and mass spectrum deconvolution.
Data analysis
LC-MS data were processed using Agilent’s MassHunter Qualitative Analysis software (version B.03.01) for feature extraction. A list of ion intensities for each detected peak was generated using a retention time (RT) index and m/z data as the identifiers for each ion. Agilent MassHunter Workstation Mass Profiler Professional software (version B.02.00) was then used for compound peak alignment. Multiple feature consolidation into a single metabolite intensity was not performed so that every (m/z,RT) feature remained separate in order to retain metabolites that were more likely to have lower interbatch variance across multiple platforms. Although individual summing of multiple features (fractionated ions, adducts, etc.) of a single metabolite is reproducible in theory, in practice inherent differences in ionization efficiency and transmission often lead to conditions where the sum of all features is not reproducible across a wide set of environmental conditions and sample matrices, and was therefore not used to avoid adding non-trivial variance. The Agilent Formula Database (Agilent, 2010) was used for compound identification by matching the accurate mass spectrum to a database of metabolite compounds and all features without a match were excluded because of the difficulties in identifying unknown compounds. The intrabatch variation of pooled replicates was examined before processing any patients to ensure ion generation and transmission variation remained low. Selected biomarker candidates were then filtered to exclude those that had more than 15% missing intensity values (Rel. Ab. = 1) within either the HCC or HCV groups. Biomarker candidates that were not excluded were then interpolated by having the missing intensity values replaced with the average of the non-trivial abundances within each group. Welch’s t-test analysis of the data was then performed to identify any significant differences in the detected compound intensities between individuals with HCC and HCV. Endogenous metabolites with low p-values (<0.05) and at least moderate mean fold changes (FC≤0.7 or FC≥1.3) for the HCC/HCV ratio were selected as potential biomarker candidates for partial least-squares discriminant analysis (PLS-DA) model generation. Five of the supervised metabolites (vida infra) were verified in separate acquisitions using commercially available standard compounds.
The interpolated MS data of the selected statistically significant metabolites were imported into Matlab (R2008a, Mathworks, Natick, MA) installed with a PLS toolbox (version 4.1, Eigenvector Research, Inc., Wenatchee, WA) for partial least squares discriminant analysis (PLS-DA). The X matrix, consisting of the MS data, was mean centered prior to all statistical analyses. Depending on the group, each subject was assigned a “0” (i.e., HCC) and “1,” (i.e., HCV) to serve as the (one-dimensional) Y matrix. Leave-one-out cross validation (CV) was chosen, and the number of latent variables (LVs) was selected according to the minimum root mean square error of CV procedure, which in this case was three for the first supervised PLS-DA model consisting of five metabolites and four for the second supervised PLS-DA model consisting of seven metabolites. PLS-DA models were created using the first three, four, five, six, and seven metabolites (ranked by p-value) with three LVs and a final model utilizing four LVs for seven metabolites. Predictions were made visually using a Y-predicted scatter plot with a cut-off value chosen to minimize errors in class membership. The R statistical package (version 2.8.0) was used to generate receiver operating characteristics (ROC) curves, calculate and compare sensitivity, specificity and area under the ROC curve (AUROC).
