Abstract
The ATP-binding cassette (ABC) proteins or transporters constitute a large protein family in plants and are involved in many different cellular functions and processes, including solute transportation, channel regulation and molecular switches, etc. Through transcriptome sequencing, a transcriptome-wide survey and expression analysis of the ABC protein genes were carried out using the laticiferous latex from Hevea brasiliensis (rubber tree). A total of 46 putative ABC family proteins were identified in the H. brasiliensis latex. These consisted of 12 ‘full-size’, 21 ‘half-size’ and 13 other putative ABC proteins, and all of them showed strong conservation with their Arabidopsis thaliana counterparts. This study indicated that all eight plant ABC protein paralog subfamilies were identified in the H. brasiliensis latex, of which ABCB, ABCG and ABCI were the most abundant. Real-time quantitative reverse transcription-polymerase chain reaction assays demonstrated that gene expression of several latex ABC proteins was regulated by ethylene, jasmonic acid or bark tapping (a wound stress) stimulation, and that HbABCB15, HbABCB19, HbABCD1 and HbABCG21 responded most significantly of all to the abiotic stresses. The identification and expression analysis of the latex ABC family proteins could facilitate further investigation into their physiological involvement in latex metabolism and rubber biosynthesis by H. brasiliensis.
Introduction
The ATP-binding cassette (ABC) protein family, especially the intrinsic membrane subfamilies, mediates a large number of fundamental cellular functions and processes that utilize ATP hydrolysis to energize the transport of solutes across membranes. These include substrate translocation, lipid trafficking, protein targeting and phytohormone transport [1–5], etc. The plant ABC proteins constitute one of the largest and most diverse protein families, and can be organized phylogenetically into eight clusters, namely the ABCA-ABCI subfamilies (ABCH is not found in plants) [6]. Two major types of ABC proteins can be characterized in plants. One is the membrane-integrated ABC protein, including the ABCA–ABCD and ABCG subfamilies, which contain both a nucleotide binding domain (NBD) and a trans-membrane domain (TMD), and the other is the soluble ABC proteins, which mainly cluster in the ABCE, ABCF and ABCI subfamilies and only contain a NBD domain [7, 8]. Whole genome sequencing has led to complete inventories of plant ABC transporters for model plants, such as Arabidopsis thaliana [9], rice (Oryza sativa L.) [10] and other plant species [11, 12], whereas little is known about ABC proteins in Hevea brasiliensis (Willd. ex Adr. de Juss.) Muell.-Arg. (para rubber tree), which is the most important natural rubber (NR)-producing plant in the world.
NR is a long chain cis-1,4-polyisoprene polymer and is a strategically indispensable industrial raw material for more than 40,000 products. H. brasiliensis, a member of the Euphorbiaceae, is almost the only species that can produce commercially viable quantities of high-quality NR [13]. NR synthesis is a typical plant secondary metabolism process that occurs in the latex vessels of rubber trees via glycolysis, followed by the mevalonate or 2-C-methyl-D-erythritol 4-phosphate pathways, which provide the direct precursor of isopentenyl diphosphate (IPP) [14]. On the rubber particle, NR is formed through sequential condensation of hundreds of thousands of IPP units and is finally compartmentalised within a special organelle that is suspended in the latex of the laticiferous cells of rubber trees [15–17]. The increasing demand for NR by the world economy has prompted investigations into the underlying molecular mechanisms behind latex metabolism and NR biosynthesis [18–20], which may lead to an improvement in the latex yield of rubber trees.
One of the latex yield-limiting factors is the regeneration potential of the latex in the H. brasiliensis laticifers between two consecutive tappings. Latex regeneration consists of several biological and molecular events, including sucrose importation into laticifers from neighboring cells [21, 22], ontogenesis, the development of rubber particles derived from endoplasmic reticulum [23] and the assembly and breakdown of complex lipids in the latex [24], etc. All the biological processes involve substrate transportation and trafficking or the turnover of intracellular components. Much less is known about the related process of solute transportation that occurs within the highly specialized and mature laticifer system, which has no plasmodesmata and is therefore apoplastically isolated from the adjacent cells in the inner bark of H. brasiliensis [25].
A hallmark biological feature of rubber trees is their reticulate network of laticiferous cells, within which NR biosynthesis is carried out. The latex collected through bark tapping is actually the pure cytoplasm of the laticiferous vessels [26]. Therefore, the latex, together with the laticiferous cells of H. brasiliensis, is a good experimental system for biochemical and molecular investigations into a particular type of cell that has specially evolved for NR biosynthesis. Characterization of the ABC transporters in the laticifers would help reveal the physiological and molecular processes underlying latex metabolism and NR biosynthesis in rubber trees. Recently, 270 ABC transporter gene contigs were identified from the bark transcriptome of rubber trees [27], but no more information is available about the ABC transporters in rubber trees. In this study, the global inventory of the ABC proteins in the laticifers of H. brasiliensis was first identified by sequencing the latex transcriptome and then the expression patterns of several ABC proteins genes were analyzed using real-time quantitative reverse transcript-polymerase chain reaction (RT-qPCR). This research provides valuable information for further investigations into the physiological roles of the ABC transporters during latex metabolism and NR biosynthesis in rubber trees.
Materials and Methods
Latex Collection and Total Latex RNA Preparation for Transcriptome Sequencing
Rubber trees (Clone Reyan 7–33–97) were planted on the experimental farm at the Chinese Academy of Tropical Agricultural Sciences in Hainan, P.R. China. Trees that had homogeneous stem girths and had been tapped for 2 years using an S/2 d/3 system (tapped every 3 days in a half spiral), were selected. The fresh latex sample was collected in a thermo bottle containing liquid nitrogen for 10 min and then immediately stored at—80°C. Total RNAs were isolated from the collected latex using our previously described method [28] and treated with DNase I (Invitrogen, Carlsbad, CA, USA) to remove genomic DNA. The quality and integrity of the RNAs were evaluated using Nanodrop 2000 (Thermo Scientific, Wilmington, DE, USA) and Bioanalyzer Chip RNA 7500 series II (Agilent, Santa Clara, USA) instruments.
Construction of the cDNA Library and Transcriptome Sequencing
Sequencing of the latex transcriptome was performed using an Illumina mRNA-Seq Sample Prep kit (Illumina Inc., San Diego, CA), according to the manufacturer’s instructions. Briefly, poly (A) mRNA was isolated from total RNA using oligo (dT) magnetic beads and fragmented into small pieces using divalent cations at an elevated temperature. The cleaved mRNA fragments were used for the first strand cDNA synthesis, together with reverse transcriptase and random hexamer-primers. The second strand cDNAs were synthesized using RNaseH and DNA polymerase I. The double-stranded cDNA fragments were purified with a QiaQuick PCR extraction kit and eluted with EB buffer for end repair and the ligation of sequencing adapters. The products were purified by agarose gel electrophoresis and the fragments that were around 200 bp in length were selected as templates and enriched by linear PCR amplification, which created the final cDNA library for transcriptome paired end sequencing via the Illumina HiSeq-2000 sequencing platform. Image analysis, base calling and quality value calculations were conducted using the Illumina/Solexa data processing pipeline.
De novo Assembly of the Latex Transcriptome
Before assembly, the raw reads that contained adaptors or unknown nucleotides larger than 5% with quality scores of Q ≤ 10 were trimmed using software found in Filter-fq (BGI, Shenzhen, China). The clean reads, which were randomly split into 25 bp K-mers for assembly into contigs using the de Bruijn graph, were then used for transcriptome de novo assembly using the Trinity software short reads assembly program (Release-20130225) [29]. The resultant contigs were further joined into scaffolds using the paired-end reads and gap filling was carried out in order to complete the scaffolds. To obtain non-redundant unigenes, the scaffolds were assembled together and clustered using the Gene Indices Clustering Tools (TGICL, version 2.1) [30] and Phrap software (Release-23.0) [31], which finally produced the assembled consensus sequences of the H. brasiliensis latex transcriptome. The transcriptome data have been submitted to the SRA database with an accession number of SRR1648124.
Functional Annotation and Classification of the Latex Transcriptome
All the assembled unigenes were used in a BLAST search and for annotation against the NCBI nr database with an E-value cut-off of 10-5. Functional categorization by Gene Ontology (GO; http://www.geneontology.org) was performed using Blast2GO software (http://www.blast2go.de/) with an E-value threshold of 10-5. Analyses of the Cluster of Orthologous Groups (COG) identifications and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations were conducted using Blastall software (http://www.ncbi.nlm.nih.gov/staff/tao/URLAPI/blastall/) against the COG database in the NCBI (COG; ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/) and the KEGG database using the BLASX algorithm with an E-value threshold of 10-5.
Identification of ABC Protein Genes in the Latex Transcriptome
A reciprocal best hits (two-direction) blast search was performed to identify the latex ABC protein genes. First, A. thaliana ABC protein sequences were retrieved from the A. thaliana Information Resource (TAIR) database. Putative ABC proteins obtained from the H. brasiliensis latex were searched by performing a tBLASTN analysis (http://www.ncbi.nlm.nih.gov/blast) against the generated latex transcriptome data using A. thaliana ABC protein sequences as queries. Second, the best hits were blasted again against the entire A. thaliana TAIR10 transcripts (http://www.arabidopsis.org/Blast/index.jsp). Non-complete latex ABC protein genes were manually assembled into complete unigenes using the CAP3 program (http://doua.prabi.fr/software/cap3). The open reading frames (ORFs) of the latex ABC protein unigenes were verified by searching the H. brasiliensis genome DNA sequences that have just been sequenced and assembled by the Rubber Research Institute of China using the Vector NTI Advance program (version 11.5.2). The polypeptide sequences corresponding to the latex ABC proteins were analyzed for the presence of ABC conserved domains using the Conserved Domain Database (CDD) at NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and the Pfam27.0 web server (http://pfam.xfam.org/) [32, 33]. The cDNA sequences of all the latex ABC protein genes have been deposited in the NCBI database with accession numbers ranging from KM035282–035324.
Sequence Analysis and Phylogenetics
The deduced amino acid sequences of the latex ABC proteins, together with those of the A. thaliana ABC proteins, were aligned by a multiple sequence comparison using the log-expectation (MUSCLE) alignment tool (http://www.ebi.ac.uk/Tools/msa/muscle) with the default program options [34]. Then they were subjected to phylogenetic analysis using the neighbor-joining method and 1000 bootstrap replicates were employed in each analysis to maximize the statistical significance [35]. The phylogenetic trees were constructed and visualized by MEGA5.05 software [36].
