SUMMARY
AgrD is the precursor for the auto-inducing peptide in a quorum sensing system regulating virulence phenotypes of the pre-eminent pathogen Staphylococcus aureus. Mass spectrometry-based methods, including molecular networking, identified formylated and non-formylated peptide variants derived from the AgrD N-terminal leader domain in S. aureus cell-free culture supernatants. Functional assessment of these peptides revealed the unexpected bioactivities including human cell-line cytotoxicity, modulation of neutrophil chemotaxis, neutrophil extracellular trap formation, and the aggravation of skin lesions in vivo.
INTRODUCTION
Nearly three decades ago, the accessory gene regulatory (Agr) quorum sensing system was identified in the leading human bacterial pathogen Staphylococcus aureus (Recsei et al., 1986). A classical model of growth phase-dependent gene regulation, Agr is studied for its important role in regulation of surface-expressed or secreted virulence factors. Natural or engineered strains of S. aureus with inactivating mutations in agr genes lose virulence in animal infection (Abdelnour et al., 1993; Villaruz et al., 2009). Thus, S. aureus infectivity depends upon an array of Agr-regulated gene products working in concert to circumvent host innate immune clearance (Novick et al., 1993; Pragman and Schlievert, 2004); yet, many mechanistic details of the Agr system remain a mystery.
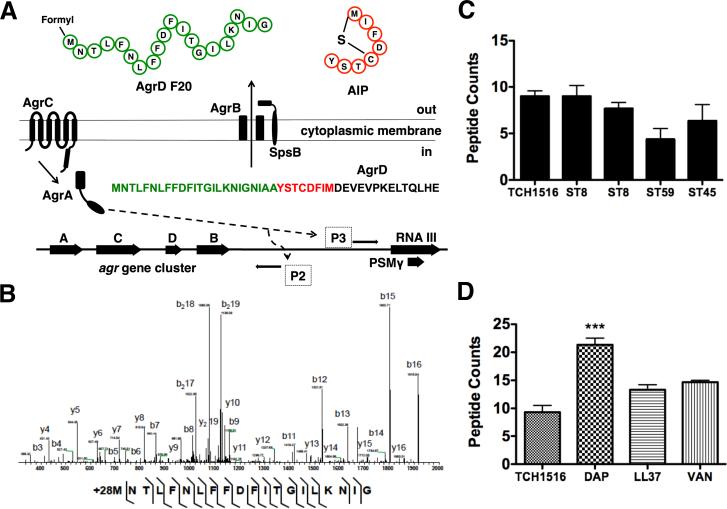
Agr quorum-sensing dynamics are governed by the posttranslationally modified, thiolactone-containing autoinducing peptide (AIP) (Enright et al., 2002). Genetically, agr is comprised of two promoters, P2 and P3, which direct the divergent transcription of RNAII and RNAIII according to AIP concentrations. RNA II encodes the four-gene operon agrABCD, and within the cluster AgrB+AgrD and AgrA+AgrC work together to carry out biosynthesis and regulatory functions. AgrD, the AIP precursor substrate, is cleaved and cyclized through thiolactone formation by cell membrane-bound AgrB. AgrA and AgrC combine to form a kinase-dependent two-component regulatory system. During critical regulatory states, the mature AIP binds extracellularly to the AgrC protein kinase, triggering activation of agrA and successive induction of divergently expressed promoters P2 and P3. RNAIII expression leads to production of two molecules, the primary effector RNA regulator and delta-toxin or PSMγ, a wide-spectrum cytolysin that is released extracellularly with either a formylated initiator methionine, a non-formylated methionine, or in truncated forms (Gonzalez et al., 2012). Taxonomically, S. aureus isolates can be divided into four agr groups based on polymorphisms within the agrABCD operon (Wright et al., 2005).
Downstream contributions of the mature AIP on S. aureus pathogenesis have been the focus of several studies (Harraghy et al., 2007). More recently, reports described the biochemistry of each individual gene product and the relationship between the enzyme AgrB and its substrate AgrD (Thoendel and Horswill, 2009). It is thought that formation of the AIP is initiated by migration of AgrD and its association with the intracellular membrane. The amphipathic AIP N-terminal region contains essential amino acids that stabilize the propeptide, allowing for association of core and C-terminal amino acids with AgrB. Endopeptidase AgrB then cleaves the highly conserved C-terminal tail of AgrD to catalyze the formation of a thiolactone ring within the core amino acids. Thereafter, through an unknown mechanism, the AIP precursor is transported across the cell membrane. Once extracellular, the type I signal peptidase SpsB (Kavanaugh et al. 2007) catalyzes the formation of two extracellular final products: (i) AIP and (ii) N-terminal peptide (Figure 1A).