Results
The LC-MS spectrum for each serum sample consisted of more than 4000 features of which nearly 800 peaks were assigned to metabolites using the Agilent database. Analytes with distinct chromatographic signatures that were missing in more than 15% of the samples from either group were omitted from further analysis. The use of this filter and the Agilent chemical library resulted in a total of 512 identified metabolites common to both groups. To identify specific metabolites that best correlated with the different disease states the library-identified metabolites were analyzed using univariate analysis. There were 48 metabolites that varied significantly (p<0.05) between the two groups and 31 of these compounds were endogenous and had at least moderate mean fold changes (FC≤0.7, FC≥1.3) for HCC/HCV as shown in Tables S1 and S2 respectively.
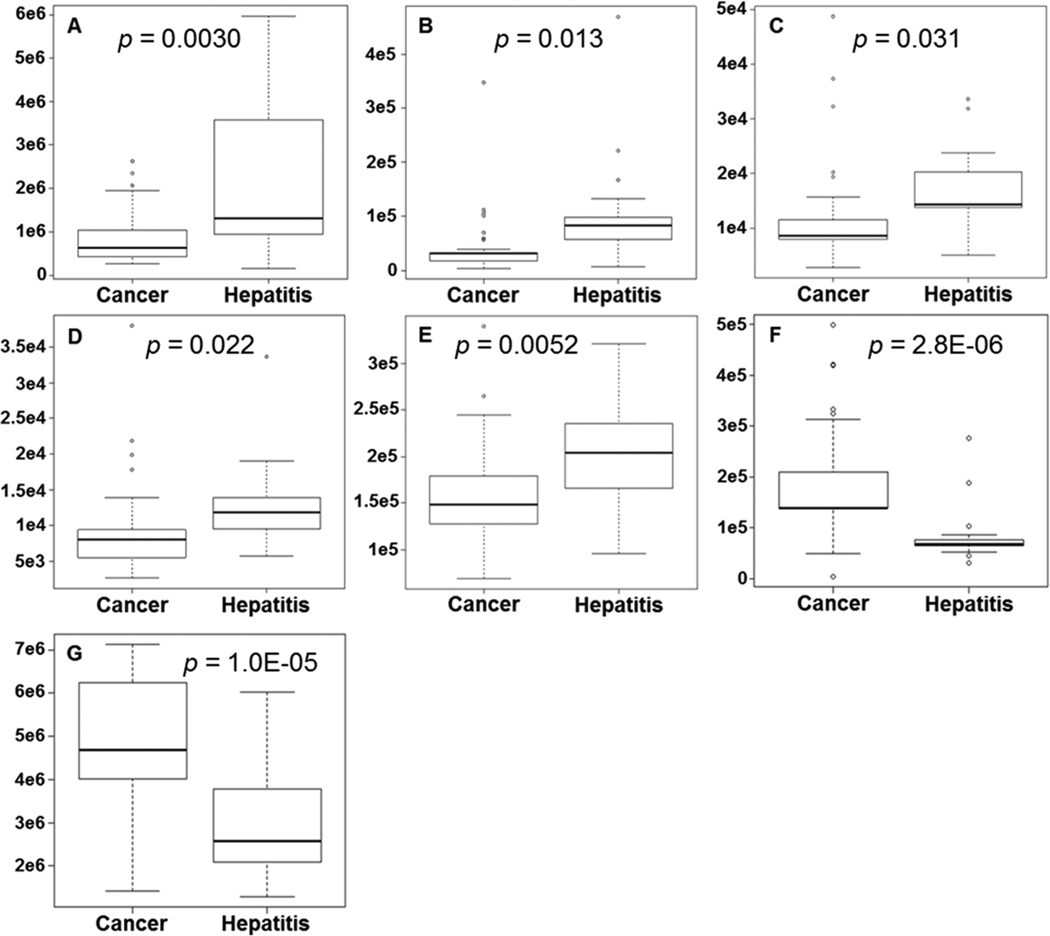
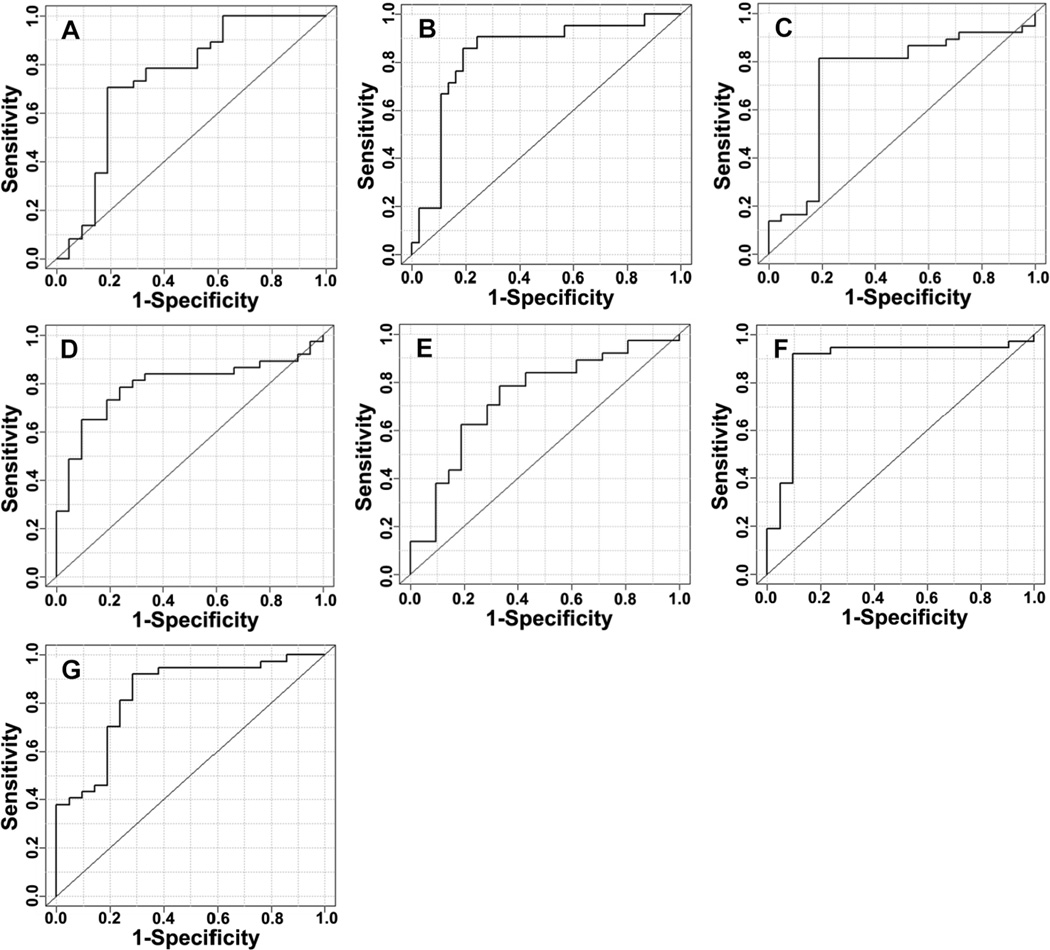
Commercially available metabolites that had: 1) previously been reported as relevant to liver diseases by other researchers and 2) had an observed mass with an accuracy of 1 ppm or less against the Agilent database were chosen as characteristic metabolites input into iterative supervised PLS-DA models. Five metabolites (xanthine, uric acid, cholyglycine, D-leucic acid, and 3-hydroxycapric acid) were calculated to yield the optimum supervised performance and validated. Two additional metabolites (arachidonyl lysolecithin and dioleoylphosphatidylcholine), which performed well in the global analysis (lowest p-values and good fold changes) but which had not been previously associated with liver diseases, were added to the list of five characteristic metabolites. Table 2 summarizes the five supervised and two additional metabolites along with their observed mass values, database delta mass values, retention times, uncorrected p-values, and fold changes. Box and whisker plots of the integral peak areas for all seven metabolites are shown in Figure 1 and illustrate the systematic decrease in observed values for HCC patients compared to individuals with HCV for the five characteristic metabolites and systematic increase in values between HCC-HCV for the two additional metabolites. The ROC curves of the cross-validated predicted class values for all seven metabolites are shown in Figure 2.
Table 2.
Identification information for LC-MS metabolites.
| Compound Name | Calculated (Da) |
Detected (Da) |
Delta (Da) |
RT (min) |
P-Value | Fold Change |
|---|---|---|---|---|---|---|
| *Uric Acid | 168.028 | 168.028 | 0.000 | 0.55 | 0.0030 | 2.6 |
| *Xanthine | 152.033 | 152.033 | 0.000 | 0.65 | 0.0052 | 1.3 |
| *Cholyglycine | 465.309 | 465.309 | 0.000 | 9.90 | 0.013 | 2.5 |
| *D-Leucic acid1 | 132.079 | 132.079 | 0.000 | 0.75 | 0.022 | 1.4 |
| *3-Hydroxycapric acid1 | 188.141 | 188.142 | 0.001 | 8.90 | 0.031 | 1.4 |
| Arachidonyl lysolecithin1 | 543.332 | 543.334 | 0.002 | 11.91 | 2.8E-06 | 0.43 |
| Dioleoylphosphatidylcholine | 785.594 | 785.596 | 0.002 | 14.87 | 1.0E-05 | 0.60 |
Na adduct corrected
Characteristic metabolite
Figure 1.
Comparisons between individuals diagnosed with HCC and HCV for each metabolite using integral peak area abundances. Box-and-whisker plots of (A) uric acid, (B) cholylglycine, (C) 3-hydroxycapric acid, (D) D-leucic acid, (E) xanthine, (F) arachidonyl lysolecithin, (G) dioleoylphosphatidylcholine.