Treatment of Rubber Trees Used for Gene Expression Analysis
Field experiments were performed using mature, 7-year-old virgin rubber trees (Clone Reyan 7–33–97) that had never been tapped. Stimulation assays with exogenous methyl jasmonate (Me-JA) or Ethrel (an ethylene releaser) were carried out according to the method described in a previous paper [28]. Briefly, 0.3% (w/w) Me-JA (Sigma–Aldrich, USA) in lanoline or 0.5% (w/w) Ethrel (Sigma–Aldrich, USA) in water was applied to the bark below the half spiral of the tapping cut. Lanoline or water was used as the mock stimulation for the control samples. The trunks of the treated trees were wrapped with black plastic film around the tapping cut and stimulated for 0, 0.5, 1.5, 4.0, 8.0 or 24.0 h, after which they were tapped for latex early in the morning on the same day. For the tapping assays, the rubber trees were sequentially tapped seven times using an S/2 d/3 system and fresh latex was collected at each tapping time for analysis. Each sample included three independent biological replicates, and the latex collected from six rubber trees was pooled to make one biological replicate for each sample.
Transcript Abundance Analysis of Latex ABC Protein Genes by RT-qPCR
Total RNA from H. brasiliensis latex was isolated using an RNeasy Plant Mini-Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Reverse transcription was performed with SuperScript III reverse transcriptase (Invitrogen, CA), followed by incubation with RNase H (Invitrogen, CA). RT-qPCR analysis was performed using the method described by Duan et al. [37]. Primers were designed for the selected genes by DNAMAN 7.02 software with a PCR product of ~200 bp. The forward and reverse primer sequences used to detect each mRNA and their efficiencies are shown in S1 Table. The cDNAs were synthesized by reverse transcriptase and quantitative gene expression analysis was carried out by RT-qPCR using a LightCycler 2.0 (Roche, Basel, Switzerland) and the following parameters: 30 s at 94°C for denaturation, followed by 45 cycles of 94°C for 5 s, 60°C for 20 s and 72°C for 20 s. Each RT-qPCR reaction was replicated three times. For normalization purposes, the H. brasiliensis 18S rRNA gene was taken as the internal reference for all RT-qPCR analyses.
Results/Discussion
Sequencing and de novo Assembly of the H. brasiliensis Latex Transcriptome
The H. brasiliensis latex transcriptome was generated using the latex collected from rubber trees that had been regularly tapped for 2 years in an S/2 d/3 system. These rubber trees had a sustainable and higher latex yield than newly tapped trees due to their regular bark tapping, which represents a mechanical stress for the rubber tree. Latex production is a natural response to mechanical wounding and studies have reported that it activates latex metabolism, which then leads to an increase in latex production [38]. The complicated biological process leading to latex production involves several biochemical reactions that occur in the laticifers and many genes or proteins may contribute to the final results. The complex biological mechanism underlying latex metabolism requires a global transcriptic survey to improve understanding of the process. Therefore, the latex transcriptome was first sequenced using the Illumina HiSeq-2000 platform. Approximately 6.17 Gb of total nucleotides were obtained, and the 6.85 Mb clean reads were assembled into 182,956 contigs and 60,909 unigenes, respectively.
Identification of ABC Proteins in the H. brasiliensis Latex Transcriptome
A reciprocal best hits (two-direction) blast search was performed in order to identify the orthologs of the ABC protein genes in the H. brasiliensis latex transcriptome. The results demonstrated that the identified latex ABC protein genes from the two blasts closely matched each other (data available in S2 Table). A total of 46 putative latex ABC protein genes were finally identified via the reciprocal search approach. The details can be found in Table 1, but only the results obtained by using the A. thaliana ABC protein sequences as queries are shown. The complete ORFs of the 46 latex ABC proteins were further verified by searching the H. brasiliensis genome DNA sequences that have recently been completed by the Rubber Research Institute of China. The genome DNA sequences for each latex ABC protein gene are shown S3 Table, which also shows the corresponding exons and introns for each latex ABC protein gene. Aligning all the transcripts of the 46 latex ABC proteins to the H. brasiliensis genome did not reveal any alternative transcripts from the same locus, since each transcript of the 46 ABC proteins was definitively located on different scaffolds of the H. brasiliensis genome, as shown in S3 Table.
Table 1. Detailed inventory of the 46 ABC transporter genes identified in the H. brasiliensis latex.
| ABC subfamily | Gene name | NCBI accession no. | Length (aa) | Topology | A. thaliana homolog | Common name | Identity (%) | |
|---|---|---|---|---|---|---|---|---|
| Subfamily A (ABCA*) | AOH; ABCA | HbABCA1 | KM035282 | 1883 | (TMD-NBD)2 | At2g41700 | AtABCA1 | 74 |
| ATH; ABCA | HbABCA2 | KM035283 | 970 | TMD-NBD | At3g47730 | AtABCA2 | 71 | |
| HbABCA7 | KM035284 | 939 | TMD-NBD | At3g47780 | AtABCA7 | 67 | ||
| Subfamily B (ABCB) | MDR; DPL(PGP) | HbABCB1 | KM035285 | 1363 | (TMD-NBD)2 | At2g36910 | AtABCB1 | 88 |
| HbABCB11 | KM035286 | 1283 | (TMD-NBD)2 | At1g02520 | AtABCB11 | 74 | ||
| HbABCB13 | KM035287 | 1135 | (TMD-NBD)2 | At1g27940 | AtABCB13 | 71 | ||
| HbABCB15 | HQ917533 | 1250 | (TMD-NBD)2 | At3g28345 | AtABCB15 | 77 | ||
| HbABCB19 | KM035288 | 1259 | (TMD-NBD)2 | At3g28860 | AtABCB19 | 89 | ||
| HbABCB20 | KM035289 | 1404 | (TMD-NBD)2 | At3g55320 | AtABCB20 | 86 | ||
| ATM; DPL(HMT) | HbABCB25 | KM035290 | 739 | TMD-NBD | At5g58270 | AtABCB25 | 83 | |
| TAP; DPL(TAP) | HbABCB26 | KM035291 | 702 | TMD-NBD | At1g70610 | AtABCB26 | 75 | |
| HbABCB28 | KM035292 | 660 | TMD-NBD | At4g25450 | AtABCB28 | 70 | ||
| DPL(LLP) | HbABCB29 | KM035293 | 650 | TMD-NBD | At5g03910 | AtABCB29 | 65 | |
| Subfamily C (ABCC) | MRP; OAD(MRP) | HbABCC2 | KM035294 | 1624 | (TMD-NBD)2 | At2g34660 | AtABCC2 | 79 |
| HbABCC5 | KM035295 | 1499 | (TMD-NBD)2 | At1g04120 | AtABCC5 | 81 | ||
| HbABCC13 | KM035296 | 1480 | (TMD-NBD)2 | At2g07680 | AtABCC13 | 66 | ||
| Subfamily D (ABCD) | PMP; FAE | HbABCD1 | KF701641 | 1337 | (TMD-NBD)2 | At4g39850 | AtABCD1 | 78 |
| HbABCD2 | KM035297 | 746 | TMD-NBD | At1g54350 | AtABCD2 | 69 | ||
| Subfamily E (ABCE) | RLI | HbABCE2 | KM035298 | 605 | NBD-NBD | At4g19210 | AtABCE2 | 93 |
| Subfamily F (ABCF) | GCN; ART(REG) | HbABCF1 | JX109943 | 605 | NBD-NBD | At5g60790 | AtABCF1 | 85 |
| HbABCF3 | KM035299 | 715 | NBD-NBD | At1g64550 | AtABCF3 | 82 | ||
| HbABCF4 | KM035300 | 726 | NBD-NBD | At3g54540 | AtABCF4 | 79 | ||
| HbABCF5 | KM035301 | 709 | NBD-NBD | At5g64840 | AtABCF5 | 79 | ||
| Subfamily G (ABCG) | WBC; EPD(WHITE) | HbABCG3 | KM035302 | 723 | NBD-TMD | At2g28070 | AtABCG3 | 81 |
| HbABCG5 | KM035303 | 626 | NBD-TMD | At2g13610 | AtABCG5 | 73 | ||
| HbABCG7 | KM035304 | 720 | NBD-TMD | At2g01320 | AtABCG7 | 78 | ||
| HbABCG11 | KM035305 | 649 | NBD-TMD | At1g17840 | AtABCG11 | 53 | ||
| HbABCG15 | KM035306 | 714 | NBD-TMD | At3g21090 | AtABCG15 | 68 | ||
| HbABCG20 | KM035307 | 715 | NBD-TMD | At3g53510 | AtABCG20 | 75 | ||
| HbABCG21 | KM035308 | 680 | NBD-TMD | At3g25620 | AtABCG21 | 68 | ||
| HbABCG22 | KM035309 | 738 | NBD-TMD | At5g06530 | AtABCG22 | 78 | ||
| HbABCG28 | KM035310 | 980 | NBD-TMD | At5g60740 | AtABCG28 | 68 | ||
| PDR; EPD(PDR) | HbABCG40 | KM035311 | 1438 | (NBD-TMD)2 | At1g15520 | AtABCG40 | 72 | |
| Subfamily I (ABCI) | NAP; CCM | HbABCI1 | KM035312 | 229 | NBD | At1g63270 | AtABCI1 | 86 |
| NAP; ISB | HbABCI6 | KM035313 | 335 | NBD | At3g10670 | AtABCI6 | 82 | |
| HbABCI7 | KM035314 | 492 | CYT | At1g32500 | AtABCI7 | 62 | ||
| HbABCI8 | KM035315 | 553 | CYT | At4g04770 | AtABCI8 | 81 | ||
| NAP; CBY(Y179) | HbABCI10 | KM035316 | 261 | NBD | At4g33460 | AtABCI10 | 71 | |
| HbABCI11 | KM035317 | 277 | NBD | At5g14100 | AtABCI11 | 72 | ||
| NAP; MKL; TGD | HbABCI13 | KM035318 | 343 | NBD | At1g65410 | AtABCI13 | 80 | |
| HbABCI14 | KM035319 | 325 | TMD | At1g19800 | AtABCI14 | 77 | ||
| HbABCI15 | KM035320 | 385 | SSA | At3g20320 | AtABCI15 | 73 | ||
| NAP; NO | HbABCI17 | KM035321 | 283 | NBD | At1g67940 | AtABCI17 | 73 | |
| HbABCI18 | KM035322 | 280 | NBD | At1g03900 | AtABCI18 | 76 | ||
| NAP; NO(ADT) | HbABCI19 | KM035323 | 300 | NBD | At1g03905 | AtABCI19 | 80 | |
| HbABCI20 | KM035324 | 329 | NBD | At5g02270 | AtABCI20 | 87 | ||
*ABCA: Similar to human abca1 protein; NBD: Nucleotide binding domain (ATP binding cassette domain); TMD: transmembrane domain; SSA: substrate binding protein; CYT: conserved soluble protein that interacts with ABC domain; DPL: Drug, peptide and lipid exporters; PGP: Similar to p-glycoprotein; LLP: Lipid A-like exporters, putative; OAD: Organic anion and drug exporters; HMT: Similar to yeast heavy metal transporters; TAP: Similar to the human transporters associated with antigen presentation; WHITE: Similar to Drosophila white protein; CCM: cytochrome C biogenesis family; ISB: iron sulfur center biogenesis family; CBY: family similar to putative cobalt uptake systems; Y179: similar to the M. janaschii Y179 protein subfamily; MKL: similar to the M. leprae MKL protein family; TGD: trigalactosyldiacyl glycerol: TGD1, 2 and 3 are components of a chloroplast phospholipid translocator; NO: proteins of unknown function, apparently unrelated to existing families and ADT: proteins of unknown function.
