Abstract
Mounting evidence has demonstrated that a specialized extracellular matrix exists in the mammalian brain, and this glycoprotein-rich matrix contributes to many aspects of brain development and function. The most prominent supramolecular assembly of these ECM glycoproteins are perineuronal nets, specialized lattice-like structures that surround the cell bodies and proximal neurites of select classes of interneurons. Perineuronal nets are composed of lecticans – a family of chondroitin sulfate proteoglycans [CSPGs] that includes aggrecan, brevican, neurocan, and versican. The presence of these lattice-like structures emerge late in postnatal brain development and coincides with the ending of critical periods of brain development. Despite our knowledge of the presence of lecticans in perineuronal nets and their importance in regulating synaptic plasticity, we know little about the development or distribution of extracellular proteases that are responsible for their cleavage and turnover. A subset of the large “A Disintegrin and Metalloproteinase with Thrombospondin Motifs” (ADAMTS) family of extracellular proteases is responsible for endogenously cleaving lecticans. We therefore explored the expression pattern of 2 aggrecan-degrading ADAMTS family members, ADAMTS15 and ADAMTS4, in hippocampus and neocortex. Here, we show that both lectican-degrading metalloproteases are present in these brain regions and each exhibits a distinct temporal and spatial expression pattern. Adamts15 mRNA is expressed exclusively by Parvalbumin-expressing interneurons during synaptogenesis, whereas, Adamts4 mRNA is exclusively generated by telencephalic oligodendrocytes during myelination. Thus, ADAMTS15 and ADAMTS4 not only exhibit unique cellular expression patterns but their developmental upregulation by these cell types coincides with critical aspects of neural development.
Keywords: Extracellular matrix, perineuronal net, hippocampus, neocortex, aggrecanase, ADAMTS, parvalbumin, aggrecan, RRID:nif-0000-00509, RRID:AB_94270, RRID:AB_2314070, RRID:AB_2068506, RRID:AB_2314535, RRID:AB_221569, RRID:AB_2278725, RRID:AB_839506, RRID:AB_570666, RRID:AB_2255374, RRID:IMSR_JAX:008069, RRID:IMSR_JAX:005630
INTRODUCTION
Proteins embedded within the extracellular matrix (ECM) impact nearly all biological functions (Rozario and DeSimone 2010). Outside of the nervous system, major constituents of the ECM include laminins, fibrillar and non-fibrillar collagens, nidogens, fibronectins, tenascins, and proteoglycans (i.e. core proteins with at least one covalently linked glycosaminoglycan side chain)(LeBleu et al. 2007; Frantz et al. 2010). A hallmark feature of these proteins is their ability to bind other ECM constituents and assemble into distinct supramolecular structures, such as fibrils, filaments, basement membranes and basal laminas. ECM molecules within these supramolecular structures bind and harbor a variety of growth factors, morphogens and metalloproteases. These metalloproteases not only liberate growth factors and morphogens from the ECM but also contribute to matrix turnover, matrix remodeling and the liberation of bio-active fragments from full-length ECM molecules (Werb and Chin 1998; Ricard-Blum and Salza 2014; Sternlicht and Werb 2001; Hynes 2009). Evidence for the importance of these molecules and supramolecular structures outside of the nervous system are ample: the disruption or dysregulation of these proteins (or the genes encoding them) results in devastating diseases (Bateman et al. 2009). However, despite early reports of ECM in brain neuropil, its presence and function in mammalian brain development and function remained controversial until only recently (Tani and Ametani 1971; Reichardt and Prokop 2011). Now, a multitude of studies have revealed regulatory roles of ECM molecules in many aspects of brain development, including neuronal and glial migration, axonal growth and targeting, synaptogenesis, synaptic plasticity, memory storage, myelination, blood brain barrier formation and maintenance, maintenance of the neural stem cell niche, and cellular responses following brain injury (Hedstrom et al. 2007; Karetko and Skangiel-Kramska 2009; Reichardt and Prokop 2011; Franco and Muller 2011; Wlodarczyk et al. 2011; Frischknecht and Gundelfinger 2012).
Although extracellular space comprises 40% of the juvenile brain parenchyma and 20% of the adult brain parenchyma (Nicholson and Sykova 1998), the supramolecular assembly of ECM components differs significantly in the mammalian brain from other tissues. Basement membranes and basal laminas are absent from brain parenchyma (except in association with blood vessels) and instead the most prominent ECM assembly in the adult mammalian brain is the perineuronal net – a specialized lattice-like structure that surrounds the soma and proximal neurites of select classes of neurons, such as Parvalbumin-expressing, fast-spiking basket cells of the hippocampus and neocortex (Celio et al. 1998; Hartig et al. 1999). Perineuronal nets appear late in postnatal development and their appearance coincides with the end of the critical period of development and activity-dependent refinement of immature neural circuits (Dityatev et al. 2010). Experimental approaches that enzymatically digest perineuronal nets reopen critical periods, indicating that these supramolecular assemblies contribute to synaptic plasticity within the brain (for review see Zimmermann and Dours-Zimmermann 2008; Karetko and Skangiel-Kramska 2009; Dityatev et al. 2010). It has also been hypothesized that by limiting neuroplasticity at synaptic sites, the relatively stable perineuronal nets may act as molecular sites for very long-term memory storage (Gogolla et al. 2009; Tsien 2013). Although the literature is rich with information regarding perineuronal nets, these structures are not the sole sites of ECM accumulation in the adult brain. Aggregates of ECM molecules are present within the synaptic clefts of the adult brain (Dityatev et al. 2006, 2010), at nodes of Ranvier (Oohashi et al. 2002; Hedstrom et al. 2007; Susuki et al. 2013), in axonal tracts (Ogawa et al. 2001), and in specialized structures that line the ventricular system, called fractones (Mercier et al. 2002; Kerever et al. 2007). During development the distribution of ECM within the brain appears even more widespread but its assembly into supramolecular structures is less obvious and therefore remains unclear (e.g., Ogawa et al. 2001; Brooks et al. 2013).