Figure 1.
The accessory gene regulatory system and N-AgrD peptide properties. (a) Four genes, agrABCD comprise the agr biosynthetic network. agrD encodes the peptide substrate undergoes PTM by membrane-bound endopeptidase agrB through cleavage and thiolactone cyclization. Once extracellular, the hybrid molecule consisting of the AIP and the N-terminal region of AgrD is further processed by peptidase SpsB into mature AIP and N-AgrD. Induction of Promoter 2 drives the transcription of the agrABCD operon. Promoter 3 induction results in the expression of RNA III that produces two final products, the primary RNA effector molecule and PSMγ. (b) MS survey of several S. aureus clinical isolates show detectable N-AgrD peptides as peptide counts. (c) Tandem MS sequencing of AgrD F24. Several contingent ion fragments, including reliable mass accuracy, localize a PTM of +28 Da to the initiator methionine of N-AgrD. (d) Antibiotic effects on the number of N-AgrD peptide counts. Challenge with a subinhibitory concentration of daptomycin increased the number of detectable peptides. Statistical analysis performed by one-way ANOVA; ***P < 0.001. Data was expressed as mean ± standard deviation.
Using molecular networking (Watrous et al., 2012), we examined mass spectrometry (MS) data derived from cell-free culture extracts of a representative community-associated methicillin-resistant S. aureus (MRSA) USA300 isolate (Kennedy et al., 2010). Analysis of the molecular network revealed extracellular AgrD N-terminal (termed N-AgrD) formylated and truncated peptide variants. Here we show these N-AgrD variant peptides have immunostimulatory properties towards neutrophils isolated from human blood, display broad-spectrum cytotoxicity, and aggravate skin lesion formation caused by Agr-deficient S. aureus. This study provides new insight into endogenous biosynthetic and virulence properties of the Agr quorum-sensing system, while revealing new functional properties of AgrD beyond its AIP-encoding domain. Furthermore, we highlight the power of the molecular networking platform as a tool to discover unforeseen bioactive peptides within a long-studied, highly important regulatory system of staphylococcal virulence.
RESULTS
Discovery of N-AgrD variant peptides by MS-based Molecular Networking
Molecular networking is an emerging platform based on the concept of mass spectral pairing, which allows for the structural grouping and successive mapping of MS data sets. Recently, our group used the platform to build a molecular network representative of extracellularly released, small molecular weight biosynthetic factors produced by MRSA (Gonzalez et al., 2014). Here, we report the continued examination of the created molecular network by de novo sequencing and peptidogenomic approaches (Kersten et al., 2011), recovering peptides pertaining to the AgrD N-terminal domain (Figure S1). Investigation into the identity of each individual mass spectrum within the AgrD grouped family-produced peptide sequence tags corresponding to N-terminal variants with mass offsets of +28 Daltons (Da), a signature for formyl-methionine modification. With the recovered chemical knowledge from the molecular networks, we used tandem MS data processing program InsPecT (Tanner et al., 2005) to support the de novo sequencing and peptidogenomics identifications. Careful manual annotation of the identified N-AgrD peptides further validated the identifications (Figure 1B; Figures S2-S6). To broaden our survey, we analyzed cell-free culture extracts of 12 additional S. aureus strains or clinical isolates (Table S1). N-AgrD peptides were produced by three USA300 lineage MRSA, two ST59 MRSA (Huang et al., 2012), and clonal colony ST45 of USA600 MRSA lineage (Figure 1C). In successive MS experiments, peptide sequences most abundantly mapped to AgrD (McIlwain et al., 2012), herein designated (i) N-AgrD F20, (ii) N-AgrD F24, and (iii) N-AgrD D20, were synthesized and isolated at >95% purity for functional assessment.