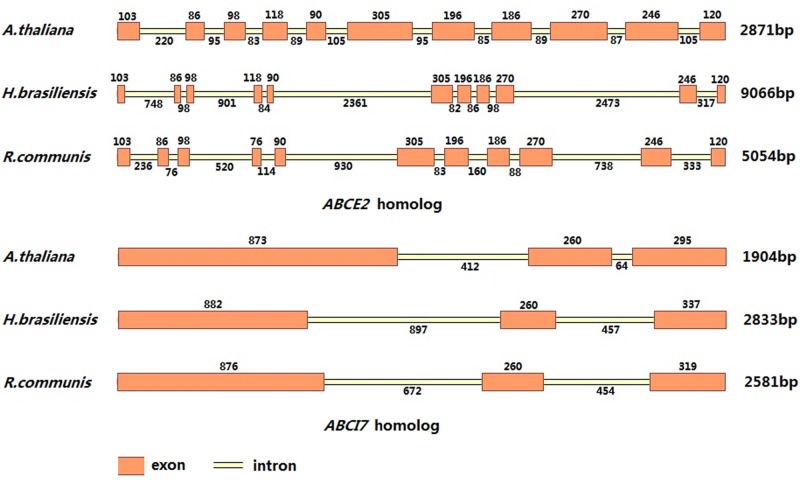
Most subfamilies of plant ABC proteins have corresponding counterparts in the human genome, so that the latex ABC transporters were also denominated using the Human Genome Organization (HUGO) nomenclature for human ABC proteins [39] in order to obtain a unified nomenclature of plant ABC proteins, as suggested by Verrier et al. in 2008 [1]. According to the HUGO nomenclature, all the eight subfamilies of plant ABC transporters were identified in the H. brasiliensis latex, among which ABCB, ABCG and ABCI were the most abundant (Table 1). By comparing the mapped reads (reads per kilobase of exon model per million mapped reads), the top five latex ABC transporters were HbABCF1, HbABCG21, HbABCI18, HbABCB15 and HbABCG11. One representative linear comparison of the exon–intron structures of two selected ABCE2 and ABCI7 genes in the genome sequences of A. thaliana, H. brasiliensis and Ricinus communis is shown in Fig. 1. The alignment analysis revealed that the H. brasiliensis latex ABC protein genes were highly conserved compared with those of a closely related species, R. communis, and a distantly related species, A. thaliana.
Figure 1. Linear representation of the exon–intron structures of the selected ABCE2 and ABCI7 genes in the genome sequences of A. thaliana, H. brasiliensis and R. communis.
Exons and introns of the ABCE2 and ABCI7 genes were predicted and mapped by the Vector NTI Advance program (version 11.5.2). The accession numbers for the ABCE2 homolog genome sequences are NC_003075.7 (10501200..10505217) for A. thaliana and NW_002994591.1 for R. communis. The accession numbers for the ABCI7 homolog genome sequences are NC_003070.9 (11749823..11752106) for A. thaliana and NW_002995814.1 for R. communis.
Of the 46 latex ABC transporters, 12 members were full-size transporters, containing two TMDs and two NBDs, 21 members were half-size transporters and contained one TMD and one NBD or two NBDs, and the other 13 members could be called quarter-size ABC molecules with only one NBD or TMD domain. All 12 full-size transporters, with the exception of one protein (HbABCG40), exhibited a forward topology orientation in which the TMDs preceded the NBDs, but most of the 21 half-size transporters had a reverse orientation where NBD preceded TMD. The cDNA sequences of the 46 latex putative ABC transporter genes were submitted to GenBank. Their accession numbers are shown in Table 1 and the predicted amino acid sequences of the putative ABC proteins are listed in S4 Table.
Phylogeny of the H. brasiliensis Latex ABC Proteins
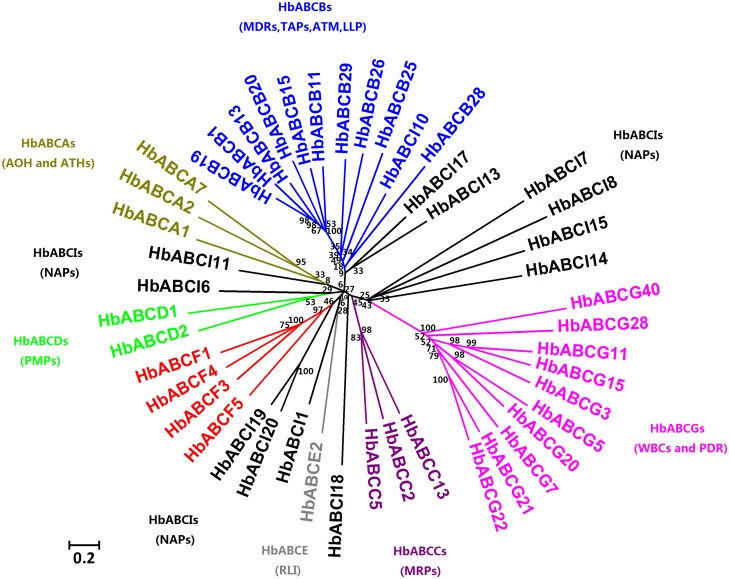
The amino acid sequences of the 46 latex putative ABC transporters were aligned using the MUSCLE alignment tool and the phylogenetic tree was generated by MEGA5.05 software as outlined in the Materials and Methods section. The phylogenetic tree for all 46 latex ABC proteins is shown in Fig. 2. With the exceptions of the ABCI and ABCE subfamilies, which only have one HbABCE2 member that was grouped on its own, the other six subfamilies of the latex ABC proteins were clustered tightly within their respective subfamilies with bootstrap values of at least 90%. The HbABCI subfamily had 13 members and they were clustered into four different clades. HbABCI7, 8, 15, 17 and 18 were grouped within the same clade, whereas HbABCI1, 6 and 11 formed their own clade and HbABCI10, 13 and 14 were in the same clade. HbABCI19 and 20 were clustered together and were closely related to the HbABCG subfamily. The lack of a coherent phylogeny within the HbABCI subfamily was consistent with the clustering patterns of the ABCI subfamilies in A. thaliana [2] and Vitis vinifera [5]. Of the 46 latex ABC proteins, 42, excluding four HbABCI members (HbABCI14, 15, 17 and 18), contained at least one NBD. The NBD phylogeny of the 42 latex ABC transporters is shown in S1 Fig.
Figure 2. Phylogenetic tree for H. brasiliensis latex ABC proteins.
The amino acid sequences of the 46 latex ABC proteins were aligned using the MUSCLE program and subjected to phylogenetic analysis by the distance with neighbor-joining method using the MEGA5.05 software. The numbers on the nodes indicate the bootstrap values after 1000 replicates. The scale bar indicates the estimated number of amino acid substitutions per site. The HUGO nomenclature was followed and the ABC protein abbreviations are as follows: AOH: ABC1 homolog; ATH: ABC-two homolog; MDR: multi-drug resistance; ATM: ABC transporter of mitochondria; TAP: transporter associated with antigen processing; LLP: Lipid A-like exporter; MRP: multi-drug resistance-associated protein; PMP: peroxisomal membrane protein; RLI: RNase L inhibitor; GCN: general control non-repressible; WBC: white-brown complex; PDR: pleiotropic drug resistance; NAP: non-intrinsic ABC protein. AOH and ATH, which belong to the ABCA subfamily; MDR, TAP, ATM and LLP, which belong to the ABCB subfamily; MRP, which belongs to the ABCC subfamily; PMP, which belongs to the ABCD subfamily; RLI, which belongs to the ABCE subfamily; GCN, which belongs to the ABCF subfamily; WBC, which belongs to the ABCG subfamily and NAP, which belongs to the ABCI subfamily.
Characteristics of the H. brasiliensis Latex ABC Family Transporters
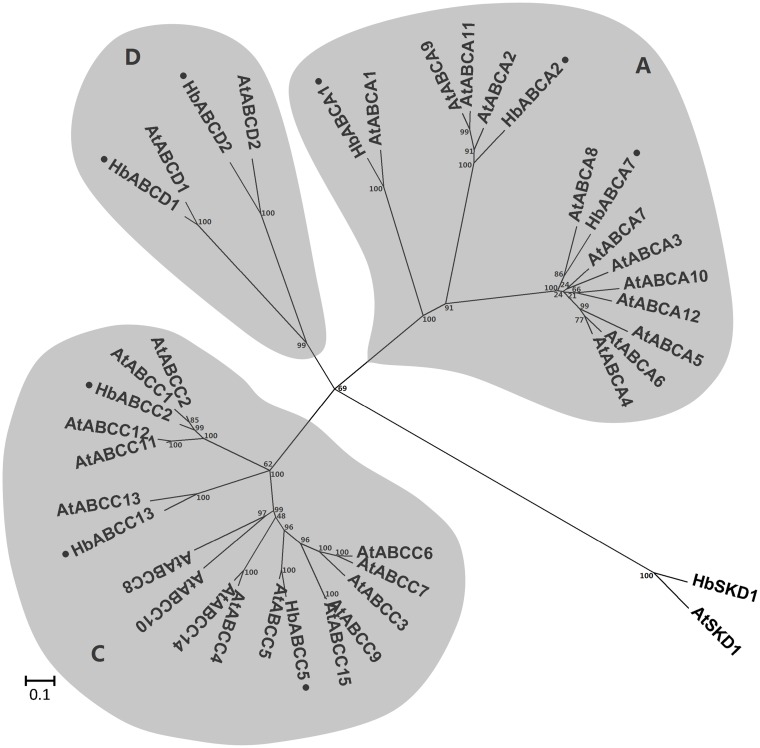
HbABCA subfamily. Three members were identified in the HbABCA subfamily. These were one full-size HbABCA1 member and two half-size members: HbABCA2 and HbABCA7. Until recently, only one full-size ABCA transporter gene has been identified in plants and was designated as AtABCA1 or AOH (the ABC one homolog) in the A. thaliana genome [2]. HbABCA1 was the largest latex ABC transporter, consisting of 1,883 amino acid residues, and shared 74% similarity with AtABCA1. The phylogenetic relationship between A. thaliana and H. brasiliensis ABCA proteins is shown in Fig. 3. The ABCA1 gene is conserved in humans and other animals. Its human counterpart (ABCA1) is involved in cellular transportation [40, 41], but the function and the localization of the plant ABCA1 transporter remains unclear.