Not only is the supramolecular assembly of the ECM different in brain, the molecular composition of ECM structures also differs significantly in brain parenchyma. Although laminins, type IV collagen, nidogens and fibronectins are present in the brain (Libby et al. 1999; Egles et al. 2007; Vasudevan et al. 2010; Labelle-Dumais et al. 2011), they are less abundant than other ECM components (Sanes 1989). The most abundant molecules in brain ECM are lecticans (a family of chondroitin sulfate proteoglycans [CSPGs] that includes aggrecan, brevican, neurocan, and versican) and their binding partners – which include link proteins (or hyaluronan and proteoglycan binding link proteins [HAPLNs]; Spicer et al. 2003; Carulli et al. 2010), tenascins, and hyaluronan (Zimmermann and Dours-Zimmermann 2008). Lecticans and their binding partners are concentrated in perineuronal nets of the adult brain (Zimmermann and Dours-Zimmermann 2008; Karetko and Skangiel-Kramska 2009). Although the specific molecular composition of perineuronal nets differs between brain regions and the cell-type ensheathed by this specialized matrix, aggrecan appears to be the most common component of perineuronal nets (Zimmermann and Dours-Zimmermann 2008).
Since increasing evidence suggests that lectican-rich perineuronal nets are regulators of synaptic plasticity and candidates for closing critical periods, we have become interested in what regulates the distribution and turnover of these molecules. Outside of the nervous system, the major mechanism to control lectican turnover is proteolysis by endogenous metalloproteases. While many families of extracellular metalloproteases cleave ECM molecules (such as Matrix Metalloproteinases [MMPs] and A Disintegrin and Metalloproteinases [ADAMs]), it is a subset of A Disintegrin and Metalloproteinases with Thrombospondin Repeats (ADAMTSs) that are responsible for cleaving lecticans: ADAMTS1,4,5,8,9, and 15 (Abbaszade et al. 1999; Tortorella et al. 1999; Tang 2001; Porter et al. 2005). Despite our knowledge of lectican distribution and function in the developing, adult and injured brain (Zimmermann and Dours-Zimmermann 2008), we know little about the spatial and temporal expression of these lectican-cleaving ADAMTS proteases. With this in mind we set out to characterize the cellular and developmental expression patterns of ADAMTSs in the hippocampus and neocortex, two telencephalic brain regions with abundant perineuronal nets. We focused our attention on ADAMTS15 and ADAMTS4 based upon previous studies documenting their expression in brain (Tang 2001; Yuan et al. 2002) and their upregulation in hippocampus and neocortex following excitotoxic lesions or pathological conditions (Yuan et al. 2002; Mayer et al. 2005; Cross et al. 2006; Venugopal et al. 2012). Here, we show that both ADAMTS15 and ADAMTS4 are developmentally regulated in the mouse hippocampus and neocortex and each are generated by distinct types of neural cells. ADAMTS15 is generated by Parvalbumin-expressing, fast-spiking basket cells (the same cells that are ensheathed by aggrecan-rich perineuronal nets), whereas, ADAMTS4 is generated exclusively by oligodendrocytes in the developing telencephalon.
METHODS
Animals
CD1 and C57/B6 mice were obtained from Harlan (Indianapolis, IN). Parv-cre (RRID:IMSR_JAX:008069) and Thy1-STOP-YFP15 (RRID:IMSR_JAX:005630) mice were obtained from Jackson Labs (Bar Harbor, ME; Stock numbers 008069 and 005630, respectively). Genomic DNA was isolated using the HotSHOT method (Truett et al. 2000) and genotyping was performed with the following primers: yfp, AAG TTC ATC TGC ACC ACC G and TCC TTG AAG AAG ATG GTG CG; cre, TGC ATG ATC TCC GGT ATT GA and CGT ACT GACGGT GGG AGA AT. The following cycling conditions were used for yfp, 35 cycles using a denaturation temperature of 94°C for 30 seconds, annealing at 55°C for one minute, and elongation at 72°C for one minute. The following cycling conditions were used for cre, 35 cycles using a denaturation temperature of 95°C for 30 seconds, annealing at 52°C for 30 seconds, and elongation at 72°C for 45 seconds. All analyses conformed to National Institutes of Health guidelines and protocols approved by the Virginia Polytechnic Institute and State University Institutional Animal Care and Use Committee.
Reagents
All chemicals and reagents were obtained from Fisher (Fairlawn, NJ) or Sigma (St. Louis, MO) unless otherwise noted. All DNA primers were obtained from Integrated DNA Technologies (Coralville, IA).
Antibodies
Monoclonal and polyclonal antibodies used in these studies are listed and described in Table 1. Fluorophore-conjugated secondary antibodies were obtained from Molecular Probes/Invitrogen (Grand Island, NY) and were applied at 1:1000 dilutions. Horseradish peroxidase (HRP)-conjugated antibodies against digoxigenin (DIG) or fluorescein (FL) were obtained from Roche (Mannheim, Germany) and were used at 1:1000.
Table 1.
Antibodies used in these studies
| Antigen | Isotype | Description of immunogen | Source/Catalogue number (RRID number) | Dilution For IHC | References documenting antibody specificity |
|---|---|---|---|---|---|
| Aggrecan (Cat315) | Mouse IgM | Feline brain proteoglycans | Millipore# MAB1581 (RRID:AB_94270) | 1:2000 | Lander et al. 1998; Brooks et al. 2013 |
| Calbindin | Rabbit polyclonal | Rat calbindin D28k | Swant,# CB-38a (RRID:AB_2314070) | 1:500 | Sloviter, 1989; Su et al. 2010 |
| Calretinin | Rabbit polyclonal | Recombinant rat calretinin | Millipore #AB5054 (RRID:AB_2068506) | 1:1000 | Liu et al. 2003; Su et al. 2010 |
| Glial fibrillary acidic protein (GFAP) | Rabbit polyclonal | GFAP isolated from cow spinal cord | DAKO #Z0334 (RRID:AB_2314535) | 1:1000 | Su et al. 2010 |
| Green Fluorescent Protein (GFP) | Rabbit polyclonal | GFP isolated from Aequorea vicotria | Life Technologies; #A-11122 (RRID:AB_221569) | 1:250 | Carlsson et al. 2010; Brooks et al. 2013 |
| Glutamate decarboxylase 67 (Gad 67) | Mouse IgG2a | Recombinant Gad67 | Millipore #MAB5406 (RRID:AB_2278725) | 1:500 | Varea et al. 2005; Su et al. 2010 |
| Ionizing calcium- binding adaptor 1 (Iba-1) | Rabbit polyclonal | Synthetic peptide corresponding to the first 29 amino acids of Iba-1 | WAKO #016-20001 (RRID:AB_839506) | 1:500 | Imai et al. 1996; Su et al. 2010 |
| Oligodendrocyte Transcription Factor 2 (Olig2) | Rabbit polyclonal | Recombinant mouse Olig2 | Millipore #AB9610 (RRID:AB_570666) | 1:50 | Heimer-McGinn and Young 2011; Goemaere et al. 2012 |
| Somatostatin | Rabbit polyclonal | Somatostatin conjugated to BSA | Millipore #AB5494 (RRID:AB_2255374) | 1:250 | Oliva et al. 2000; Su et al. 2010 |
Antibody characterizations
Aggrecan
The mouse anti-chondroitin sulfate proteoglycan (CSPG) protein antibody (Cat315)(RRID:AB_94270) detects a >500 kDa band in Western blots and specifically recognizes an oligosaccharide epitope on aggrecan in the developing and adult brain (Matthews et al. 2002). Analysis in aggrecan-deficient mutant mice confirmed the specificity of Cat315 for aggrecan (Matthews et al. 2002; Brooks et al. 2013).