Figure 2.
ROC curves for the cross-validated predicted class values of each metabolite. (A) uric acid (AUROC = 0.74), (B) cholylglycine (AUROC = 0.83), (C) 3-hydroxycapric acid (AUROC = 0.73), (D) D-leucic acid (AUROC = 0.79), (E) xanthine (AUROC = 0.74), (F) arachidonyl lysolecithin (AUROC = 0.88), (G) dioleoylphosphatidylcholine (AUROC = 0.84).
Comparing metabolic profiles of Hepatocellular Carcinoma and Hepatitis C patients
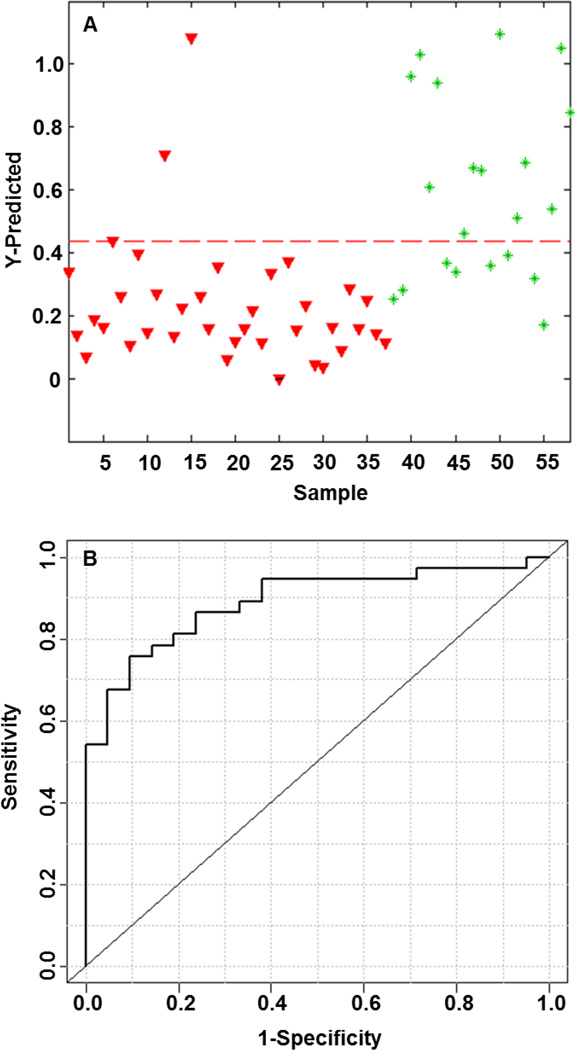
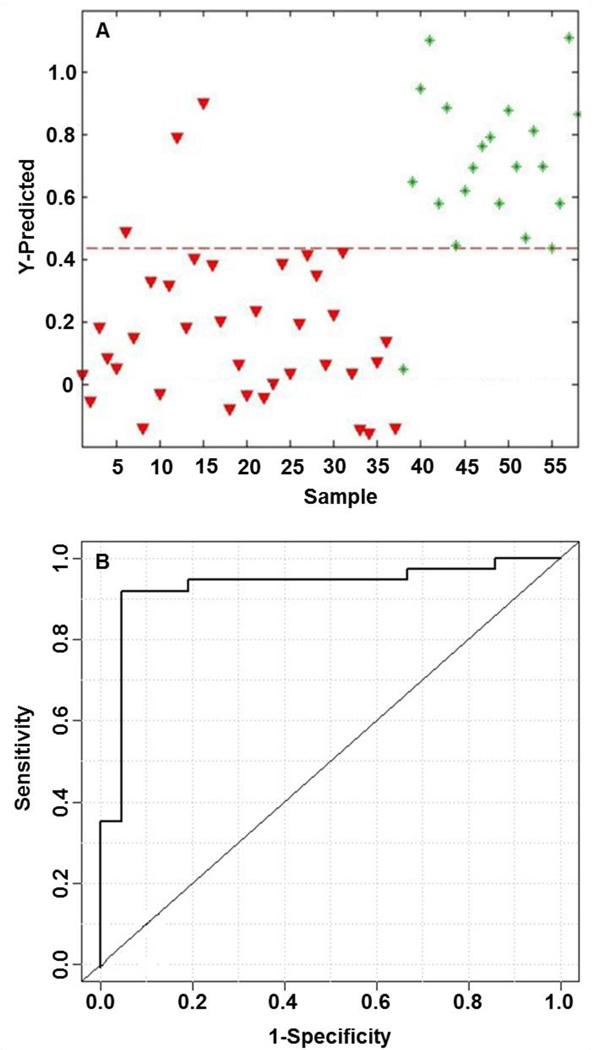
A supervised PLS-DA model was built using the integral peak area intensities for the five biologically significant and characteristic metabolites to test the classification accuracy for the two patient groups as shown in Figure 3. The model derived from five supervised metabolites provided a sensitivity and specificity of 92% and 62%, respectively, with a calculated AUROC of 0.89. Levels of the metabolites between HCC patients compared to individuals with HCV were compared using box-and-whisker plots. All characteristic metabolites showed that levels were elevated in individuals with HCV and were lower for those diagnosed with HCC. The second supervised PLS-DA model shown in Figure 4 was created using the same five metabolites plus the two additional species listed in Table 2. The results of this second model are improved substantially and show a sensitivity and specificity of 92% and 95%, respectively, with a calculated AUROC of 0.93. Table 3 summarizes the PLS-DA model results of cumulative addition of endogenous metabolites by p-value alone. Each row of the table is the cumulative inclusion of the results of a PLS-DA model that includes that metabolite and the metabolites listed above it. The results show a quantitative comparison of the diminishing returns of adding additional metabolites to yield optimum supervised performance. The optimum performance of the PLS-DA model built from metabolites chosen by p-values alone had a sensitivity of 73%, specificity of 95%, and an AUROC of 0.92 for seven metabolites, two of which are in common with the first approach.
Figure 3.
Metabolic profile analysis of five characteristic LC-MS metabolites for PLS-DA model discrimination of individuals with HCC or HCV. (A) PLS-DA predicted model denoted with a dashed red line (sensitivity = 0.92, specificity = 0.62) between individuals with HCC (red triangles) on the left and individuals with HCV (green asterisks) on the right. (B) ROC curve for the cross-validated predicted class values (AUROC = 0.89).
Figure 4.