Figure 3. Phylogenetic relationship between A. thaliana and H. brasiliensis latex ABCA, ABCC and ABCD proteins.
The amino acid sequences of all the A. thaliana ABCA, ABCC, ABCD and H. brasiliensis latex proteins were aligned using the MUSCLE program and subjected to phylogenetic analysis by the distance with neighbor-joining method using MEGA5.05 software. The numbers on the nodes indicate the bootstrap values after 1000 replicates. The scale bar indicates the estimated number of amino acid substitutions per site. Accession numbers for the A. thaliana sequences are AtABCA1 (NP_850354.2), AtABCA2 (NP_190357.2), AtABCA3 (NP_190358.3), AtABCA4 (NP_190359.5), AtABCA5 (NP_190360.2), AtABCA6 (NP_190361.2), AtABCA7 (NP_190362.2), AtABCA8 (NP_190363.3), AtABCA9 (NP_200981.1), AtABCA10 (NP_200982.1), AtABCA11 (NP_200977.2), AtABCA12 (NP_200978.1), AtABCC1 (NP_174329.1), AtABCC2 (NP_181013.1), AtABCC3 (NP_187915.1), AtABCC4 (NP_182301.1), AtABCC5 (NP_171908.1), AtABCC6 (NP_187916.3), AtABCC7 (NP_187917.3), AtABCC8 (NP_001189944.1), AtABCC9 (NP_191575.2), AtABCC10 (NP_191473.2), AtABCC11 (NP_174331.2), AtABCC12 (Q9C8H0.1), AtABCC13 (Q9SKX0.3), AtABCC14 (NP_191829.1), AtABCC15 (NP_191656.2), AtABCD1 (NP_568072.1) and AtABCD2 (NP_175837.2). AtSKD1 (AEC08019.1) and HbSKD1 (AIN75626.1) were used as outgroups. The H. brasiliensis latex ABC proteins are marked with a dot.
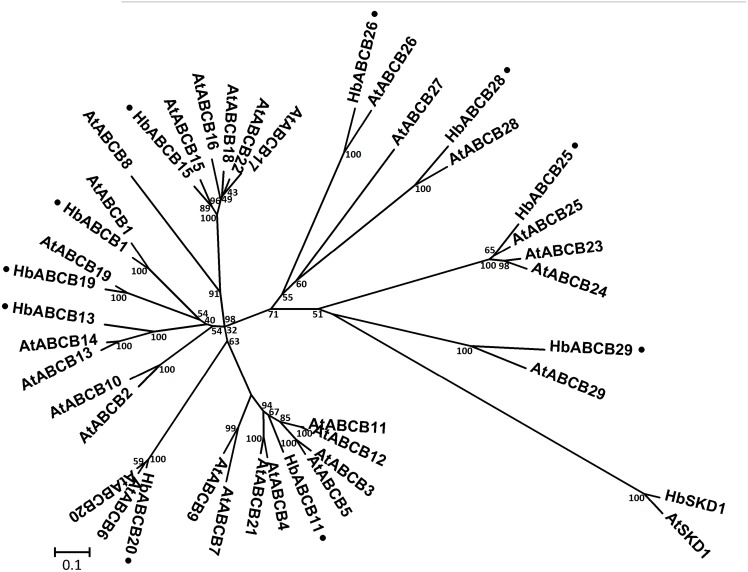
HbABCB subfamily. The latex HbABCB subfamily contains ten members. Six are full-size molecules, conventionally named multi-drug resistance (MDRs) [2] or drug, peptide and lipid exporters (DPLs)/PGP (similar to p-glycoprotein) [3] and four are half-size proteins, consisting of one ABC transporter in mitochondria (ATM) [2], two TAPs (similar to the human transporter associated with antigen presentation) [2] and one LLP (lipid A-like exporters, putative) [3]. All 21 full-size ABCB transporters in the A. thaliana genome have been shown to be localized on the plasma membrane [6, 42, 43] and AtABCB1, AtABCB4, AtABCB14, AtABCB15 and AtABCB19, have been characterized as auxin transporters or are associated with polar auxin transport in A. thaliana [44–46]. Furthermore, ABCB14-mediated auxin transport was recently reported to be involved in Fe homeostasis in rice [47]. Orthologs of AtABCB1, AtABCB15 and AtABCB19 were also identified in H. brasiliensis latex (Fig. 4), but it remains to be determined whether they are implicated in auxin transport in the H. brasiliensis laticifers that are apoplastically isolated from the neighboring cells [25].
Figure 4. Phylogenetic tree for A. thaliana and H. brasiliensis latex ABCB protein sequences.
The amino acid sequences of all A. thaliana ABCB and H. brasiliensis latex proteins were aligned using the MUSCLE program and subjected to phylogenetic analysis by the distance with neighbor-joining method using MEGA5.05 software. The numbers on the nodes indicate the bootstrap values after 1000 replicates. The scale bar indicates the estimated number of amino acid substitutions per site. Accession numbers for the A. thaliana sequences are AtABCB1 (NP_181228.1), AtABCB2 (NP_194326.2), AtABCB3 (NP_192091.1), AtABCB4 (NP_182223.1), AtABCB5 (NP_192092.1), AtABCB6 (NP_181480.1), AtABCB7 (NP_199466.1), AtABCB8 (Q9LHK4.1), AtABCB9 (NP_193539.6), AtABCB10 (NP_172538.1), AtABCB11 (NP_171753.1), AtABCB12 (NP_171754.1), AtABCB13 (NP_174115.1), AtABCB14 (NP_174122.1), AtABCB15 (NP_189475.1), AtABCB16 (NP_189477.4), AtABCB17 (NP_189479.1), AtABCB18 (NP_189480.1), AtABCB19 (NP_189528.1), AtABCB20 (NP_191092.1) AtABCB21 (NP_191774.2), AtABCB22 (NP_683599.1), AtABCB23 (NP_567813.1), AtABCB24 (NP_194591.1), AtABCB25 (NP_200635.1), AtABCB26 (NP_177218.3), AtABCB27 (NP_198720.2), AtABCB28 (NP_194275.2) and AtABCB29 (NP_196011.1). AtSKD1 (AEC08019.1) and HbSKD1 (AIN75626.1) were used as outgroups. The H. brasiliensis latex ABC proteins are marked with a dot.
Less information is available about the function and the localization of the half-size members of the plant ABCB subfamily. Among the four half-size ABCB proteins in the H. brasiliensis latex, one member, HbABCB25, is a close homolog of A. thaliana ABCB25/ATM3, which is a mitochondrial ABC transporter involved in the biogenesis of iron-sulfur proteins and molybdenum-containing enzymes in plants [48, 49]. Proteomic data from previous studies have demonstrated that A. thaliana ABCB26/TAP1 is localized to the chloroplast [50] and the results of this study suggest that the latex HbABCB26, a homolog of At ABCB26/TAP1, might possibly be localized to the Frey-Wyssling particles, which are very specialized chromoplasts in rubber tree latex [51]. However, this needs to be verified.
HbABCC subfamily. In plants, ABCC subfamily transporters belong to the multidrug resistance-associated proteins (MRPs) that were originally identified in human drug-resistant cell lines [52]. They only have full-size ABC proteins that contain an additional N-terminal extension (NTE or TMD0) of around 220 amino acids [53]. Three members of the ABCC subfamily were identified in the H. brasiliensis latex transcriptome, whereas 15 ABCC protein genes were determined in the A. thaliana genome [2]. Their sequence similarity was confirmed by the phylogenetic analysis (as shown in Fig. 3). Until recently, only the full-size ABCC subfamily transporters have been shown to be localized on the vacuolar membrane in plants [54]. Furthermore, their localization to the tonoplasts gives ABCC subfamily transporters an important function in the general vacuolar sequestration of conjugated metabolites, such as anthocyanidin 3-O-glucosides [55], abscisic acid (ABA) glucosyl ester [56] and phenolic compounds [57], etc. In the case of the H. brasiliensis laticifers, no typical central vacuoles have been observed, although polydispersed microvacuoles called lutoids are present, and laticifers have been implicated in pH regulation and solute accumulation [58, 59]. It has not been confirmed whether the latex ABCC transporters are intrinsic lutoid membrane proteins that are involved in the maintenance of ion homeostasis between lutoids and cytosol, which is crucial for lutoid-mediated rubber particle aggregation and sequential latex coagulation.
HbABCD subfamily. The ABCD subfamily transporters are conventionally designated as PMPs (peroxisomal membrane protein), which are intrinsic membrane proteins of peroxisomes and are mainly involved in fatty acid β-oxidation through the importation of acyl-CoA esters into the peroxisome [60]. Only half-size ABCD proteins have been detected in mammals and fungi, whereas both half-size and full-size ABCD transporters have been detected in plants. The prototypical plant member of the ABCD subfamily is the A. thaliana protein, Comatose (CTS; also known as AtABCD1) [61, 62]. Until recently, only one half-size and one full-size member of the ABCD subfamily have been identified in the A. thaliana genome and their orthologs were also present in the H. brasiliensis latex transcriptome (as shown in Fig. 3). Loss-of-function AtABCD1 mutants are significantly impaired in several important metabolic and developmental processes, such as germination, fertility, seedling establishment and root growth. These studies provided evidence that, besides the involvement in fatty acid β-oxidation, the plant ABCD transporters are also involved in auxin and jasmonic acid (JA) metabolism via the importation of the auxin precursor, indolbutyric acid (IBA), and the JA precursor, 12-oxophytodienoic acid (OPDA), into the peroxisome [63, 64]. Several attempts have now been made to elucidate the physiological and molecular characteristics of JA in H. brasiliensis since recent reports demonstrated that JA and its conjugate, methyl jasmonic acid (Me-JA), are key inducers of laticifer differentiation [65] and are regulators of rubber biosynthesis-related genes in rubber trees [28]. Further investigation is needed to ascertain whether the latex ABCD transporters mediate JA biosynthesis in the H. brasiliensis laticifers, which are the natural rubber-producing cells of rubber trees.