Calbindin
The calbindin D-28k antibody (RRID:AB_2314070) recognizes a single band of ~28 kDa on Western blots of mouse and rat brain extracts (see manufacturer’s datasheet) and stained cells in the rodent hippocampus that were identical to previous reports (Sloviter 1989; Danglot et al. 2006; Klausberger and Somogyi 2008). Immunoreactivity of anti-Calbindin in IHC was marginally enhanced following in situ hybridization (ISH) (Su et al. 2010).
Calretinin
The calretinin antibody (RRID:AB_2068506) recognizes a single band of ~30 kDa on Western blots with mouse hippocampal extracts (unpublished observation, J.S. and M.A.F.) and stained cells in the mouse hippocampus and neocortex that were identical to previous reports (Liu et al. 2003; Xu et al. 2004; Danglot et al. 2006; Klausberger and Somogyi 2008). Immunoreactivity of anti-Calretinin in IHC was marginally enhanced following in situ hybridization (ISH) (Su et al. 2010).
GFAP
The GFAP antibody (RRID:AB_2314535) recognizes several bands around ~50 kDa in brain extracts which correspond to different isoforms of GFAP (Kamphuis et al. 2012) and, as expected, fails to detect protein in synaptically-enriched biochemical fractions purified from cerebellar extracts (Su et al. 2012). Previous IHC studies have demonstrated that this antibody specifically labels GFAP in transfected cells in vitro or in astrocytes in vivo (Su et al. 2010; Kamphuis et al. 2012).
Gad67
The Gad67 antibody (RRID:AB_2278725) detects a single band of ~68 kDa in rat and mouse brain extracts and has no detectable cross-reactivity with Gad65 (manufacturer’s datasheet; see also Varea et al. 2005). Previous IHC studies demonstrated this antibody labels interneurons and inhibitory synapses in mouse cortex (Xu et al. 2006). Here, IHC with anti-Gad67 labeled similar, albeit less, interneurons and synaptic structures in hippocampus as anti-Gad65/67 (Su et al. 2010).
Iba-1
The Iba-1 antibody (RRID:AB_839506) detects microglia and macrophages but not neural cells (see manufacturer’s datasheet). In our hands this antibody labeled small cells in mouse hippocampus that resembled microglia, which matched previous descriptions (Imai et al. 1996; Su et al. 2010).
Olig2
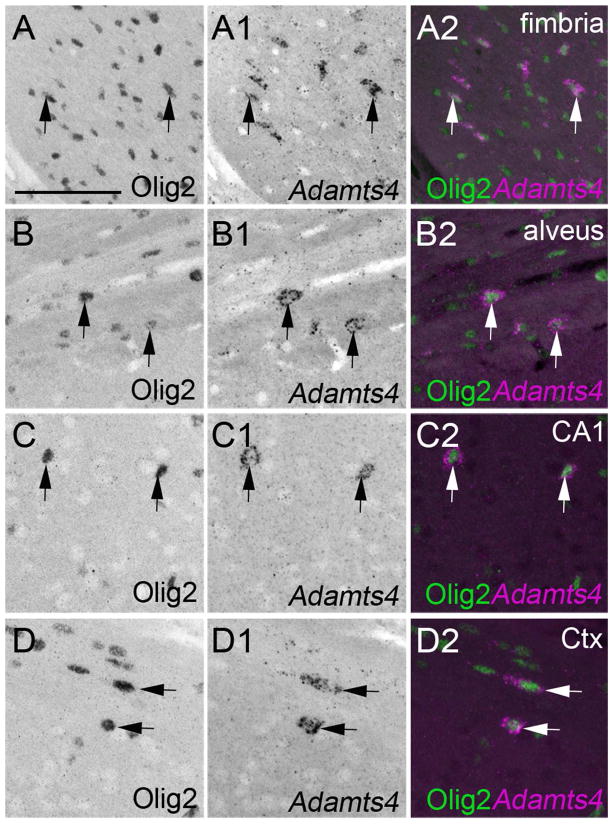
The Olig2 antibody (RRID:AB_570666) detects a 32 kDa protein in mouse and rat brain lysates (manufacturer’s datasheet). Immunoreactive cells labeled with this antibody co-expressed O1, a marker of differentiated oligodendrocytes (Xiao et al. 2012) and in this study were abundant in regions of the telencephalon that contained myelinated axons and abundant oligodendrocytes (Figure 7).
Figure 7.