Metabolic profile analysis of seven LC-MS metabolites for PLS-DA model discrimination of individuals with HCC or HCV. (A) PLS-DA predicted model denoted with a dashed red line (sensitivity = 0.92, specificity = 0.95) between individuals with HCC (red triangles) on the left and individuals with HCV (green asterisks) on the right. (B) ROC curve for the cross-validated predicted class values (AUROC = 0.93).
Table 3.
Iterative PLS-DA modeling cumulative results.
| Metabolite | HCC vs HCV | Cumulative PLS-DA | |||
|---|---|---|---|---|---|
| p-valuea | Sensitivity | Specificity | AUROCb | LVc | |
| Arachidonyl lysolecithin | 2.8E-06 | N/A | N/A | N/A | N/A |
| Dioleoylphosphatidylcholine | 1.0E-05 | N/A | N/A | N/A | N/A |
| 17beta-Estradiol 17-(beta-D-glucuronide) | 1.3E-04 | 0.70 | 0.81 | 0.88 | 3 |
| 3-Deoxyvitamin D3 | 2.2E-04 | 0.70 | 0.81 | 0.88 | 3 |
| Myristoyl L-a-lysophosphatidylcholine | 2.6E-04 | 0.73 | 0.81 | 0.89 | 3 |
| Choline | 6.0E-04 | 0.68 | 0.91 | 0.89 | 3 |
| Uric Acid | 3E-03 | 0.73, 0.73 | 0.95, 0.95 | 0.89, 0.92 | 3,4 |
p-value determined from Welch’s t-test
area under the ROC curve
number of latent variables
Discussion
This study focuses on identifying metabolites for the establishment of improved clinical biomarkers for HCC and insights into the altered metabolism in HCC patients compared to HCV patients. It is well known AFP is not a highly accurate marker, as is clear even from the somewhat limited data available on patients in this study (see Table 1). Thus it is of interest to examine the metabolome for possibly better clinical biomarkers.
In this study, we built supervised PLS-DA statistical models based on two approaches. First we focused on metabolites with low p-values that had previously shown a biological connection with either liver metabolism or cancer. As a result of our global metabolomics analysis, thirty-one endogenous metabolites showed low p-values and significant fold changes in distinguishing these patients. Eleven commercially available metabolites had already been shown to be directly or indirectly associated with liver metabolism, distress, and/or general cancer mechanisms not localized to a specific tissue: 17beta-estradiol 17-(beta-D-glucuronide) (p = 1.3E-04) [57], choline (p = 6.0E-04) [19, 52], uric acid (p = 3.0E-04) [36, 37, 38], xanthine (p = 5.2E-03) [28, 37, 38], 5-hydroxytryptophan (p = 8.5E-03) [59], Indole-3-ethanol (p = 1.2E-02) [57, 58], cholylglycine (p = 1.3E-02) [39, 40], D-leucic acid (p = 2.2E-02) [43, 44], glycolaldehyde (p = 2.3E-02) [53], 1-methylguanine (p = 2.5E-02) [59, 60], and 3-hydroxycapric acid (p = 3.1E-02). [41, 42] We hypothesize that the simplest and most ubiquitous metabolites present in serum will be more robust representatives of any model across technical platforms, operator skill, and biological sets obtained at different clinical locations. The second approach uses the endogenous metabolites with lowest p-values as possible inputs to the statistical model.
The first approach has some significant advantages: 1) metabolite compounds with pre-established connections to specific tissues or diseases have an increased probability of representing biochemical changes associated with the disease of interest and 2) adoption of robust clinical tests often requires implementation on multiple technical platforms for complementary validation in the current regulatory environment. Leveraging pre-existing clinical and academic research to select and exclude metabolites increases the probability that a discovery model would validate and translate to the clinical setting. We note that testing existing biomarkers in this context can improve the statistical power and reduce the false discovery rate, as we are essentially testing known biomarkers in a set of new patient samples.
Two species, 1-methylguanine and 17-beta-estradiol 17-(beta-D-glucuronide), have not been observed with sufficient appearance frequency in LC-QTOF acquisitions across clinical sample sets to confidently choose them as initial contributors to a supervised model as other more ubiquitously observed metabolites in ESI would be more appropriate. Of the nine remaining candidates, choline has clearly been demonstrated to be representative of direct dietary intake of choline containing foods. [19, 52] Although choline was accurately identified using a commercial standard and has a low p-value of 6.0E-04, including it in the model would likely bias the clinical results to a significant degree based on the dietary intake of patients in addition to any effect caused by disease; therefore we chose to exclude this species. Both 5-hydroxytryptophan and indole-3-ethanol were excluded as candidates because the commercial standards eluted at different retention times and produced different MS/MS spectra leading to the conclusion that these species were incorrectly identified.
The remaining six biologically significant metabolites were ordered by increasing p-values and PLS-DA models using three, four, and five metabolites were created. The first five rows of Table 4 summarize the PLS-DA model results of cumulative addition of the endogenous metabolites by p-value. The inclusion of uric acid, xanthine, and cholylglycine resulted in model performance with a sensitivity and specificity of 95% and 62%, respectively, with a calculated AUROC of 0.89. The addition of D-leucic acid did not change the performance of the model. The addition of glycolaldehyde to the first four metabolites did improve the specificity (67%) and AUROC (92%) but decreased the sensitivity (92%). The addition of 3-hydroxycapric acid to the first four metabolites from Table 4 only reduced the sensitivity to 92% with no improvement in specificity (62%) or AUROC (89%). Although glycolaldehyde has been shown to induce apoptosis in breast cancer cells and has an association with general cancer metabolic pathways its small size, ~61 m/z, means that it may be a poor choice to include in a panel of biomarkers to monitor for a clinical test given the popularity of QqQ platforms. [53] It is well known that high abundant clusters present below 80 m/z are often unmanaged and internal or external standards cannot compensate for poor interface conditions resulting in solvent interference. Choline was excluded based on dietary bias and glycolaldehyde is to be excluded based on robust practical ESI considerations required for a regulated clinical environment. Retaining this metabolite would require that targeted QqQ methodology dampen solvent and background ions that may obscure the analyte’s abundance. This would be challenging to implement unless laboratories either had controlled ambient relative humidity and particulate conditions or heightened ESI expertise to modify the interface voltages, emitter distances, ESI-plume sampling angle, chromatographic flow rate, etc. to reduce the appearance of overlapping non-resolved background ions obscuring the signal. In order to reduce anticipated variance of the biomarker test glycolaldehyde was excluded to increase the viability of widespread adoption of the panel.