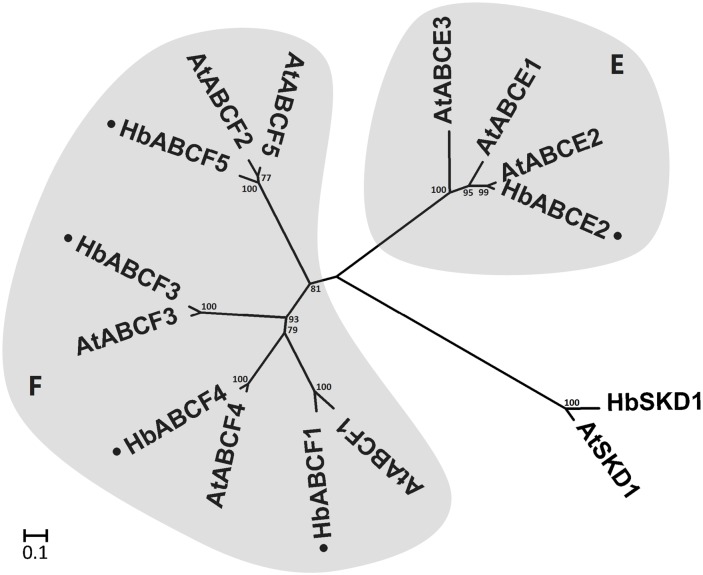
HbABCE and HbABCF subfamilies. The members of the ABCE and the ABCF subfamilies are thought to be soluble proteins that contain only two NBDs and no TMDs. The A. thaliana genome contains three ABCE and five ABCF members, whereas only one ABCE protein and four ABCF proteins were identified in the H. brasiliensis latex transcriptome (Fig. 5). However, information is relatively scarce about these soluble ABC proteins compared with other intrinsic membrane ABC transporters. They are probably involved in non-transport processes, as is the case for their yeast and human orthologs, which participate in ribosome recycling and translational control [54]. Only one ABC transporter, which is a member of the ABCF subfamily, was identified in rubber particles through proteome analysis [66], but it is unclear whether this protein is related to biogenesis or to the biosynthesis of rubber particles.
Figure 5. Phylogenetic relationship between A. thaliana and H. brasiliensis latex ABCE/ABCF proteins.
The amino acid sequences of A. thaliana ABCE/ABCF proteins and H. brasiliensis latex proteins were aligned using the MUSCLE program and subjected to phylogenetic analysis by the distance with neighbor-joining method using MEGA5.05 software. The numbers on the nodes indicate the bootstrap values after 1000 replicates. The scale bar indicates the estimated number of amino acid substitutions per site. Accession numbers for the A. thaliana sequences are AtABCE1 (NP_187973.1), AtABCE2 (NP_193656.2), AtABCE3 (NP_194759.1), AtABCF1 (NP_200887.1), AtABCF2 (NP_196555.2), AtABCF3 (NP_176636.1), AtABCF4 (NP_567001.1) and AtABCF5 (NP_201289.1). AtSKD1 (AEC08019.1) and HbSKD1 (AIN75626.1) were used as outgroups. The H. brasiliensis latex ABC proteins are marked with a dot.
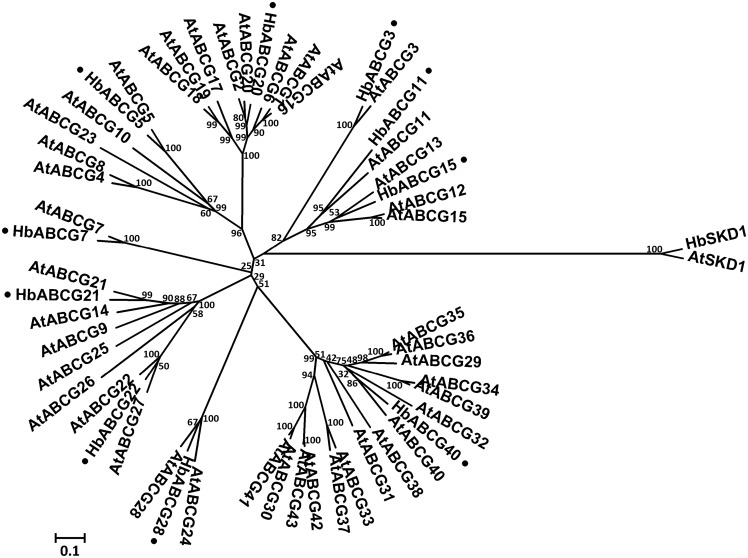
HbABCG subfamily. The largest ABC subfamily is ABCG, which consists of 28 half-size (WBC) and 15 full-size (PDR) transporters in the A. thaliana genome. All the ABCG subfamily transporters have a unique reverse “NBD-TMD” orientation and are reported to be plasma membrane-localized proteins, except for AtABCG19, which has been localized to the vacuolar membrane and confers kanamycin-resistance in plants [67]. In the H. brasiliensis latex transcriptome, nine WBC-type ABCG proteins and one PDR-type ABCG protein were detected. The phylogenetic analyses of A. thaliana and H. brasiliensis latex ABCG proteins are shown in Fig. 6.
Figure 6. Phylogenetic relationship between A. thaliana and H. brasiliensis latex ABCG proteins.
The amino acid sequences of all the A. thaliana ABCG proteins and H. brasiliensis latex proteins were aligned using the MUSCLE program and subjected to phylogenetic analysis by the distance with neighbor-joining method using MEGA 5.05 software. The numbers on the nodes indicate the bootstrap values after 1000 replicates. The scale bar indicates the estimated number of amino acid substitutions per site. Accession numbers for the A. thaliana sequences are AtABCG1 (NP_181467.1), AtABCG2 (NP_181272.1), AtABCG3 (NP_850111.1), AtABCG4 (NP_194305.1), AtABCG5 (NP_178984.1), AtABCG6 (NP_196862.1), AtABCG7 (NP_178241.1), AtABCG8 (NP_200098.1), AtABCG9 (NP_194472.3), AtABCG10 (NP_175734.1), AtABCG11 (NP_173226.2), AtABCG12 (NP_175561.1), AtABCG13 (NP_175557.1), AtABCG14 (NP_564383.1), AtABCG15 (NP_188746.2), AtABCG16 (NP_191069.2), AtABCG17 (NP_191070.1), AtABCG18(NP_191071.1), AtABCG19 (NP_191073.1), AtABCG20 (NP_190919.1), AtABCG21 (NP_189190.2), AtABCG22 (NP_568169.1), AtABCG23 (NP_197442.1), AtABCG24 (NP_175745.4), AtABCG25 (NP_565030.1), AtABCG26 (NP_187928.2), AtABCG27 (NP_190799.2), AtABCG28 (NP_200882.4), AtABCG29 (NP_566543.1), AtABCG30 (NP_193258.3), AtABCG31 (NP_180555.2), AtABCG32 (NP_180259.1), AtABCG33 (NP_181265.1), AtABCG34 (NP_181179.2), AtABCG35 (NP_172973.1), AtABCG36 (NP_176196.1), AtABCG37 (NP_190916.1), AtABCG38 (Q7PC85.1), AtABCG39 (NP_176867.2), AtABCG40 (NP_173005.1), AtABCG41 (NP_680692.1), AtABCG42 (NP_680693.5) and AtABCG43 (NP_680694.2). AtSKD1 (AEC08019.1) and HbSKD1 (AIN75626.1) were used as outgroups. The H. brasiliensis latex ABC proteins are marked with a dot.
ABCG half-transporters have been implicated in several plant functions. A. thaliana AtABCG25 and AtABCG40 are involved in abscisic acid (ABA) transport and responses [68, 69]. Recent studies have indicated that ABCG9, ABCG11 and ABCG14 are involved in lipid/sterol homeostasis regulation, which is required for proper vascular development in A. thaliana [70]. Finally, some members of ABCG subfamily confer plant resistance to various biotic and abiotic stresses [71, 72].
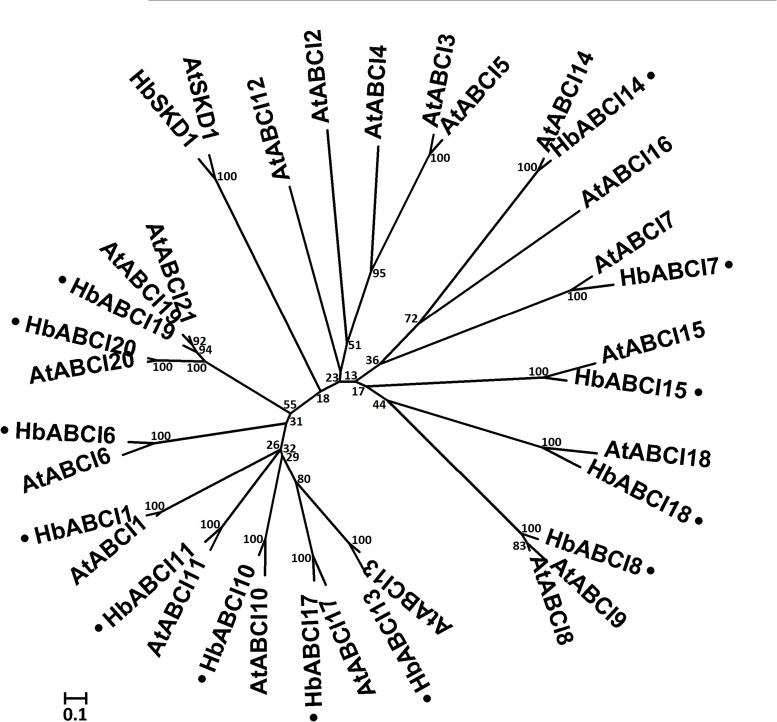
HbABCI subfamily. The ABCI subfamily proteins consist of bacteria-type soluble or peripheral ABC components with a single NBD and are designated as non-intrinsic ABC proteins (NAPs) [2]. Genetic evidence has demonstrated that the ABCI subfamily protein subunits include not only NBD and TMD domains, but also homologues of established soluble cytosolic proteins that interact with NBDs and putative substrate-binding proteins and are similar to the periplasmic-binding proteins in bacteria [3, 73, 74]. A total of 13 ABCI proteins were identified in the H. brasiliensis latex transcriptome and there were 21 orthologs in the A. thaliana genome (Fig. 7). Three A. thaliana ABCI proteins: AtABCI19, 20 and 21 have been shown to translate into cytosolic proteins [75], whereas several other ABCI subfamily members are predicted to be localized to the chloroplast or mitochondria [1, 76].