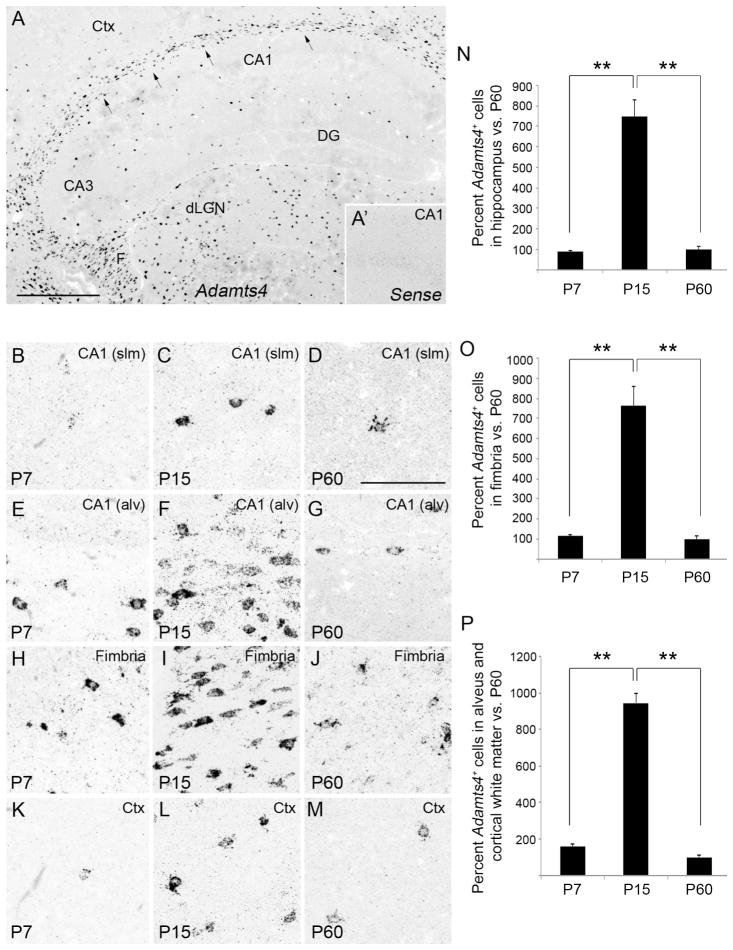
Developmental expression of Adamts4 mRNA in mouse hippocampus. A. Localization of Adamts4 mRNA by in situ hybridization in coronal sections of P15 mouse hippocampus. Arrows highlight Adamts4 expression in the alveus and cortical white matter. A1. Shows an image of in situ hybridization with sense control probes in the CA1 region of a P15 mouse hippocampus. B–M. High magnification images of Adamts4 mRNA in the stratum lacunosum-molecular of CA1 (B–D), the alveus of CA1 (E–G), the fimbria (H–J) and layer V of neocortex (K–M) of P7, P15 and P60 mice. N. Percent of Adamts4-expressing cells in hippocampus, labeled by in situ hybridization, compared to numbers of Adamts4-expressing cells in adult (P60) hippocampus. **Differs from indicated samples at P<0.001 by Tukey-Kramer Difference Between Means Test. Error bars indicate SEM. O,P. Percent of Adamts4-expressing cells in the fimbria (D) or alveus and cortical white matter (E), compared to adult (P60) numbers. **Differs from indicated samples at P<0.001 by Tukey-Kramer Difference Between Means Test. Error bars indicate SEM. CA, Cornu Ammonis areas; Ctx, Neocortex; DG, Dentate Gyrus; dLGN, Dorsal Lateral Geniculate Nucleus; F, fimbria; slm, stratum lacunosum-moleculare. Scale bar in A = 400 μm and in B = 100 μm.
Som
In IHC, the Som antibody (RRID:AB_2255374) labels subsets of interneurons that were distributed as previously reported in mouse hippocampus, cortex, and retina (Oliva et al. 2000; Xu et al. 2004, 2006; Danglot et al. 2006; Klausberger and Somogyi 2008; Acosta et al. 2008). Previous studies demonstrated that immunoreactivity was inhibited by preincubation with non-labeled somatostatin (Arimura et al. 1975). Importantly, this antibody worked poorly with our standard IHC protocol, however immunolabeling was enhanced after ISH (Su et al. 2010).
Fluorescent in situ hybridization
Fluorescent in situ hybridization was performed on 16μm cryosectioned coronal sections as previously described (Su et al. 2010, 2011, 2013; Brooks et al. 2013). Sense and anti-sense riboprobes were generated against Syt1, Parv, Adamts4 and Adamts15. Image Clones of the entire coding region of Syt1, Adamts4, and Adamts15 were purchased from Open Biosystems (Huntsville, AL; Clone ID Syt1: 5363062; Adamts4: 5345809; Adamts15: 30619053). An 800 bp fragment of Parv (corresponding to nucleotides 2–825) was PCR-cloned into pGEM Easy T vector (Promega, Madison, WI) with the following primers: TCT GCT CAT CCA AGT TGC AG and TCC TGA AGG ACT CAA CCC C. Riboprobes were synthesized using digoxigenin (DIG)- or fluorescein (FL)- labeled UTP (Roche, Mannheim, Germany) and the MAXIscript in vitro Transcription Kit (Ambion, Austin, TX). Riboprobes were hydrolyzed into ~0.5 kb fragments. Coronal brain sections were prepared and hybridized as described previously (Fox and Sanes 2007; Su et al. 2010). Bound riboprobes were detected by horseradish peroxidase (Hrp)-conjugated anti-DIG or anti-FL antibodies and fluorescent staining with Tyramide Signal Amplification (TSA) systems (PerkinElmer, Shelton, CT). As controls, sense riboprobes were generated and tested on postnatal day 14 (P14) coronal brain sections. No signal was detected for sense riboprobes against sequences from Syt1, Parv, Adamts4 or Adamts15 Image Clones. For cell counts all images were obtained with identical parameters and to ensure similar regions were analyzed in all ages and regions, we only analyzed coronal sections of hippocampus that also contained central regions of visual thalamus – which includes both dorsal and ventral lateral geniculate nuclei (dLGN and vLGN, respectively). For quantification purposes all riboprobe-labeled cells in the entire hippocampus of these sections were counted manually and at least two sections were analyzed per animal (from both hemispheres). In each animal the mean number of riboprobe-positive cells per hippocampal section was calculated. For statistical analysis, the mean number of riboprobe-positive cells per hippocampal section was averaged and analyzed from at least three animals per age. Limiting analysis to identical regions of hippocampus reduced potential errors from counting different sized regions of hippocampus. Likewise, all images and analysis of expression in neocortex were obtained from layer V of regions of neocortex dorsal to hippocampus in coronal sections that contained dLGN and vLGN.