Properties of the N-AgrD Variant Peptides
To estimate the production and release of the N-AgrD variants, MRSA USA300 cell-free cultures were extracted and examined by tandem MS. N-AgrD peptides were only detected in late exponential phase extracts, in agreement with previous reports on the temporal production of the AIP (Junio et al., 2013). Estimated AIP culture concentrations vary from 2-10 μM (Junio et al., 2013), therefore we predicted that N-AgrD peptide variants would be detected in stoichiometric concentrations. We detected 2-3 μM N-AgrD F24 in extracted cultures of USA300 strain TCH1516 and 1-2 μM in extracted cultures of the ST59 strain. There was increased abundance of N-AgrD peptides upon exposure of MRSA to subinhibitory concentrations of cell-wall active antibiotics daptomycin (0.125 mg/L) or vancomycin (0.125 mg/L), or human cationic defense peptide LL-37 (1.2 mg/L) (Figure 1D).
PMN Chemotaxis Induced by N-AgrD Formylated Variants
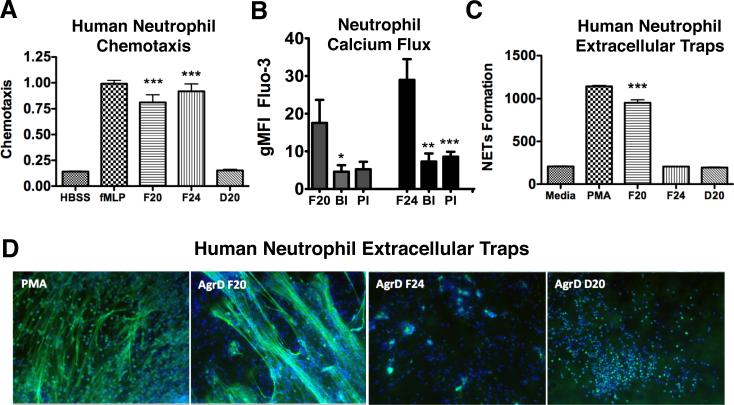
Leukocytes of the host innate immune system recognize formylated bacterial products as pathogen-associated molecular patterns (PAMPs) (Mogensen, 2009), as mediated through formylpeptide receptors (FPR), a class of G protein-coupled receptors associated with chemotaxis. The human FPR family is comprised of three members, FPR1, FPR2/ALX and FPR3; only the first two are expressed in neutrophils. Recently, members of the S. aureus-derived phenol-soluble modulin (PSM) peptide family were shown to selectively activate FPR2/ALX (Kretschmer et al., 2010). Because N-AgrD F20 and N-AgrD F24 peptides contained formyl groups, we tested their abilities to act as chemoattractants for human neutrophils. Nanomolar quantities of N-AgrD F20, N-AgrD F24 and N-AgrD D20 were placed in a chamber adjacent to a suspension of freshly isolated neutrophils, and chemotaxis was quantified. Both formylated peptides N-AgrD F20 and F24 showed strong chemoattractant properties comparable to the well-known chemattractant formyl-Methionyl-Leucyl-Phenylalanine (fMLP), whereas nonformylated N-AgrD D20 did not stimulate chemotaxis (Figure 2A). Using specific antagonists of the FPR1 and FPR2 receptors and calcium influx as a surrogate measure of receptor activation (Dixit et al., 2012), significant decreases in receptor activation were observed by pharmacological blockade, indicating the N-AgrD peptides function though a FPR-dependent mechanism (Figure 2B).
Figure 2.
N-AgrD peptide variants stimulate PMN chemotaxis and NET formation. (a) As an indicator of migration, elastase enzymatic activity in neutrophil supernatants was measured at 405 nm. Formylated peptides N-AgrD F20 and N-AgrD F24 stimulated increased chemotaxis. The N-AgrD D20 peptide, which is non-formylated, did not exhibit chemoattractant properties. (b) PMN calcium influx indicating receptor activation. A significant decrease in activation was observed by each pharmacological blockade, indicating the N-AgrD peptide family functions though a formyl peptide receptor-dependent mechanism. BI = BOC, selective inhibitor of FPR1; PI: PBP, selective inhibitor of FPR2. (c) The Quant-iT Picogreen assay was used to quantify extracellular DNA release upon stimulation by the N-AgrD peptides. Only formylated peptide NAgrD F20 stimulated a significant increase in NET production. (d) Microscopy confirmed only peptide N-AgrD D20 stimulated formation of web-like DNA structures similar to the PMA positive control. Statistical analysis by one-way ANOVA; *** P <0.001. Data was expressed as mean ± standard deviation.