Figure 7. Phylogenetic relationship between A. thaliana and H. brasiliensis latex ABCI proteins.
The amino acid sequences of all the A. thaliana ABCI proteins and H. brasiliensis latex proteins were aligned using the MUSCLE program and subjected to phylogenetic analysis by the distance with neighbor-joining method using MEGA 5.05 software. The numbers on the nodes indicate the bootstrap values after 1000 replicates. The scale bar indicates the estimated number of amino acid substitutions per site. Accession numbers for the A. thaliana sequences are AtABCI1 (NP_176516.1), AtABCI2 (NP_085482.1), AtABCI3 (NP_085546.2), AtABCI4 (NP_565344.2), AtABCI5 (NP_973435.1), AtABCI6 (NP_187678.1), AtABCI7 (NP_564404.1), AtABCI8 (NP_192386.1), AtABCI9 (NP_680390.1), AtABCI10 (NP_195072.2), AtABCI11 (NP_196914.1), AtABCI12 (NP_566688.4), AtABCI13 (NP_564850.1), AtABCI14 (NP_973869.1), AtABCI15 (NP_566659.1), AtABCI16 (NP_181270.1), AtABCI17 (NP_176961.1), AtABCI18 (NP_563693.1), AtABCI19 (NP_563694.1), AtABCI20 (NP_195847.1), and AtABCI21 (NP_199224.1). AtSKD1 (AEC08019.1) and HbSKD1 (AIN75626.1) were used as outgroups. The H. brasiliensis latex ABC proteins are marked with a dot.
The functional significance of the ABCI proteins remains to be determined. Previous studies have suggested that A. thaliana ABCI subfamily proteins can be assembled into multi-subunit ABC transporters, which is similar to the way that ABC proteins are formed in prokaryotes. Five members of the A. thaliana ABCI proteins: AtABCI1, 2, 3, 4 and 5, have been reported to be involved in cytochrome c biogenesis [77], AtABCI6, 7 and 8 were responsible for Fe-S cluster assembly and iron homeostasis regulation [78, 79] and AtABCI13, 14 and 15 have been shown to be involved in plastid lipid formation [80–83].
Expression Profiling of the Latex ABC Protein Genes
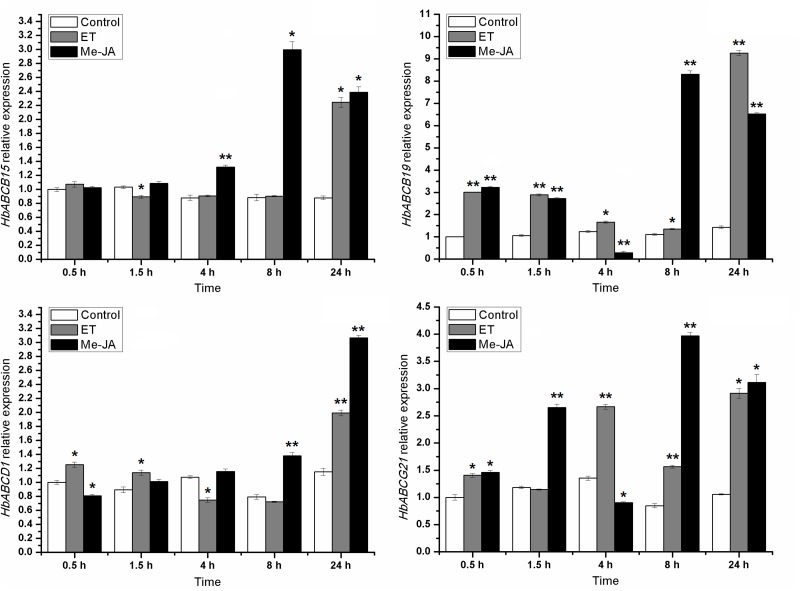
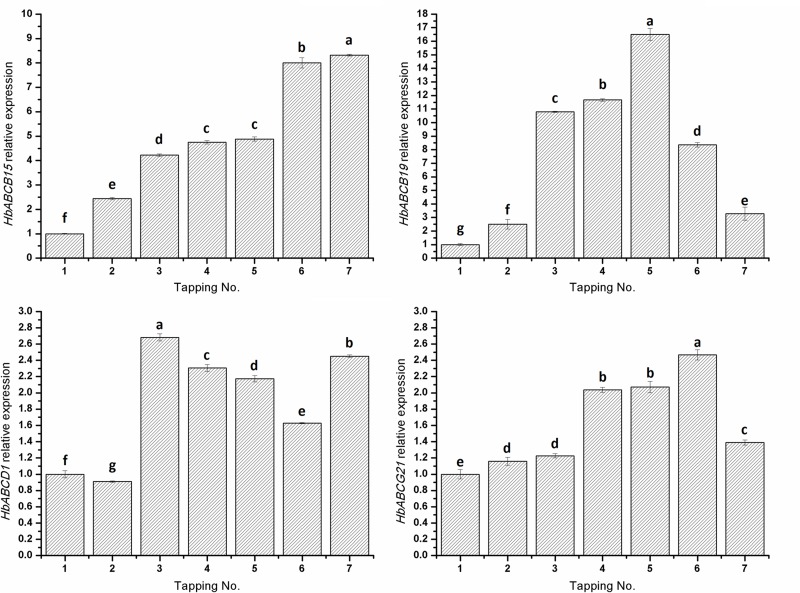
Plant ABC family proteins have been shown to be involved in many physiological processes that allow the plant to adapt to changing environments and to cope with biotic and abiotic stresses. To investigate further the possible roles played by latex ABC transporters during latex metabolism in rubber trees, the gene expression patterns of all the latex ABC transporters were first ascertained using rubber trees, which were treated with Ethrel or Me-JA for 24 h or tapped at the seventh times. The results demonstrated that the most of the latex ABC transporter genes were controlled by Ethrel, Me-JA or bark tapping to varying degrees (S5 Table), among which HbABCB15, HbABCB19, HbABCD1 and HbABCG21 were significantly up-regulated by the three different stimulations.
The gene expression time courses for HbABCB15, HbABCB19, HbABCD1 and HbABCG21 were further analyzed using rubber trees that had been treated with Ethrel or Me-JA at different time points. The data indicated that although the four HbABC genes were highly regulated by Ethrel or Me-JA, their individual expression profiling was different from each other (Fig. 8). Bark tapping, as a simulated wound stress, could also induce expression of the four selected ABC transporter genes, but in different ways (Fig. 9). Several studies reported that salicylic acid (SA), Me-JA/JA and ABA could modulate the gene expression of plant ABC transporters [84–86]. However, information about the responses of ABC protein genes to ethylene and bark tapping (a wound stress) is still relatively limited.
Figure 8. Regulatory effects of ethylene and JA on the gene expressions of the four HbABC transporters in the H. brasiliensis latex.
Mature, virgin (untapped) rubber trees were treated with Ethrel (ET) or methyl-jasmonate (Me-JA) for 0.5, 1.5, 4.0, 8.0 or 24.0 h. The controls were rubber trees that had not been treated with stimulants. Three independent biological replicates were included in each sample. Fresh latex from each sample was collected and used to isolate the total RNA. The transcript abundance of each gene was detected by RT-qPCR and the values are shown as the mean ± S.D. Statistical significance was determined using Students t-test using SPSS 19.0 software (Chicago, U.S.A.). When compared with the control, one asterisk shows a significant difference with a P-value < 0.05 and two asterisks show a very significant difference with a P-value < 0.01.
Figure 9. Effects of bark tapping on the gene expressions of the four HbABC transporters in the H. brasiliensis latex.
Mature, virgin rubber trees were tapped sequentially seven times using an S/2 d/3 tapping system. Each sample contained three independent biological replicates. Fresh latex from each tapping was collected and used to isolate the total RNA. The transcript abundance of each gene was detected by RT-qPCR and the values are shown as the mean ± S.D. One-way ANOVA was performed using SPSS 19.0 software (Chicago, U.S.A.). The Student–Newman–Keuls test was used for multiple comparisons testing to investigate the significant differences between groups. Bars with different letters are significantly different at the p < 0.05 level. The values represent the mean ± SD of three biological replicates.
Ethrel, Me-JA and bark tapping are three efficient stimulants of latex production and laticifer differentiation in rubber trees, but the underlying molecular mechanisms remain poorly understood. The identification and expression analyses of the latex ABC transporter genes provide some valuable insights into how the related latex ABC transporters regulate latex metabolism/regeneration and rubber biosynthesis by H. brasiliensis.
Concluding Remarks
The physiological functions of plant ABC transporters have only been marginally characterized compared to human ABC transporters. The whole latex transcriptome survey has produced an inventory of the laticifer-specific ABC transporters of H. brasiliensis. Laticiferous cells are where natural rubber is synthesized and stored. To date, the transportation of various substrates related to latex metabolism and rubber biosynthesis inside the laticiferous cells has been poorly understood. Several latex ABC transporters were transcriptionally induced by ethylene, JA and bark tapping (a wound stress), all of which activate latex metabolism or laticifer differentiation in rubber trees. The identification and expression analysis of the latex ABC transporters may facilitate further studies into their possible involvement in latex metabolism and rubber biosynthesis by both H. brasiliensis and other latex-producing plants.
Supporting Information
The conserved NBD and TMD domains in the H. brasiliensis latex ABC transporters were predicted by Pfam 27.0 (http://pfam.xfam.org/). The amino sequences of the latex ABC protein NBDs were aligned using the MUSCLE program and subjected to phylogenetic analysis by the distance with neighbor-joining method (1000 bootstrap replicates) using MEGA5.05 software. The scale bar indicates the estimated number of amino acid substitutions per site.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
We appreciate the technical support for the Illumina HiSeq-2000 sequencing and initial data analysis by the Beijing Genomics Institute-Shenzhen (BGI, Shenzhen, China) and are grateful to Dr. Fang Yongjun and Cheng Han for their help with some of the data analysis.
Data Availability
All relevant cDNA sequences are available within the text and at the NCBI database. Accession numbers are KM035282-035324 and etc., as shown in the text.