For double FISH (D-FISH), after the first TSA reaction sections were washed in TBS, incubated in 0.3% H2O2 for 30 min, and reacted with the second POD-conjugated antibody. For fluorescent in situ hybridization coupled with immunohistochemistry (IHC), standard IHC was performed after completing the TS amplification step described above. Images were obtained on a Zeiss AxioImager A2 fluorescent microscope or a Zeiss LSM700 confocal microscope. A minimum of three animals per age was compared in ISH experiments.
Immunohistochemistry (IHC)
IHC was performed as previously described (Su et al. 2010, 2011). After mice were transcardially perfused with phosphate-buffered saline (PBS; pH 7.4) and 4% paraformaldehyde in PBS (PFA; pH 7.4), brains were removed and post-fixed in PFA for 12 hours at 4°C. Brains were cryopreserved in 25–30% sucrose solution for 24 hours, embedded in Tissue Freezing Medium (Electron Microscopy Sciences, Hatfield, PA), and cryosectioned (16μm coronal sections). Sections were air-dried onto Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA) before incubating in blocking buffer (2.5% BSA, 5% Normal Goat Serum [NGS], 0.1% Triton-X in PBS for at least 30 minutes). Primary antibodies were diluted in blocking buffer and incubated on the tissue sections for >12 hours at 4°C. After washing in PBS, fluorophore-conjugated secondary antibodies diluted 1:1000 in blocking buffer were incubated on sections for 1 hour at room temperature. After thorough washing in PBS, sections were stained with DAPI (1:10,000 in water) and were mounted with Vectashield (Vector Laboratories, Burlingame, CA). Images were acquired on a Zeiss AxioImager A2 fluorescent microscope a Zeiss LSM 700 confocal microscope.
RNA isolation and quantitative reverse transcriptase polymerase chain reactions (qPCR)
RNA was isolated using the BioRad Total RNA Extraction from Fibrous and Fatty Tissue Kit (BioRad, Hercules, CA). For hippocampus and neocortex, each sample contained RNA isolated from an individual wild-type mouse (and from the entire hippocampus or neocortex). Since three samples were analyzed per age, a total of 3 mice were analyzed per age. For dLGN, each sample contained RNA isolated from three littermate wild-type mice and 3 samples were analyzed per age (so a total of 9 mice per age were used to generate dLGN samples). RNA concentrations were determined spectro-photometrically with a NanoDrop ND-1000 (Wilmington, DE). cDNAs were generated from 200 ng of RNA with the Superscript II Reverse Transcriptase First Strand cDNA Synthesis Kit (Life Technologies, Carlsbad, CA). qPCR was performed on a CFX Connect Real-Time PCR Detection System (BioRad, Hercules, CA) using iTaq Universal SYBR Green Supermix (BioRad, Hercules, CA) as described previously (Su et al. 2010; Brooks et al. 2013). The following primer pairs were used: Adamts1, GGA CAC AAA TCG CTT CTT CC and CAG AAC ACC CGG AAC CAG T; Adamts4, CCA GGA AAA GTC GCT GGT AG and AGT GCC CGA TTC ATC ACT G; Adamts5, GTC ACA TGA ATG ATG CCC AC and CAA ATG GCA GCA CCA ACA TA; Adamts8, ATC ACC GTG AGG ATG TGG TT and CAA GAG GTT TGT GTC CGA GG, Adamts15, ACA CTG CCA TCC TCT TCA CC and TCT TGG GGT CAC ACA TGG TA; Gapdh, CGT CCC GTA GAC AAA ATG GT and TTG ATG GCA ACA ATC TCC AC. The following cycling conditions were used with 20 ng of RNA: 95°C for 30 seconds, followed by 40 cycles of amplification (94°C for 5 seconds, 60°C for 30 seconds, 72°C for 20 seconds, read plate) and a melting curve analysis. Relative quantities of RNA were determined using the ΔΔ-CT method (Livak and Schmittgen, 2001) and were normalized to levels of Gapdh in each sample. To ensure specificity of cDNA amplification in qPCR we: 1). designed primers to span large introns; 2). performed melting curve analysis following each qPCR reaction; 3). ran qPCR products on agarose gels to ensure amplification of single products. A minimum of 3 independent samples (each in triplicate) were run for each gene and at each age examined.
RESULTS
Neuronal expression of Adamts15 in hippocampus and neocortex
To assess the distribution of Adamts15 mRNA in mouse telencephalon we generated riboprobes against Adamts15 and performed in situ hybridization on coronal sections of P14 mouse brains. In contrast to the robust distribution of Adamts15-expressing cells in dorsal thalamus (dT) and the dorsal lateral geniculate nucleus (dLGN)(Brooks et al. 2013), we observed a sparse distribution of Adamts15-expressing cells in hippocampus and neocortex (Figure 1B). To quantitate levels of Adamts15 expression in hippocampus and neocortex we performed quantitative reverse transcriptase polymerase chain reactions (qPCR) on RNA extracted from P7 and P15 mouse hippocampus and neocortex. At both ages, there was no significant differences in the level of Adamts15 mRNA in hippocampus or neocortex (at P7 Adamts15 mRNA expression in neocortex was 1.28+/−0.2 [SEM] fold higher than that in hippocampus, n=3, P=0.28 by T-test; at P15 Adamts15 mRNA expression in neocortex was 1.7+/−0.4 [SEM] fold higher than that in hippocampus, n=3, P=0.11 by T-test). In contrast, qPCR confirmed Adamts15 mRNA expression was dramatically higher in regions of dorsal thalamus compared with these regions of the telencephalon (at P7 Adamts15 mRNA expression was 22.0+/−2.6 [SEM] fold higher in dLGN than in hippocampus and 17.3+/− 2.0 [SEM] folder higher in dLGN than neocortex. n=3, P<0.0001 by T-test).
Figure 1.