NET Induction by the N-AgrD F20 Formylated Peptide
Neutrophil extracellular trap (NET) formation is a specialized cell death process wherein nuclear DNA is extruded to ensnare and kill pathogens (Brinkmann et al., 2004). As quantified by DNA release, AgrD F20 strongly stimulated NET formation (Figure 2C), whereas no stimulatory effects were observed with peptides N-AgrD F24 and N-AgrD D20. N-AgrD F20 stimulation of NETs was corroborated by fluorescence microscopy, which showed the hallmark web-like NET structures were formed by neutrophils upon N-AgrD F20 stimulation (Figure 2D).
N-AgrD Variant Peptides Cytotoxicity Against Human Cell Lines and Erythrocytes
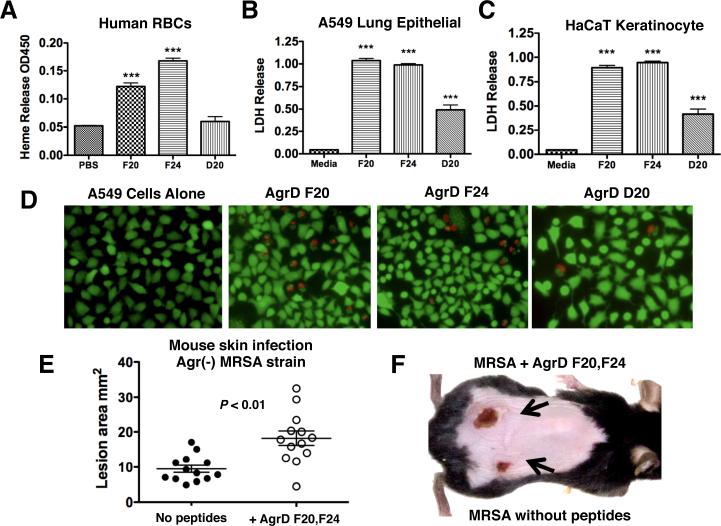
Hemolytic ability of the N-AgrD peptides was tested against erythrocytes from freshly washed human blood. N-AgrD F20 and N-AgrD F24 showed significant hemolytic activity, but N-AgrD D20 peptide did not induce erythrocyte lysis (Figure 3A). USA300 MRSA are associated with complicated skin and soft tissue infections and severe necrotizing pneumonia, as the pathogen breaks down host epithelial tissue barriers (Lim et al., 2012; Wang et al., 2007). We tested the effect of micromolar concentrations of N-AgrD peptides on a human alveolar epithelial cell line (A549) and a human keratinocyte cell line (HaCaT). As measured by lactate dehydrogenase release (LDH), N-AgrD F20 and N-AgrD F24 formylated peptides exhibited twice the cytolytic activity against both tissue cell lines as the non-formylated N-AgrD peptide (Figure 3B, 3C). A549 and HaCaT cytotoxicity was corroborated by live/dead cell counts (Figure S7A, S7B) and visual assessment by microscopy (Figure 3D; Figure S7C). Additionally, a scrambled-sequence peptide was included as a negative control and showed no activity, verifying N-AgrD bioactivity is sequence-specific.
Figure 3.
Erythrocyte hemolysis, cytotoxtic properties of N-AgrD against epithelial cells and murine models of infection. (a) Heparinized human whole blood was processed and treated with N-ArgD peptides or saline control. N-AgrD F20 and N-AgrD F24 showed moderate hemolysis activity. The N-AgrD D20 peptide did not cause hemolysis. (b) A549 human alveolar epithelial cell and (c) HaCaT human keratinocyte cell lines were incubated with N-AgrD peptides for 120 min; cell damage was monitored by LDH release. All three peptides displayed significant cytolytic activity. Statistical analysis by one-way ANOVA; ***P <0.001. (d) Fluorescent microscopy of A549 cells after live/dead cell staining. (e) In vivo contribution of the N-AgrD F20 and N-AgrD F24 peptides as assessed in murine lesion infection models. Subcutaneous infection of C57Bl/6 mice with an S. aureus agr null strain was tested +/− addition of a 1:1 mixture of AgrD F20 and AgrD F24 (10 μg/ml) on contralateral flanks. Statistical analysis was performed by Student's t-test; **P <0.01. (f) Appearance of a representative infected mouse flank shows addition of the N-AgrD F20 + N-AgrD F24 peptides to the S. aureus agr null strain results in an approximate doubling of lesion area. All statistical data was expressed as mean ± standard deviation.