Funding Statement
This work was supported by the National Natural Science Foundation with the website of (http://www.nsfc.gov.cn). The grant numbers are 31070607 and 31270713, which were received by the authors of NZY and ZRZ, respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rees DC, Johnson E, Lewinson O (2009) ABC transporters: the power to change. Nat Rev Mol Cell Biol 10: 218–227. 10.1038/nrm2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schmitz G, Kaminski WE, Orse E (2000) ABC transporters in cellular lipid trafficking. Curr Opin Lipidol 11: 493–501. 10.1097/00041433-200010000-00007 [DOI] [PubMed] [Google Scholar]
- 3. Kim S, Yamaoka Y, Ono H, Nishida I, Lee Y, et al. (2013) AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proc Natl Acad Sci USA 110: 773–778. 10.1073/pnas.1214159110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boursiac Y, Keran S, Corratge-Faillie C, Gojon A, Krouk G, et al. (2013) ABA transport and transporter. Trends Plant Sci 18: 325–333. 10.1016/j.tplants.2013.01.007 [DOI] [PubMed] [Google Scholar]
- 5. Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, et al. (2005) Cellular efflux of auxin catalyzed by the Arabdopsis MDR/PGP transporter AtPGP1. Plant J 44: 179–194. 10.1111/j.1365-313X.2005.02519.x [DOI] [PubMed] [Google Scholar]
- 6. Verrier PJ, Bird D, Burla B, Dassa E, Forestier C, et al. (2008) Plant ABC proteins: a unified nomenclature and updated inventory. Trends Plant Sci 13: 151–159. 10.1016/j.tplants.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 7. Rea PA (2007) Plant ATP‑binding cassette transporters. Annu Rev Plant Biol 58: 347–375. 10.1146/annurev.arplant.57.032905.105406 [DOI] [PubMed] [Google Scholar]
- 8. Yazaki K, Shitan N, Sugiyama A, Takanashi K (2009) Cell and molecular biology of ATP-binding cassette proteins in plants. Int Rev Cell Mol Biol 276: 263–299. 10.1016/S1937-6448(09)76006-X [DOI] [PubMed] [Google Scholar]
- 9. Sanchez-Fernandez R, Davis TGE, Coleman JOD, Rea PA (2001) The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem 276: 30231–30244. 10.1074/jbc.M103104200 [DOI] [PubMed] [Google Scholar]
- 10. Carcia O, Bouige P, Forestier C, Dassa E (2004) Inventory and comparative analysis of Rice and Arabidopsis ATP-binding cassette (ABC) systems. J Mol Biol 343: 249–265. 10.1016/j.jmb.2004.07.093 [DOI] [PubMed] [Google Scholar]
- 11. Pang K, Li Y, Liu M, Meng Z, Yu Y (2013) Inventory and general analysis of the ATP-binding cassette (ABC) gene superfamily in maize (Zea mays L.).Gene 526: 411–428. 10.1016/j.gene.2013.05.051 [DOI] [PubMed] [Google Scholar]
- 12. Çakır B, Kılıçkaya O (2013) Whole-genome survey of the putative ATP-binding cassette transporter family genes in Vitis vinifera. PLoS One 8: e78860 10.1371/journal.pone.0078860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Beilen JB, Poirier Y (2007) Establishment of new crops for the production of natural rubber. Trends Biotechnol 25: 522–529. 10.1016/j.tibtech.2007.08.009 [DOI] [PubMed] [Google Scholar]
- 14. Chow KS, Mat-Isa MN, Bahari A, Ghazali AK, Alias H, et al. (2012) Metabolic routes affecting rubber biosynthesis in Hevea brasiliensis latex. J Exp Bot 63: 1863–1871. 10.1093/jxb/err363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cornish K, Wood DF, Windle JJ (1999) Rubber particles from four different species, examined by transmission electron microscopy and electron-paramagnetic-resonance spin labeling, are found to consist of a homogenous rubber core enclosed by a contiguous monolayer biomembrane. Planta 210: 85–96. 10.1007/s004250050657 [DOI] [PubMed] [Google Scholar]
- 16. Cornish K (2001) Similarities and differences in rubber biochemistry among plant species. Phytochemistry 57: 1123–1134. 10.1016/S0031-9422(01)00097-8 [DOI] [PubMed] [Google Scholar]
- 17. Kekwick RGO (1989) The formation of isoprenoids in Hevea latex. In Physiology of Rubber Tree Latex (eds d’Auzac J, Jacob JL & Chrestin H), Boca Raton, FL: CRC Press; pp: 146–164. [Google Scholar]
- 18. Hillebrand A, Post JJ, Wurbs D, Wahler D, Lenders M, et al. (2012) Down-regulation of small rubber particle protein expression affects integrity of rubber particles and rubber content in Taraxacum brevicorniculatum . PLoS ONE 7: e41874 10.1371/journal.pone.0041874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Post J, van Deenen N, Fricke J, Kowalski N, Wurbs D, et al. (2012) Laticifer-specific cis-prenyltransferase silencing affects the rubber, triterpene, and inulin content of Taraxacum brevicorniculatum . Plant Physiol 158: 1406–1417. 10.1104/pp.111.187880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang C, Xiao X, Li H, Fan Y, Yang J, et al. (2013) Comparative analysis of latex transcriptome reveals putative molecular mechanisms underlying super productivity of Hevea brasiliensis . PLoS One 8: e75307 10.1371/journal.pone.0075307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dusotoit-Coucaud A, Brunel N, Kongsawadworakul P, Viboonjun U, Lacointe A, et al. (2009) Sucrose importation into laticifers of Hevea brasiliensis, in relation to ethylene stimulation of latex production. Ann Bot 104: 635–647. 10.1093/aob/mcp150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tang C, Huang D, Yang J, Liu S, Sakr S, et al. (2010) The sucrose transporter HbSUT3 plays an active role in sucrose loading to laticifer and rubber productivity in exploited trees of Hevea brasiliensis (para rubber tree). Plant Cell Environ 33: 1708–1720. 10.1111/j.1365-3040.2010.02175.x [DOI] [PubMed] [Google Scholar]
- 23. Herman EM, Schmidt MA (2004) Endoplasmic reticulum to vacuole trafficking of endoplasmic reticulum bodies provides an alternate pathway for protein transfer to tht vacuole. Plant Physiol 136: 3440–3446. 10.1104/pp.104.051722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hasma H, Subramaniam A (1986) Composition of lipids in latex of Hevea brasiliensis clone RRIM 501. J Nat Rubb Res 1: 30–40. [Google Scholar]
- 25. de Fay E, Jacob JL (1989) Anatomical organization of the laticiferous system in the bark. In Physiology of Rubber Tree Latex (eds d’Auzac J, Jacob JL & Chrestin H), Boca Raton, FL: CRC Press: pp: 3–14. [Google Scholar]
- 26. d’ Auzac J, Jacob JL (1989) The composition of latex from Hevea brasiliensis as a laticiferous cytoplasm. In Physiology of Rubber Tree Latex (eds d’Auzac J, Jacob JL & Chrestin H), Boca Raton, FL: CRC Press: pp: 89–94. [Google Scholar]
- 27. Mantello CC, Cardoso-Silva CB, da Silva CC, de Souza LM, Scaloppi Junior EJ, et al. (2014) De novo assembly and transcriptome analysis of the rubber tree (Hevea brasiliensis) and SNP markers development for rubber biosynthesis pathways. PLoS One 9(7):e102665 10.1371/journal.pone.0102665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zeng RZ, Duan CF, Li XY, Tian WM, Nie ZY, et al. (2009) Vacuolar-type inorganic pyrophosphatase located on the rubber particle in the latex is an essential enzyme in regulation of the rubber biosynthesis in Hevea brasiliensis . Plant Sci 176: 602–607. 10.1016/j.plantsci.2009.01.009 [DOI] [Google Scholar]
- 29. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pertea G, Huang X, Liang F, Antonescu V, Sultana R, et al. (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19: 651–652. 10.1093/bioinformatics/btg034 [DOI] [PubMed] [Google Scholar]
- 31. Green P (1996) PHRAP documentation. University of Washington, Seattle, USA. [Google Scholar]
- 32. Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. (2009) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 37: D205–210. 10.1093/nar/gkn845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, et al. (2012) Finn RD. The Pfam protein families database. Nucleic Acids Res 40: D290–301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Edger RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 36. Tamura K, Peterson D, Peterson N, Stecher G, Kumar S, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duan CF, Rio M, Leclercq J, Bonnot F, Montoro P, et al. (2010) Gene expression pattern in response to wounding, methyl jasmonate and ethylene in the bark of Hevea brasiliensis . Tree Physiol 30: 1349–1359. 10.1093/treephys/tpq066 [DOI] [PubMed] [Google Scholar]
- 38. Tupy J (1989) Sucrose supply and utilization for latex production. In Physiology of Rubber Tree Latex (eds d’Auzac J, Jacob JL & Chrestin H), Boca Raton, FL: CRC Press; pp: 179–199. [Google Scholar]
- 39. Dean M, Rzhetsky A, Allikmets R (2001) The human ATP-binding cassette (ABC) transporters superfamily. Genome Res 11: 1156–1166. 10.1101/gr.GR-1649R [DOI] [PubMed] [Google Scholar]
- 40. Lee JY, Parks JS (2005) ATP-binding cassette transporter AI and its role in HDL formation. Curr. Opin. Lipidol 16: 19–25. 10.1097/00041433-200502000-00005 [DOI] [PubMed] [Google Scholar]
- 41. Chao CJ, Yin K, Fu YC, Tang CK (2012) The interaction of ApoA-I and ABCA1 triggers signal transduction pathways to mediate efflux of cellular lipids. Mol Med 18: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Peer WA, et al. (2007) Interactions among PIN-FORMED and P-Glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147. 10.1105/tpc.106.040782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee M, Choi Y, Burla B, Kim YY, Lee Y, et al. (2008) The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat Cell Biol 10: 1217–1223. 10.1038/ncb1782 [DOI] [PubMed] [Google Scholar]
- 44. Titapiwatanakun B, Murphy AS (2009) Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J Exp Bot 60: 1093–1107. 10.1093/jxb/ern240 [DOI] [PubMed] [Google Scholar]
- 45. Kaneda M, Schuetz M, Lin BS, Chanis C, Samuels AL (2011) ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J Exp Bot 62: 2063–2077. 10.1093/jxb/erq416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cho M, Lee ZW, Cho HT (2012) ATP-binding cassette B4, an auxin-efflux transporter, stably associates with the plasma membrane and shows distinctive intracellular trafficking from that of PIN-FORMED proteins. Plant Physiol 159: 642–654. 10.1104/pp.112.196139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu Y, Zhang S, Guo H, Qi Y, Jiang DA, et al. (2014) OsABCB14 functions in auxin transport and iron homeostasis in rice (Oryza sativa L.). Plant J. 10.1111/tpj.12544 [DOI] [PubMed] [Google Scholar]