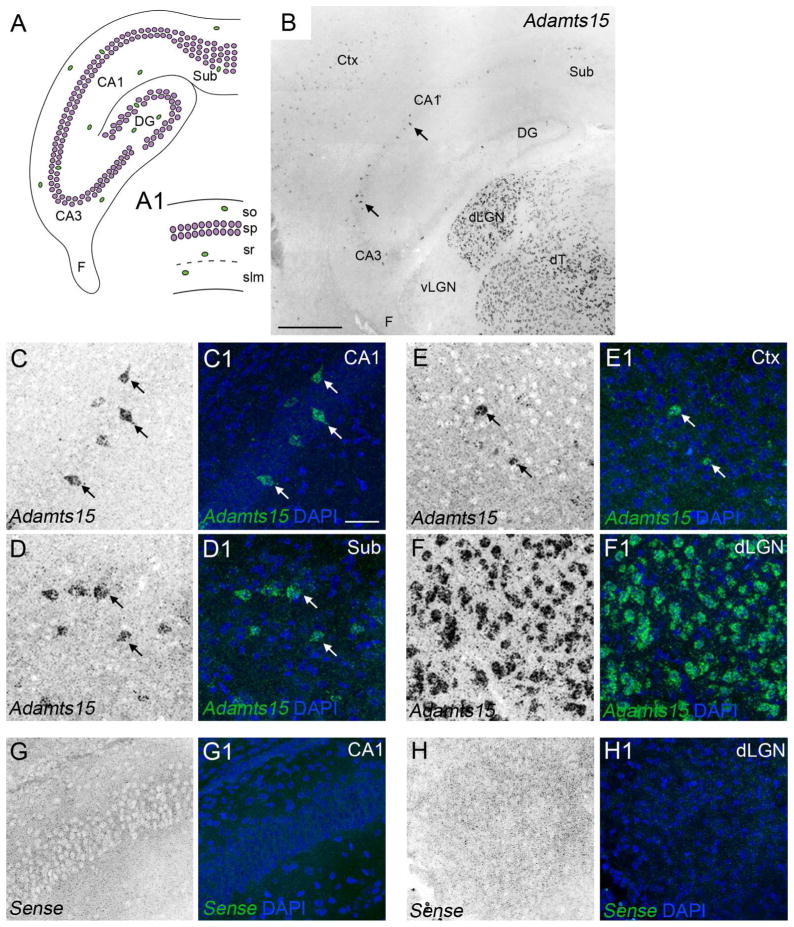
Sparse expression of Adamts15 mRNA in hippocampus and neocortex. A. Schematic representation of the mouse hippocampus. Excitatory neurons are depicted in magenta. Inhibitory neurons are depicted in green. A1 depicts a higher magnification view of the 4 distinct layers in the CA1 region of hippocampus. B. Localization of Adamts15 mRNA by in situ hybridization in coronal sections of P14 mouse hippocampus. C–F. Shows high magnification images of Adamts15 mRNA in CA1 (C), subiculum (D), layer V of neocortex (E), and dLGN (F) in P14 mouse brain. Cell nuclei are labeled by DAPI staining. Arrows indicate Adamts15 mRNA expression by sparse populations of cells in CA1, Sub, and Ctx. G,H. Shows high magnification images of in situ hybridization with sense control probes in coronal sections of P14 mouse hippocampus (G) and dLGN (H). CA, Cornu Ammonis areas; Ctx, Neocortex; DG, Dentate Gyrus; dLGN, Dorsal Lateral Geniculate Nucleus; dT, Dorsal Thalamus; F, Fimbria; slm, stratum lacunosum-moleculare; so, stratum oriens; sp, stratum pyramidale; sr, stratum radiatum; Sub, Subiculum. Scale bar in B = 400 μm and in C = 100 μm for C–H.
We next examined the distribution of Adamts15 mRNA expression to better understand its cellular origin. We focused much of our initial attention on Adamts15 expression in hippocampus because of its unique cytoarchitecture: the hippocampus is not only a layered structure but is subdivided into the dentate gyrus (DG), several Cornu Ammonis areas (CA1-4), and the subiculum (Sub)(Figure 1A). In each of these regions excitatory neurons are densely clustered into a cell-rich layer, which is called stratum pyramidale in CA1-4 and subiculum and is called stratum granulosum in DG. While inhibitory interneurons also reside in these cell-dense layers, they also populate the other hippocampal layers, which include strata oriens, radiatum and lacunosum-moleculare of CA1-4 (Figure 1A1), the molecular layer and hilus of the DG, and the polymorphic and molecular layer of the subiculum. Although sparsely distributed, Adamts15-expressing cells were observed in all hippocampal regions (Figure 1B), but the highest density appeared in, or adjacent to, stratum pyramidale in CA3, CA1, and subiculum (Figure 1C–F).
Next, we set out to determine whether Adamts15 mRNA was generated by telencephalic neurons. For this, we performed a double fluorescent in situ hybridization with DIG-labeled Adamts15 riboprobes and FL-conjugated riboprobes against Syt1, the gene that encodes Synaptotagmin 1 – a synaptic-vesicle associated calcium sensor expressed by neurons. We previously reported widespread Syt1 mRNA expression by both excitatory and inhibitory neurons in mouse hippocampus (Su et al. 2010). Here, we show that in all regions and layers of the P14 mouse hippocampus, cells expressing Adamts15 mRNA also contained Syt1 mRNA (Figure 2A–F). However, as expected from the sparse distribution of Adamts15 (Figure 1B), while nearly all Adamts15-expressing cells contained Syt1 mRNA (99.85%+/−0.1% [SEM] of Adamts15-expressing cells contained Syt1 mRNA; n=1294 cells from P7, P14, P21 and P56 hippocampi), not all Syt1-expressing cells contained Adamts15 mRNA (Figure 2A–F). These findings suggest that Adamts15 mRNA is generated by a small set of neurons in hippocampus.
Figure 2.