N-AgrD Peptides Aggravate Skin Lesions in a Murine Model
S. aureus is the most common cause of skin and soft tissue infections in humans, and the majority of the isolates cultured from these infections are predominately agr positive. We sought to determine the contribution of N-AgrD peptides to disease progression in vivo independent of the well-established role of the Agr system in virulence. To that end, we inoculated mice subcutaneously with the agr null strain S. aureus RN6911 in the presence or absence of a micromolar mixture of the N-AgrD F20 and N-AgrD F24. Presence of the N-AgrD peptides significantly increased ensuing lesion sizes (Figure 3E). After a 72 hrs, the lesion sizes caused by the RN6911 strain harvesting the N-AgrD peptides were approximately double in size (Figure 3E, 3F) despite equal bacterial loads (Figure S8), consistent with the endogenous proinflammatory and cytotoxic properties of these peptides observed in our in vitro testing.
DISCUSSION
The current functional assignment for the N-terminal region of AgrD indicates the amphipathic peptide governs intercellular membrane anchoring and successive processing by AgrB (Zhang et al., 2004). Our study shows the extracellular release of N-AgrD as a set of bioactive structurally diverse peptides, including formylated variants. These findings indicate that S. aureus has evolved to maximize or chemically salvage its protein repertoire in order to gain ecological fitness.
N-AgrD structurally resembles the PSM toxin family in S. aureus. PSMs are cytolytic, formylated and non-formylated peptide variants that are upregulated in community-associated MRSA and genetically linked to the agr system (PSMγ is embedded within RNA III) (Wang et al., 2007; Gonzalez et al., 2014). Because of the structural similarities between peptides, N-AgrD could play a potential role in biofilm structural formation as demonstrated for PSMs (Periasamy et al., 2012). In concurrence with the work herein, N-AgrD was recently shown to be a constituent of amyloid fibrils of biofilms and possess further cellular functions akin to PSMs (Schwartz et al., 2014).
Mechanisms governing the amounts of N-AgrD participating in amyloid fibrils (biofilm) versus free to the surrounding environment is likely a dynamic process governed by environmental stimuli. This notion is supported by our observation that subinhibitory concentrations of cell-wall inhibiting antibiotics or LL-37 increased abundance of N-AgrD peptides in a similar manner to PSMs (Gonzalez et al., 2012). The interplay between promiscuous S. aureus exoproteases and the AgrD leader domain may lead to the wide repertoire of N-AgrD variants. Proteolysis studies on the AIP showed S. aureus supernatants produced variant cleavage products of the N-terminal AgrD domain independent of SpsB (Kavanaugh et al., 2007). Our accumulated studies on the AgrD leader domain show a new, potentially important component of the now classical agr pathway.
In summary, we applied MS-based tools to identify N-AgrD variants that possess cytotoxic and proinflammatory activities. These peptides potentially modulate S. aureus disease manifestations, and suggest that the N-terminal leader domain of this protein is not simply an innocuous castoff during processing of the mature AIP. Like many pattern recognition molecules with mixed proinflammatory and toxic activities, their effect on the host is complex and likely to vary dependent on the site, stage, and magnitude of infection. Therefore, the effects N-AgrD variants have on S. aureus gene regulation (i.e. AgrA-AgrC signaling) merits further study. Ultimately, this work exemplifies the use of an emerging tool like molecular networking can aid in elucidating molecules that influence the establishment of infection, leading to better understanding their broader impact on the pathogen-host interaction.