- 48. Bernard DG, Cheng Y, Zhao Y, Balk J (2009) An allelic mutant series of ATM3 reveals its key role in the biogenesis of cytosolic ironsulfur proteins in Arabidopsis. Plant Physiol 151: 590–602. 10.1104/pp.109.143651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Teschner J, Lachmann N, Schulze J, Mendel RR, Bittner F, et al. (2010) A novel role for Arabidopsis Mitochondrial ABC transporter ATM3 in molybdenum cofactor biosynthesis. Plant Cell 22: 468–480. 10.1105/tpc.109.068478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ferro M, Brugiere S, Salvi D, Seigneurin-Berny D (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084. 10.1074/mcp.M900325-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de Fay E, Hebant C, Jacob JL (1989) Cytology and cytochemistryof the laticiferous system. In Physiology of Rubber Tree Latex (eds d’Auzac J, Jacob JL & Chrestin H), Boca Raton, FL, CRC Press; pp: 15–30. [Google Scholar]
- 52. Cole SPC, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, et al. (1992) Overexpression of a transporter gene in a multidrug-resistant human lung-cancer cell-line. Science 258: 1650–1654. 10.1126/science.1360704 [DOI] [PubMed] [Google Scholar]
- 53. Klein M, Burla B, Martinoia E (2006) The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. FEBS Letters 580: 1112–1122. 10.1016/j.febslet.2005.11.056 [DOI] [PubMed] [Google Scholar]
- 54. Kang J, Park J, Choi H, Burla B, Kretzschmar T, et al. (2011). Plant ABC transporters. The Arabidopsis Book: e0153, doi: 10.1199/tab.0153 [DOI] [PMC free article] [PubMed]
- 55. Francisco RM, Regalado A, Ageorges A, Burla BJ, Nagy R, et al. (2013) ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-Glucosides. Plant Cell 25: 1840–1854. 10.1105/tpc.112.102152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Burla B, Pfrunder S, Nagy R, Francisco RM, Lee YS, et al. (2013) Vacuolar transport of abscisic acid glucosyl ester is mediated by ATP-binding cassette and proton-antiport mechanisms in Arabidopsis. Plant Physiol 163: 1446–1458. 10.1104/pp.113.222547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yazaki K (2006) ABC transporters involved in the transport of plant secondary metabolites. FEBS Letters 580: 1183–1191. 10.1016/j.febslet.2005.12.009 [DOI] [PubMed] [Google Scholar]
- 58. d’Auzac J, Prevot JC, Jacob JL (1995) What’s new about lutoids? A vacuolar system model from Hevea latex. Plant Physiol Biochem 33: 765–777. [Google Scholar]
- 59. Amalou Z, Bangratz J, Chrestin H (1992) Ethrel (ethylene releaser)-induced increases in the adenylate pool and transtonoplast ΔpH within Hevea latex cells. Plant Physiol 98: 1270–1276. 10.1104/pp.98.4.1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Theodoulou FL, Holdsworth M, Baker A (2006) Peroxisomal ABC transporters. FEBS Lett 580: 1139–1155. 10.1016/j.febslet.2005.12.095 [DOI] [PubMed] [Google Scholar]
- 61. Zolman BK, Silva ID, Bartel B (2001) The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol 127: 1266–1278. 10.1104/pp.010550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Footitt S, Slocombe SP, Larner V, Kurup S, Holdsworth M, et al. (2002) Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J 21: 2912–2922. 10.1093/emboj/cdf300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Theodoulou FL, Job K, Slocombe SP, Footitt S, Holdsworth M, et al. (2005) Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisomes. Plant Physiol 137: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mendiondo GM, Medhurst A, Roermund van, Theodoulou FL, Holdsworth MJ, et al. (2014) Barley has two peroxisomal ABC transporters with multiple functions in β-oxidation. J Exp Bot 65: 4833–4847. 10.1093/jxb/eru243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hao BZ, Wu JL (2000) Laticifer differentiation in Hevea brasiliensis: induced by exogenous jasmonic acid and linolenic acid. Ann Bot 85: 37–43. 10.1006/anbo.1999.0995 [DOI] [Google Scholar]
- 66. Dai LJ, Kang GJ, Li Y, Nie ZY, Zeng RZ, et al. (2013) In-depth proteome analysis of the rubber particles of Hevea brasiliensis (para rubber tree). Plant Mol Biol 82: 155–168. 10.1007/s11103-013-0047-y [DOI] [PubMed] [Google Scholar]
- 67. Mentewab A, Stewart CN (2005) Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat Biotechnol 23: 1177–1180. 10.1038/nbt1005-1315b [DOI] [PubMed] [Google Scholar]
- 68. Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, et al. (2010) ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci. USA 107: 2361–2366. 10.1073/pnas.0912516107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, et al. (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid Proc Natl Acad Sci USA 107: 2355–2360. 10.1073/pnas.0909222107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hir R, Sorin C, Chakraborti D, Moritz T, Schaller H, et al. (2013) ABCG9, ABCG11 and ABCG14 ABC transporters are required for vascular development in Arabidopsis. Plant J 76: 811–824. 10.1111/tpj.12334 [DOI] [PubMed] [Google Scholar]
- 71. Kim DY, Jin JY, Alejandro S, Martinoia E, Lee Y (2010) Overexpression of AtABCG36 improves drought and salt stress resistance in Arabidopsis. Physiol Plantarum 139: 170–180. 10.1111/j.1399-3054.2010.01353.x [DOI] [PubMed] [Google Scholar]
- 72. Stukkens Y, Bultreys A, Grec S, Trombik T, Boutry M, et al. (2005) NpPDR1, a pleiotropic drug resistance-type ATP-binding cassette transporter from Nicotiana plumbaginifolia, plays a major role in plant pathogen defense. Plant Physiol 139: 341–352. 10.1104/pp.105.062372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dassa E, Bouige P (2001) The ABC of ABCs: a phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol 152: 211–229. 10.1016/S0923-2508(01)01194-9 [DOI] [PubMed] [Google Scholar]
- 74. Bouige P, Laurent D, Piloyan L, Dassa E (2002) Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr Protein Pept Sci 3: 541–559. 10.2174/1389203023380486 [DOI] [PubMed] [Google Scholar]
- 75. Marin E, Divol F, Bechtold N, Vavasseur A, Nussaume L, et al. (2006) Molecular characterization of three Arabidopsis soluble ABC proteins which expression is induced by sugars. Plant Sci 171: 84–90. 10.1016/j.plantsci.2006.02.014 [DOI] [Google Scholar]
- 76. Shimoni-Shor E, Hassidim M, Yuval-Naeh N, Keren N (2010) Disruption of Nap14, a plastid localized non-intrinsic ABC protein in Arabidopsis thaliana results in the over-accumulation of transition metals and in aberrant chloroplast structures. Plant Cell Environ 33: 1029–1038. 10.1111/j.1365-3040.2010.02124.x [DOI] [PubMed] [Google Scholar]
- 77. Rayapuram N, Hagenmuller J, Grienenberger JM, Giege P, Bonnard G (2007) AtCCMA interacts with AtCcmB to form a novel mitochondrial ABC transporter involved incytochrome C maturation in Arabidopsis. J Biol Chem 282: 21015–21023. 10.1074/jbc.M704091200 [DOI] [PubMed] [Google Scholar]
- 78. Xu XM, Adams S, Chua NH, Moller SG (2005) AtNAP1 represents an atypical SufB protein in Arabidopsis plastids. J Biol Chem 280: 6648–6654. 10.1074/jbc.M413082200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Xu XM, Moller SG (2004) AtNAP7 is a plastidic SufC-like ATP-binding cassette/ATPase essential for Arabidopsis embryogenesis. Proc Natl Acad Sci USA 101: 9143–9148. 10.1073/pnas.0400799101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xu C, Fan J, Riekhof W, Froehlich JE, Benning C (2003) A permease-like protein involved in ER to thylakoidlipid transfer in Arabidopsis. EMBO J: 22, 2370–2379. 10.1093/emboj/cdg234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xu C, Fan J, Froehlich JE, Awai K, Benning C (2005) Mutation of the TGD1 chloroplast envelope proteinaffects phosphatidate metabolism in Arabidopsis. Plant Cell 17: 3094–3110. 10.1105/tpc.105.035592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Awai K, Xu C, Tamot B, Benning C (2006) A phosphatidic acid-binding protein of thechloroplast inner envelope membrane involved in lipid trafficking.Proc. Natl Acad Sci U S A 103: 10817–10822. 10.1073/pnas.0602754103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lu B, Xu C, Awai K, Jones AD, Benning C (2007) A small ATPase protein of Arabidopsis, TGD3, involved in chloroplast lipid import. J Biol Chem 282: 35945–35953. 10.1074/jbc.M704063200 [DOI] [PubMed] [Google Scholar]
- 84. Grc S, Vanham D, de Ribaucourt JC, Purnelle B, Boutry M (2003) Identification of regulatory sequence elements within the transcription promoter region of NpABC1, a gene encoding a plant ABC transporter induced by diterpenes. Plant J 35: 237–250. 10.1046/j.1365-313X.2003.01792.x [DOI] [PubMed] [Google Scholar]
- 85. Eichhorn H, Klinghammer M, Becht P, Tenhaken R (2006) Isolation of a novel ABC-transporter gene from soybean induced by salicylic acid. J Exp Bot 57: 2193–2201. 10.1093/jxb/erj179 [DOI] [PubMed] [Google Scholar]
- 86. Zhang R, Zhu J, Cao HZ, An YR, Huang JJ, et al. (2013) Molecular cloning and expression analysis of PDR1-like gene in ginseng subjected to salt and cold stresses or hormonal treatment. Plant Physiol Biochem 71: 203–211. 10.1016/j.plaphy.2013.07.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The conserved NBD and TMD domains in the H. brasiliensis latex ABC transporters were predicted by Pfam 27.0 (http://pfam.xfam.org/). The amino sequences of the latex ABC protein NBDs were aligned using the MUSCLE program and subjected to phylogenetic analysis by the distance with neighbor-joining method (1000 bootstrap replicates) using MEGA5.05 software. The scale bar indicates the estimated number of amino acid substitutions per site.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant cDNA sequences are available within the text and at the NCBI database. Accession numbers are KM035282-035324 and etc., as shown in the text.