Table 4.
Iterative PLS-DA modeling cumulative results.
| Metabolite | HCC vs HCV | Cumulative PLS-DA | |||
|---|---|---|---|---|---|
| p-valuea | Sensitivity | Specificity | AUROCb | LVc | |
| Uric acid | 3.0E-03 | N/A | N/A | N/A | N/A |
| Xanthine | 5.2E-03 | N/A | N/A | N/A | N/A |
| Cholylglycine | 1.3E-02 | 0.95 | 0.62 | 0.89 | 3 |
| D-Leucic acid | 2.2E-02 | 0.95 | 0.62 | 0.89 | 3 |
| Glycolaldehyde, 3-Hydroxycapric acid | 2.3 E-02, 3.2E-02 | 0.92, 0.92 | 0.67, 0.62 | 0.92, 0.89 | 3,3 |
| Arachidonyl lysolecithin | 2.8E-06 | 0.89 | 0.91 | 0.91 | 3 |
| Dioleoylphosphatidylcholine | 1.0E-05 | 0.89, 0.92 | 0.95, 0.95 | 0.93, 0.93 | 3,4 |
p-value determined from the Welch’s t-test
area under the ROC curve
number of latent variables
We noted that three of the metabolites had short retention times. To address the possibility that ion suppression might adversely affect the observed biological relationships derived from the void volume elution of three of these metabolites a separate chromatographically optimized targeted acquisition on a QqQ was performed to separate metabolites out of the void volume. [54] The results were unchanged.
The optimum number of metabolites to include is traditionally the minimum required to create a model with the highest sensitivity, selectivity, and AUROC which in this case would be uric acid, xanthine, and cholylglycine. In clinical settings, minimizing monitored metabolites has tangible practical value but in this set we have the opportunity to provide redundancy by including both D-leucic acid and 3-hydroxycapric acid. By retaining the two extra metabolites that have biologically relevant importance but have little impact on the model performance we position the test to monitor additional metabolic pathways. This should increase robustness of the model in clinical patient sets that may have drug, diet, or genetic perturbed purine metabolism which would detrimentally affect a three metabolite model where both uric acid and xanthine could be adversely affected. The first complete set of supervised metabolites selected included uric acid, xanthine, cholylglycine, D-leucic acid, and 3-hydroxycapric acid with a sensitivity and specificity of 92% and 62%, respectively, with a calculated AUROC of 0.89. Alternatively, with a lower threshold value the sensitivity is 78% with a specificity of 86%.
We hypothesized that iteratively increasing the number of biomarkers beyond the biologically relevant five could improve the model discriminating HCV and HCC by utilizing the best candidates from Table 3, arachidonyl lysolecithin (p = 2.8E-06) and dioleoylphosphatidylcholine (p = 1.0E-05). The last two rows of Table 4 summarizes the PLSDA model results of cumulative addition of these metabolites to our existing five supervised metabolites. A hybrid supervised-global approach using seven metabolites yielded a model with a sensitivity and specificity of 92% and 95%, respectively, with a calculated AUROC of 0.93. If D-leucic acid and 3-hydroxycapric acid were excluded leaving only uric acid, xanthine, cholylglycine, arachidonyl lysolecithin, and dioleoylphosphatidylcholine the model would yield a sensitivity and specificity of 89% and 95%, respectively, with a calculated AUROC of 0.93.
Comparison of the individual metabolites between the two patient groups showed clear differences as shown in Figure 1, with all five biologically relevant metabolites being decreased in individuals with HCC and the levels of two additional metabolites increasing in individuals with HCC. The good discrimination between HCC and HCV based on a PLS-DA model that uses five specific liver disease related metabolites detected by LC-MS indicates that this methodology may have good potential for identifying the early onset of liver cancer in HCV patients that might not be detectable by cross sectional imaging or tumor markers (i.e., AFP) currently available. The addition of two other metabolites to the model provides a clinically significant 0.95 specificity.
Two of the five characteristic metabolites, uric acid and xanthine, are linked metabolically and are both involved in the purine degradation pathway. Xanthine is formed from guanine by guanine deaminase, hypoxanthine by xanthine oxidoreductase, or xanthosine by purine nucleoside phosphorylase (PNP). [36] Uric acid is subsequently formed from xanthine by xanthine oxidase which plays an important role in human purine catabolism. [37, 38] Purine metabolism has already been shown to be altered in chemically induced transplantable hematomas in rats and that xanthine oxidoreductase activity decreases in individuals with cancer. [39] The lower levels of both these purine pathway metabolites in HCC fit previous observations of reduced xanthine oxidoreductase activity. The presence of two metabolically linked metabolites that have a strong positive affiliation between the two patient groups supports the hypothesis that it is possible to monitor the perturbed metabolism of liver cancer via LC-MS.
The appearance of cholylglycine and 3-hydroxycapric acid as potential biomarkers is not that unexpected as both metabolites have previously been used to gauge liver dysfunction from serum levels employing non-LC-MS techniques. [40, 41, 42] Disturbance in the metabolic pathways of bile acid derivatives such as sulfolithocholylglycine could contribute to the changes in cholylglycine, while variation in 3-hydroxyacyl-CoA dehydrogenase activity might be responsible for relative differences of 3-hydroxycapric acid we observe in HCC/HCV patients. The general performance of liver fatty acid oxidation can in part be gauged by the output of the 3-hydroxy dicarboxylic acid family of metabolites. [43] In this case, the overproduction of both of these metabolites in the HCV patients may indicate that sustained accelerated metabolism in hepatic cells for long periods of time places individuals at risk for developing HCC. Comparing levels of these metabolites in a group of ‘a priori’ healthy individuals in future studies would be informative. This investigation would address an interesting hypothesis, namely whether HCV ramps up metabolism from the normal state and only returns to more normal values after hepatic cells can no longer sustain that level of output after the onset of HCC.
D-leucic acid has less established evidence in the literature that directly links it with liver dysfunction. Nevertheless, a complex relationship has been shown between mTOR activity and cancer, and that leucine or a leucine derivative may be involved in mTOR activity. [44, 45] Dleucic acid (D-2-Hydroxyisocaproate) also has a corresponding enzyme, L-2-hydroxyisocaproate dehydrogenase. This enzyme is orthologous to the L-lactate dehydrogenase enzyme which converts pyruvate to lactate and back. Pyruvate and lactic acid ratios have already been shown to be a significant determiner of cirrhosis [46] and given our HCC cohort it is unsurprising that an orthologous small molecule, D-leucic acid, is shown to be reflective of this tissue damage. Dleucic acid is also related to more general cellular mechanisms of cancer and the addition of this metabolite when creating a PLS-DA model increased robustness as cirrhosis can be indicative of cancerous tissue, which was a primary driver for its retention.