SIGNIFICANCE
Our results reveal that the N-terminal leader end of the AIP substrate AgrD is an extracellularly released peptide existing in variant forms with unexpected bioactivities in vitro and in vivo. These peptides may play a role in MRSA virulence; before this study, only the core amino acids of mature AgrD were believed to contribute to bacterium's pathogenic cycle. This project arose by the application of the new and emerging tool of mass spectrometry-based molecular networking, which highlights the value of integrating cutting-edge analytical tools to the study of biological systems. Because the complex array of virulence factors produced by MRSA is not fully understood, it is critical to apply new strategies that extend beyond the classical paradigm of proteomic identification. These new technologies can lead to unforeseen information on the molecular repertoire of human pathogens.
EXPERIMENTAL PROCEDURES
Identification of N-AgrD by Mass Spectrometry
Staphylococcus aureus cultures (Table S1) were grow in Todd-Hewitt Broth (THB) at 37°C to late exponential-early stationary growth phase, pelleted at 4,000 rpm × 10 min, and 1 ml supernatant collected, extracted with an equal volume of 1-butanol, and dried by SpeedVac. SDS-PAGE gels showed no Coomassie staining indicative of larger proteins; therefore the extracts were considered to contain peptides and other small molecular weight species. Solid extracts were resuspended and processed by Thermo-Finnegan linear trap quadruple liquid chromatography tandem MS (LC MS/MS). Peptidogenomic annotation of the collected tandem MS data indicated sequence tags of the N-AgrD, and mass offsets of +28 Da were localized to the N-terminal methionine residue. Targeted use of the MS processing program InsPecT identified several N-AgrD variant peptides with formylated initiator methionine residues. Here we focused on the three most abundant and highest scoring peptides obtained from InsPecT, after validation of the assignments by manual annotation.
Synthetic peptides
The following peptides were synthesized by American Peptide Company (Sunnyvale, CA): (i) N-AgrD F20: formyl-MNTLFNLFFDFITGILKNIG (ii) N-AgrD F24: formyl-MNTLFNLFFDFITGILKNIGNIAA (iii) N-AgrD D20: FNLFFDFITGILKNIGNIAA. (iii) Scramble peptide N-AgrD D20: NIAKFFLTIILFNNDFGAGI. Additional experimental procedures can be found in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Mass spectrometry guides the discovery of unexpected variant N-terminal AgrD peptides of MRSA.
N-terminal AgrD peptides exhibit cytotoxic and pro-inflammatory bioactivities in vitro and in vivo.
ACKNOWLEDGEMENTS
D.J.G. was supported through fellowships from the A.P. Giannini Medical Research Foundation, the UC President's Postdoctoral Fellowship Program, and the UCSD NIH IRACDA K12 program (GM068524). Research was supported by NIH grants HD071600 (V.N.), AI057153 (V.N.) and GM097509 (P.C.D) and GMS10RR029121 (P.C.D.). The mass spectrometry data is deposited in GNPS-MASSIVE (gnps.ucsd.edu). We thank Jack E. Dixon for allowing access to analytical instrumentation used to generate data in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes eight figures, one table, and Supplemental Experimental Procedures can be found with this article online at doi:________________.
REFERENCES
- Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61:3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- Dixit N, Kim MH, Rossaint J, Yamayoshi I, Zarbock A, Simon SI. Leukocyte function antigen-1, kindlin-3, and calcium flux orchestrate neutrophil recruitment during inflammation. J Immunol. 2012;189:5954–5964. doi: 10.4049/jimmunol.1201638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez DJ, Okumura CY, Hollands A, Kersten R, Akong-Moore K, Pence MA, Malone CL, Derieux J, Moore BS, Horswill AR, et al. Novel phenol-soluble modulin derivatives in community-associated methicillin-resistant Staphylococcus aureus identified through imaging mass spectrometry. J Biol Chem. 2012;287:13889–13898. doi: 10.1074/jbc.M112.349860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez DJ, Vuong L, Gonzalez IS, Keller N, McGrosso D, Hwang JH, Hung J, Zinkernagel A, Dixon JE, Dorrestein PC, et al. Phenol soluble modulin (PSM) variants of community-associated methicillin-resistant Staphylococcus aureus (MRSA) captured using mass spectrometry-based molecular networking. Mol Cell Proteomics. 2014;13:1262–1272. doi: 10.1074/mcp.M113.031336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harraghy N, Kerdudou S, Herrmann M. Quorum-sensing systems in staphylococci as therapeutic targets. Anal Bioanal Chem. 2007;387:437–444. doi: 10.1007/s00216-006-0860-0. [DOI] [PubMed] [Google Scholar]