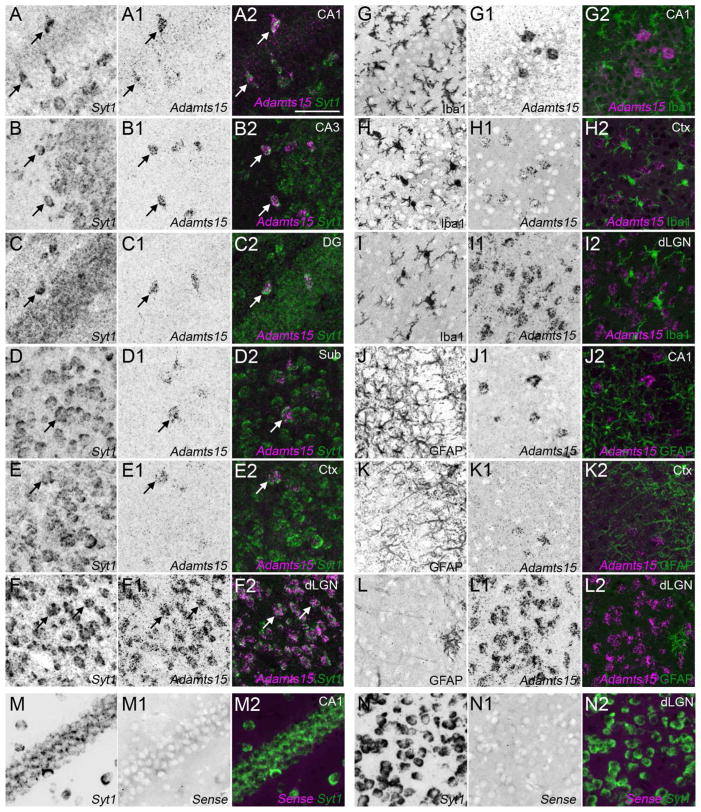
Neuronal expression of Adamts15 mRNA in hippocampus and neocortex. A–F. Double fluorescent in situ hybridization with riboprobes directed against Adamts15 and Syt1 show that in all regions of the P14 hippocampus (A–D), layer V of neocortex (E) and in dLGN (F) cells expressing Adamts15 also generate Syt1 mRNA. G–L. Fluorescent in situ hybridization coupled with immunostaining for Iba1 (G–I) or GFAP (J–L) revealed that neither Iba1-positive microglia or GFAP-positive astrocytes generate Adamts15 mRNA. M,N. Shows high magnification images of double in situ hybridization with Adamts15 sense control probes and Syt1 riboprobes in coronal sections of P14 mouse hippocampus (M) and dLGN (N). CA, Cornu Ammonis areas; Ctx, Neocortex; DG, Dentate Gyrus; dLGN, Dorsal Lateral Geniculate Nucleus; Sub, Subiculum. Scale bar in A = 100 μm for all panels.
To rule out expression of Adamts15 by glial cells we coupled in situ hybridization for Adamts15 mRNA with immunostaining for protein markers of astrocytes and microglial cells (i.e. glial fibrillary acidic protein [GFAP] and ionized calcium binding adaptor molecule 1 [Iba1], respectively). No Adamts15-expressing cells were observed to be immunoreactive for GFAP or Iba1 (Figure 2G–L), indicating that neither astrocytes nor microglial cells generate Adamts15 mRNA in mouse hippocampus. Taken together, our results indicate that Adamts15 mRNA expression is exclusively generated by a small subset of neurons in the hippocampus.
Based on these results, we also assessed the cellular origin of Adamts15 mRNA expression in neocortex. Identical expression patterns of Adamts15 mRNA were observed in P14 mouse neocortex as described above for hippocampus: all cortical Adamts15-expressing cells co-expressed Syt1 mRNA, not all Syt1-expressing cells generated Adamts15 mRNA (Figure 2A–F), and Adamts15-expressing cortical cells were not immunoreactive for astrocytic or microglial markers (Figure 2G–L).
Finally, we compared these telencephalic expression patterns with the distribution of Adamts15-expressing cells in dLGN. Just as we observed in neocortex and hippocampus, all Adamts15-expressing cells co-expressed Syt1 mRNA in dLGN and these cells were not immunoreactive for standard astrocytic or microglial markers (Figure 2G–L). However, in dLGN, most, if not all, Syt1-expressing cells generated Adamts15 mRNA (Figure 2A–F). Taken together these data suggest that in all regions examined Adamts15 mRNA is generated solely by neurons, but that in each regions different populations of neurons may generate Adamts15.
Parvalbumin-expressing hippocampal neurons generate Adamts15 mRNA
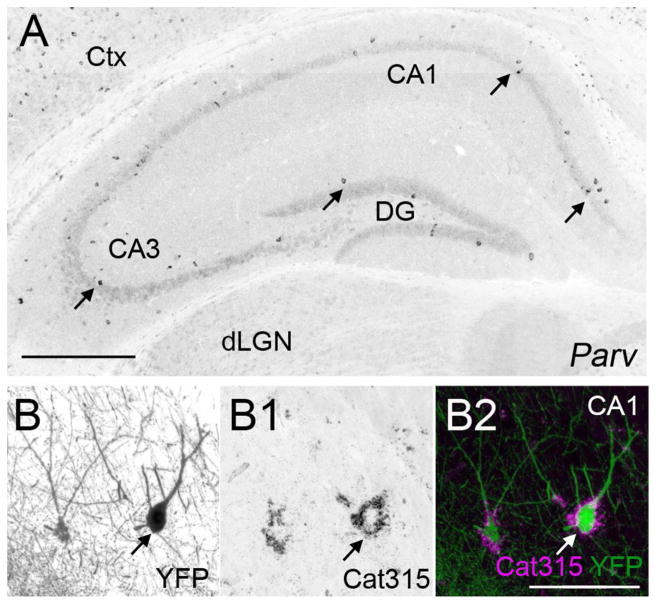
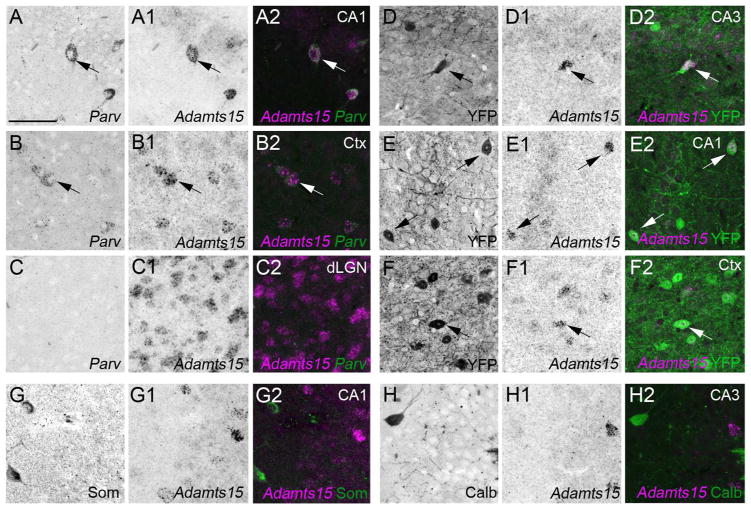
The sparse distribution and location of Adamts15 mRNA in hippocampus and neocortex suggested that its expression may be limited to interneurons, or even a single type of interneuron. Hippocampal and cortical interneurons can be divided into distinct subtypes based upon their morphology, location, electrophysiology, connectivity and neurochemical gene expression (Klausberger and Somogyi, 2008; Freund and Buzsaki 1996; McBain and Fisahn, 2001; Danglot et al. 2006). The enrichment of Adamts15 mRNA in select populations of hippocampal neurons in, or adjacent to, stratum pyramidale and in sparse populations of cortical neurons resembled the well-described distribution of fast-spiking, basket cells – a class of GABAergic interneurons that generate inhibitory axo-somatic synapses onto pyramidal cells. Fast-spiking basket cells in hippocampus and neocortex express the calcium binding protein Parvalbumin. In situ hybridization for Parv, the gene that encodes Parvalbumin, revealed strikingly similar patterns of expression of Adamts15 and Parv mRNA in hippocampus and neocortex (Figure 1B and 3A). The possibility that Parv+ interneurons expressed Adamts15, a known Aggrecan cleaving protease (Porter et al. 2005), was intriguing since these cells not only generate Aggrecan (Okaty et al. 2009) but are ensheathed in lectican-rich perineuronal nets (McRae et al 2010; Yamada and Jinno, 2013). Here, we demonstrate that genetically labeled Parv+ interneurons are ensheathed by lecticans detected with the monoclonal Cat315 antibody (Figure 3B) In postnatal brain Cat315 recognizes aggrecan protein (Matthews et al. 2002; Brooks et al. 2013).