The phosphatidylcholine and the lysolecithin metabolites, dioleoylphosphatidylcholine and arachidonyl lysolecithin, have not previously been associated with hepatic disorders but have surfaced in recent colorectal, breast, and ovarian cancer research. Altered choline metabolism and higher observed concentrations of phosphatidylcholines in cancer cells is not believed to be a simple indicator of increased phospholipid production but instead results from activation of different enzymes in growth factor-mediated cell signaling pathways. [47–50] Plasma lysophosphatidylcholine levels have been observed to be lower in colorectal cancer patients while lysophosphatidic acid has been shown to be elevated in patients with ovarian cancer. [51, 52] HCC patients have elevated levels of both dioleoylphosphatidylcholine and arachidonyl lysolecithin compared to individuals with HCV.
The PLS-DA model built using the characteristic supervised metabolite integral peak area intensities provided a sensitivity and specificity of 92% and 62%, respectively, with a calculated AUROC of 0.89. Changing the cutoff threshold produces a sensitivity of 78% and a specificity of 86% using the same model. However, the addition of arachidonyl lysolecithin and dioleoylphosphatidylcholine improves the model significantly with a new calculated sensitivity and specificity of 92% and 95%, respectively, and an AUROC of 0.93. This model was very effective in identifying individuals with HCC and had a low false positive rate (1 patient) in the 21 HCV patients under study. Nevertheless, the single misclassification of an individual with HCV has two possible interpretations: Either these markers are not strong candidates to discriminate early onset HCC in individuals with HCV or these markers are so effective at discrimination that they are indicating which patients in the HCV group have developed preclinical or occult HCC. Prospective serum samples from HCV patients under surveillance known to later develop HCC would address the apparent false positive. Those tissue acquisition protocols are ongoing.
There are several limitations of this study, including the limited clinical information available on these 58 patients. We are unable to address possible metabolic differences driven by changes in fibrosis state or cirrhosis. All HCC patients were stage 4 and therefore we are unable to comment on changes as HCC progresses using this cohort. Ideal clinical information that would be optimal to include for a larger validation cohort would include AFP levels on all patients, hepatopathies, METAVIR scores (inflammation and fibrosis stage), viral loading, and a selection of HCC patients undergoing identical drug treatments in order to address the inherent heterogeneities within these diseases. Although improved depth of available clinical information can provide a greater impact as to the particular mechanism of disease progression, absence of this knowledge does not exclude the capacity of a metabolite panel to report changes between the two disease states. As long as high impact clinical confounders (such as chemotherapy) are excluded, biomarkers can discriminate between two states without knowledge of all possible clinical confounders in general cohorts. The clinical cohort we have available is sufficient for discovery and demonstration that a larger more intensive study with a more ideal patient pool would be worth the substantial effort required to organize the necessary collaboration between clinicians and scientists. Future, mechanistic studies, perhaps on animal models would of course add additional insight and possibly lead to improved biomarker panels.
In conclusion, we have shown that the metabolic profiling of serum using LC-MS along with multivariate statistical methods allows a detailed picture of metabolic changes in HCC compared with HCV. Due to the significant alteration of a number of metabolites, HCC patients can be discriminated from HCV patients and vice versa. The high levels observed for the characteristic metabolites derived from the hybrid supervised global approach in the HCV patients compared to HCC patients could be particularly noteworthy if levels in disease free individuals could be established directly and simultaneously compared in a future validation cohort.
Supplementary Material
Highlights.
Metabolite based biomarkers associated with cancer risk were identified.
Patients with HCV can be distinguished from those with HCC.
Biologically relevant metabolite selection is more effective than unbiased global selection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceived and designed the experiments: DR JB MM NS. Performed the experiments: JB EH. Analyzed the data: JB EH DR. Contributed reagents/materials/analysis tools: DR MM NS. Wrote the paper: JB DR MM. Contributed clinical expertise: MM NS.
Potential Conflict of Interest
DR reports holding an executive position and holding equity in Matrix-Bio, Inc.
References
- 1.World Health Organization. Cancer. 2006
- 2.Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, Huang GT, Iloeje UH. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295(1):65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Hepatocellular carcinoma: an epidemiologic view. J Clin Gastroenterol. 2002;35:72–78. doi: 10.1097/00004836-200211002-00002. [DOI] [PubMed] [Google Scholar]
- 4.Hickman IJ, Clouston AD, Macdonald GA, Purdie DM, Prins DM, Ash S, Jonsson JR, Powell EE. Effect of weight reduction on liver histology and biochemistry in patients with chronic hepatitis C. Gut. 2002;51:89–94. doi: 10.1136/gut.51.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlach JT, Diepolder HM, Zachoval R, Gruener NH, Jung MC, Ulsenheimer A, Schraut WW, Schirren CA, Waechtler M, Backmund M, Pape GR. Acute hepatitis C: high rate of both spontaneous and treatment-induced viral clearance. Gastroenterology. 2003;125:80–88. doi: 10.1016/s0016-5085(03)00668-1. [DOI] [PubMed] [Google Scholar]