- Huang TW, Chen FJ, Miu WC, Liao TL, Lin AC, Huang IW, Wu KM, Tsai SF, Chen YT, Lauderdale TL. Complete genome sequence of Staphylococcus aureus M013, a PVL-positive, ST59-SCCmec type V strain isolated in Taiwan. J Bacteriol. 2012;194:1256–1257. doi: 10.1128/JB.06666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junio HA, Todd DA, Ettefagh KA, Ehrmann BM, Kavanaugh JS, Horswill AR, Cech NB. Quantitative analysis of autoinducing peptide I (AIP-I) from Staphylococcus aureus cultures using ultrahigh performance liquid chromatography-high resolving power mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;930:7–12. doi: 10.1016/j.jchromb.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh JS, Thoendel M, Horswill AR. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol Microbiol. 2007;65:780–798. doi: 10.1111/j.1365-2958.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- Kennedy AD, Porcella SF, Martens C, Whitney AR, Braughton KR, Chen L, Craig CT, Tenover FC, Kreiswirth BN, Musser JM, et al. Complete nucleotide sequence analysis of plasmids in strains of Staphylococcus aureus clone USA300 reveals a high level of identity among isolates with closely related core genome sequences. J Clin Microbiol. 2010;48:4504–4511. doi: 10.1128/JCM.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten RD, Yang YL, Xu Y, Cimermancic P, Nam SJ, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer D, Gleske AK, Rautenberg M, Wang R, Koberle M, Bohn E, Schoneberg T, Rabiet MJ, Boulay F, Klebanoff SJ, et al. Human formyl peptide receptor 2 senses highly pathogenic Staphylococcus aureus. Cell Host Microbe. 2010;7:463–473. doi: 10.1016/j.chom.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WH, Lien R, Huang YC, Lee WJ, Lai JY. Community-associated methicillin-resistant Staphylococcus aureus necrotizing pneumonia in a healthy neonate. J Microbiol Immunol Infect. 2012 doi: 10.1016/j.jmii.2012.07.001. S1684-1182(12)00145-4. [DOI] [PubMed] [Google Scholar]
- McIlwain S, Mathews M, Bereman MS, Rubel EW, MacCoss MJ, Noble WS. Estimating relative abundances of proteins from shotgun proteomics data. BMC Bioinformatics. 2012;13:308. doi: 10.1186/1471-2105-13-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasami S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, Cheung GY, Otto M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci USA. 2012;109:1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pragman AA, Schlievert PM. Virulence regulation in Staphylococcus aureus: the need for in vivo analysis of virulence factor regulation. FEMS Immunol Med Microbiol. 2004;42:147–154. doi: 10.1016/j.femsim.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick RP. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- Schwartz K, Sekedat MD, Syed AK, O'Hara B, Payne DE, Lamb A, Boles BR. The AgrD N-terminal leader peptide of Staphylococcus aureus has cytolytic and amyloidogenic properties. 2014;82:3837–3844. doi: 10.1128/IAI.02111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- Thoendel M, Horswill AR. Identification of Staphylococcus aureus AgrD residues required for autoinducing peptide biosynthesis. J Biol Chem. 2009;284:21828–21838. doi: 10.1074/jbc.M109.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaruz AE, Bubeck Wardenburg J, Khan BA, Whitney AR, Sturdevant DE, Gardner DJ, DeLeo FR, Otto M. A point mutation in the agr locus rather than expression of the Panton-Valentine leukocidin caused previously reported phenotypes in Staphylococcus aureus pneumonia and gene regulation. J Infect Dis. 2009;200:724–734. doi: 10.1086/604728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci U S A. 2012;109:E1743–1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JS, 3rd, Traber KE, Corrigan R, Benson SA, Musser JM, Novick RP. The agr radiation: an early event in the evolution of staphylococci. J Bacteriol. 2005;187:5585–5594. doi: 10.1128/JB.187.16.5585-5594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lin J, Ji G. Membrane anchoring of the AgrD N-terminal amphipathic region is required for its processing to produce a quorum-sensing pheromone in Staphylococcus aureus. 2004;279:19448–19456. doi: 10.1074/jbc.M311349200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.