- 6.Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, Kaslow RA, Margolis HS. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–562. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- 7.Seeff LB. Natural history of chronic hepatitis C . Hepatology. 2002;36:35–46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 8.Alter M. Epidemiology of hepatitis c virus infection. WJG. 2007;13(17):2436–2441. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkins T, Malcolm JK, Raina D, Schade RR. Hepatitis C. diagnosis and treatment. Am Fam Physician. 2010;81(11):1351–1357. [PubMed] [Google Scholar]
- 10.Strader DB, Wright T, Thomas DL, Seef LB. Diagnosis, management, and treatment of hepatitis C . Hepatology. 2004;39:1147–1171. doi: 10.1002/hep.20119. [DOI] [PubMed] [Google Scholar]
- 11.Maylin S, Martinot-Peignoux M, Moucari R, Boyer N, Ripault MP, Cazals-Hatem D, Giuily N, Castelnau C, Cardoso AC, Asselah T, Feray C, Nicolas-Chanoine MH, Bedossa P, Marcellin P. Eradication of hepatitis C virus in patients successfully treated for chronic hepatitis C . Gastroenterology. 2008;135(3):821–829. doi: 10.1053/j.gastro.2008.05.044. [DOI] [PubMed] [Google Scholar]
- 12.Everson GT, Shiffman ML, Hoefs JC, Morgan TR, Sterling RK, Wagner DA, Desanto JL, Curto TM, Wright EC. Quantitative tests of liver function measure hepatic improvement after sustained virological response: results from the HALT-C trial. Aliment Pharmacol Ther. 2008;29(5):589–601. doi: 10.1111/j.1365-2036.2008.03908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward RP, Kugelmas M, Libsch KD. Management of hepatitis C: evaluating suitability for drug therapy. Am Fam Physician. 2004;69(6):1429–1438. [PubMed] [Google Scholar]
- 14.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 15.Tiziani S, Lopes V, Gunther UL. Early stage diagnosis of oral cancer using 1H NMR-based metabolomics. Neoplasia. 2009;11:269–276. doi: 10.1593/neo.81396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431–440. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thysell E, Surowiec I, Hornberg E, Crnalic S, Widmark A, Johansson AI, Stattin P, Bergh A, Moritz T, Antti H, Wikstrom P. Metabolomic characterization of human prostate cancer bone metastases reveals increased levels of cholesterol. PLoS One. 2010;5:e14175. doi: 10.1371/journal.pone.0014175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asiago VM, Alvarado LZ, Shanaiah N, Gowda GA, Owusu-Sarfo K, Ballas RA, Raftery D. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res. 2010;70:8309–8318. doi: 10.1158/0008-5472.CAN-10-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, Britt EB, Fu X, Chung Y, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholson JK, Lindon JC, Holmes E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 21.Gowda GA, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics-based methods for early disease diagnostics. Expert Rev Mol Diagn. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crews B, Wikoff WR, Patti GJ, Woo HK, Kalisiak E, Heideker J, Siuzdak G. Variability analysis of human plasma and cerebral spinal fluid reveals statistical significance of changes in mass spectrometry-based metabolomics data. Anal Chem. 2009;81(20):8538–8544. doi: 10.1021/ac9014947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant Mol Biol. 2002;48(1–2):155–171. [PubMed] [Google Scholar]
- 24.Serkova NJ, Niemann CU. Pattern recognition and biomarker validation using quantitative 1H-NMR-based metabolomics. Expert Rev Mol Diagn. 2006;6(5):717–731. doi: 10.1586/14737159.6.5.717. [DOI] [PubMed] [Google Scholar]
- 25.Davis VW, Bathe OF, Schiller DE, Slupsky CM, Sawyer MB. Metabolomics and surgical oncology: Potential role for small molecule biomarkers. J Surg Oncol. 2010;21831:1–9. doi: 10.1002/jso.21831. 2010. [DOI] [PubMed] [Google Scholar]
- 26.Tan Y, Yin P, Tang L, Xing W, Huang Q, Cao D, Zhao X, Wang W, Lu X, Xu Z, Wang H, Xu G. Metabolomics study of stepwise hepatocarcinogenisis from the model rats to the patients potential biomarkers for small hepatocellular diagnosis. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.010694. M111.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson AD, Maurhofer O, Beyoglu D, Lanz C, Krausz KW, Pabst T, Gonzalez FJ, Dufour JF, Idle JR. Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer Res. 2011;71:6590. doi: 10.1158/0008-5472.CAN-11-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voet D, Voet J, Pratt C. Fundamentals of Biochemistry: Life at the Molecular Level. 2008. p. 840. [Google Scholar]
- 29.Ressom HW, Xiao JF, Tuli L, Varghese RS, Zhou B, Tsai TH, Ranjbar MR, Zhao Y, Wang J, Di Poto C, Cheema AK, Tadesse MG, Goldman R, Shetty K. Utilization of metabolomics to identify serum biomarkers for hepatocellular carcinoma in patients with liver cirrhosis. Anal Chim Acta. 2012;743:90–100. doi: 10.1016/j.aca.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Q, Tan Y, Yin P, Ye G, Gao P, Lu X, Wang H, Xu G. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res. 2013;73(16):4992–5002. doi: 10.1158/0008-5472.CAN-13-0308. [DOI] [PubMed] [Google Scholar]
- 31.Shangfu L, Hongxia L, Yibao J, Shuhai L, Zongwei C, Yuyang J. Metabolomics study of alcohol-induced liver injury and hepatocellular xenografts in mice. J Chrom B. 2011;879(24):2369–2375. doi: 10.1016/j.jchromb.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 32.Becker S, Kortz L, Helmschrodt C, Thiery J, Ceglarek U. LC-MS-based metabolomics in the clinical laboratory. J Chrom B. 2012;883:68–75. doi: 10.1016/j.jchromb.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 33.Chen F, Xue J, Zhou L, Wu S, Chen Z. Identification of serum biomarkers of hepatocarcinoma through liquid chromatography/mass spectrometry-based metabonomic method. Anal Bioanal Chem. 2011;401(6):1899–1904. doi: 10.1007/s00216-011-5245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christians U, Klawitter J, Hornberger A, Klawitter J. How Unbiased is Non-Targeted Metabolomics and is Targeted Pathway Screening the Solution? Current Pharmaceutical Biotechnology. 2011;12(7):1053–1066. doi: 10.2174/138920111795909078. [DOI] [PubMed] [Google Scholar]
- 35.Dunn WB, Wilson ID, Nicholls AW, Broadhurst D. The Importance of Experimental Design in Large-Scale and MS-Driven Untargeted Metabolonic Studies of Humans. Bioanalysis. 2012;4(18):2249–2264. doi: 10.4155/bio.12.204. [DOI] [PubMed] [Google Scholar]
- 36.Hill R. Molybdenum-containing hydroxylases. Arch Biochem Biophys. 2005;433(1):107–116. doi: 10.1016/j.abb.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Harrison R. Structure and function of xanthine oxidoreductase: where are we now. Free Radic Biol Med. 2002;33(6):774–797. doi: 10.1016/s0891-5849(02)00956-5. [DOI] [PubMed] [Google Scholar]
- 38.Weber G. Enzymes of purine metabolism in cancer. Clin Biochem. 1983;16(1):57–63. doi: 10.1016/s0009-9120(83)94432-6. [DOI] [PubMed] [Google Scholar]
- 39.Tanggo Y. Clinical usefulness of serum cholylglycine determination in various liver diseases. Gastroentrol Jpn. 1982;17(5):447–452. doi: 10.1007/BF02774722. [DOI] [PubMed] [Google Scholar]
- 40.Gilmore T, Thompson RP. Kinetics of 14C-glychocholic acid clearance in normal man and in patients with liver disease. Gut. 1978;19(12):1110–1115. doi: 10.1136/gut.19.12.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niwa T, Yamada K, Ohki T, Furukawa H. 3-Hydroxyhexanoic acid: an abnormal metabolite in urine and serum of diabetic ketoacidotic patients. J Chrom B Biom Sci Appl. 1985;333:1–7. doi: 10.1016/0378-4347(85)80001-3. [DOI] [PubMed] [Google Scholar]
- 42.Tserng K, Jin SJ. Metabolic origin of urinary 3-hydroxydicarboxylic acids. Biochem. 1991;30(9):2508–2514. doi: 10.1021/bi00223a030. [DOI] [PubMed] [Google Scholar]
- 43.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 44.Hidayat S, Yoshino K, Tokunaga C, Hara K, Matsuo M, Yonezawa K. Inhibition of amino acid m-TOR signaling by a leucine derivative induces G1 arrest in Jurkat cells. Biochem Biophys Res Commun. 2003;301:417–423. doi: 10.1016/s0006-291x(02)03052-8. [DOI] [PubMed] [Google Scholar]
- 45.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–84. [PubMed] [Google Scholar]
- 46.Mizobuchi N, Kuwao F, Takeda I, Takemura T, Morita S, Horimi T, Takahashi I. Changes in ketone body ratio and levels of pyruvate and lactate in arterial blood of patients with hepatocellular carcinoma after transcatheter arterial embolization. Rinsho Byori. 1990;38(7):825–829. [PubMed] [Google Scholar]
- 47.Pelech SL, Vance DE. Signal transduction via phosphatidylcholine cycles. Trends Biochem Sci. 1989;14:28–30. [Google Scholar]
- 48.Glunde K, Serkova NJ. Therapeutic targets and biomarkers identified in cancer choline phospholipid metabolism. Pharmacogenomics. 2006;7:1109–1123. doi: 10.2217/14622416.7.7.1109. [DOI] [PubMed] [Google Scholar]
- 49.Podo F, Francesco S, Iorio E, Canese R, Carpinelli G, Fausto A, Canevari S. Inhibition of amino acid m-TOR signaling by a leucine derivative induces G1 arrest in Jurkat cells. Biochem Biophys Res Commun. 2007;301:417–423. doi: 10.1016/s0006-291x(02)03052-8. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Z, Xiao Y, Elson P, Tan H, Plummer SJ, Berk M, Aung PP, Lavery IC, Achkar JP, Li L, Casey G, Xu Y. Plasma lysophosphatidylcholine levels: potential biomarkers for colorectal cancer. J Clinical Onclogy. 2007;25(19):2696–2701. doi: 10.1200/JCO.2006.08.5571. [DOI] [PubMed] [Google Scholar]
- 51.Kim K, Sengupta S, Berk M, Kwak Y, Escobar PF, Belinson J, Mok SC, Xu Y. Hypoxia enhances lysophophatidic acid responsiveness in ovarian cancer cells and lysophosphatidic acid induces ovarian tumor metastasis in vivo . Cancer Res. 2006;66(16):7983–7990. doi: 10.1158/0008-5472.CAN-05-4381. [DOI] [PubMed] [Google Scholar]
- 52.Veenema K, Solis C, Li R, Wang W, Maletz CV, Abratte CM, Caudill MA. Adequate intake levels of choline are sufficient for preventing elevations in serum markers of liver dysfunction in Mexican American men but are not optimal for minimizing plasma total homocysteine increases after a methionine load. Am J Clin Nutr. 2008;88(3):685–692. doi: 10.1093/ajcn/88.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Al-Maghrebi MA, Al-Mulla F, Benov LT. Glycolaldehyde induces apoptosis in a human breast cancer cell line. Arch Biochem Biophys. 2003;417(1):123–127. doi: 10.1016/s0003-9861(03)00334-5. [DOI] [PubMed] [Google Scholar]
- 54.Baniasadi H, Nagana Gowda GA, Gu H, Zeng A, Zhuang S, Skill N, Maluccio M, Raftery D. Targeted Metabolic Profiling of Hepatocellular Carcinoma and Hepatitis C using LC-MS/MS. Electrophoresis. 2013;34:2910–2917. doi: 10.1002/elps.201300029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen ZS, Robey RW, Belinsky MG, Shchaveleva I, Ren XQ, Sugimoto Y, Ross DD, Bates SE, Kruh GD. Transport of methotrexate, methotrexate polyglutamates, and 17beta-estradiol 17-(beta-D-glucuronide) by ABCG2: effects of acquired mutations at R482 on methotrexate transport. Cancer Res. 2003;63(14):4048–4054. [PubMed] [Google Scholar]
- 56.Bouchard S, Bousquet C, Roberge AG. Characteristics of dihydroxyphenylalanine/5-hydroxytryptophan decarboxylase activity in brain and liver of cat. J Neurochem. 1981;37(3):781–787. doi: 10.1111/j.1471-4159.1982.tb12555.x. [DOI] [PubMed] [Google Scholar]
- 57.Kosalec I, Ramic S, Jelic D, Antolovic R, Pepeljnjak S, Kopjar N. Assessment of tryptophol genotoxicity in four cell lines in vitro: a pilot study with alkaline comet assay. Archives of Industrial Hygiene and Toxicology. 2011;62(1):41–49. doi: 10.2478/10004-1254-62-2011-2090. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka K, McConnell B, Niemczura WP, Mower HF. Characterization and mutagenicity of 1-nitrosotryptophol and 6-nitrotryptophol possible genotoxic substances associated with smoking and alcohol consumption. Cancer Letters. 1989;44(2):109–116. doi: 10.1016/0304-3835(89)90005-0. [DOI] [PubMed] [Google Scholar]
- 59.McDonald JJ, Stohrer G, Brown GB. Conversion of the Oncogenic 1-Methylguanine 3-Oxide to 3-Hydroxy-1-methylxanthine. Cancer Research. 1976;36:3604–3607. [PubMed] [Google Scholar]
- 60.Kerr J. Interaction of normal and tumor transfer RNA methyltransferases with ethionine-induced methyl-deficient rat liver transfer RNA. Cancer Letters. 1975;44(2):109–116. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.