Figure 3.
Parv+ interneurons share a similar expression pattern to Adamts15 mRNA and are ensheathed in aggrecan-rich perineuronal nets. A. Localization of Parv mRNA by in situ hybridization in coronal sections of P21 mouse hippocampus. B. Immunostaining revealed that Parv+ neurons in hippocampus are ensheathed by Aggrecan (Acan)-rich perineuronal nets in P37 hippocampus of Parv-Cre::Thy1-STOP-YFP15 mice. Parv+ neurons are genetically labeled with YFP in these mice. Scale bar in A = 400 μm and in B = 100 μm.
To test whether Adamts15 mRNA was expressed by Parv+ interneurons we performed double fluorescent in situ hybridization with FL-conjugated Adamts15 riboprobes and DIG-conjugated Parv riboprobes. Since Parv expression is developmentally regulated in the telencephalon and becomes upregulated by the beginning of the third postnatal week of mouse development (Del Rio et al. 1994; Jiang and Swann 2005), we began by assessing co-expression in P56 hippocampus. Nearly all Adamts15-expressing hippocampal neurons co-expressed Parv (in adult, 99% +/− 1% [SEM] Adamts15-expressing neurons co-expressed Parv; n=103 cells from 5 mice) (Figure 4A–C). We confirmed these results by assessing Adamts15 mRNA expression in transgenic mice in which Parv+ interneurons are selectively labeled with Yellow Fluorescent Protein (YFP) (Parv-Cre::Thy1-STOP-YFP15). In hippocampi from P37 Parv-Cre::Thy1-STOP-YFP15 mice, Adamts15 mRNA expressing neurons contained YFP, although not all YFP neurons contained Adamts15 mRNA (Figure 4D–E). Similarly, we observed a high degree of co-expression of Adamts15 and Parv in neocortex (by both double in situ hybridization and in Parv-Cre::Thy1-STOP-YFP15 brains)(Figure 4B,F). Although similar subtypes of neurons generate Adamts15 mRNA in the hippocampus and neocortex, these experiments also confirmed that different types of neurons are capable of generating Adamts15 in different brain regions: neurons expressing Adamts15 in dLGN do not co-express Parv mRNA (Figure 4C) and are not fluorescently labeled in Parv-Cre::Thy1-STOP-YFP15 mice (data not shown).
Figure 4.
Co-expression of Adamts15 and Parv mRNA by hippocampal and cortical interneurons. A–C. Double fluorescent in situ hybridization with riboprobes directed against Adamts15 and Parv show that in all regions of the P21 hippocampus (A) and neocortex (B) cells expressing Adamts15 also generate Parv mRNA (see arrows). In dLGN (C), Parv mRNA does not colocalize with Adamts15-expressing cells. D–F. Localization of Adamts15 mRNA by in situ hybridization in hippocampus and neocortex of P37 Parv-Cre::Thy1-STOP-YFP15 mice confirmed expression of Adamts15 mRNA by Parv+ neurons (see arrows). G,H. Adamts15 mRNA did not colocalize with markers for other types of hippocampal interneurons at P14, such as Calbindin (Calb) or Somatostatin (Som). CA, Cornu Ammonis areas; Ctx, Neocortex; DG, Dentate Gyrus; dLGN, Dorsal Lateral Geniculate Nucleus; Sub, Subiculum. Scale bar in A = 100 μm for all panels.
In the developing hippocampus and neocortex similar trends were observed as described above for adult tissues, however the percentage of co-expression of Adamts15 and Parv was slightly reduced (at P14, 81.4% +/− 10.5% [SEM] Adamts15-expressing neurons co-expressed Parv: n=280 cells from 6 mice). This may be due to the late upregulation of Parv in hippocampus and neocortex (Del Rio et al. 1994; Jiang and Swann 2005). Alternatively, the reduced co-expression of Adamts15 and Parv mRNAs may be due to transient expression of Adamts15 mRNA by other classes of interneurons early in development. To test the second of these possibilities, we assessed the co-expression of Adamts15 mRNA with Calbindin (Calb) and Somatostatin (Som), two neurochemicals generated by subtypes of interneurons that reside in regions of hippocampus containing Adamts15 expression (Figure 1B, Klausberger and Somogyi, 2008; McBain and Fisahn, 2001; Danglot et al. 2006). We observed little, if any, colocalization of Adamts15 mRNA with these classes of interneurons in the P10-P14 hippocampus (0% Adamts15-expressing neurons co-expressed Som: n=424 cells; 1.6% Adamts15-expressing neurons co-expressed Calb: n=503 cells)(Figure 4G,H). These data lend support to the notion that reduced co-expression of Adamts15 and Parv mRNAs by hippocampal interneurons during early development reflect the late onset of Parv expression, although it remains possible that other classes of interneurons generate Adamts15 mRNA early in development.
Developmental regulation of Adamts15 expression in hippocampus
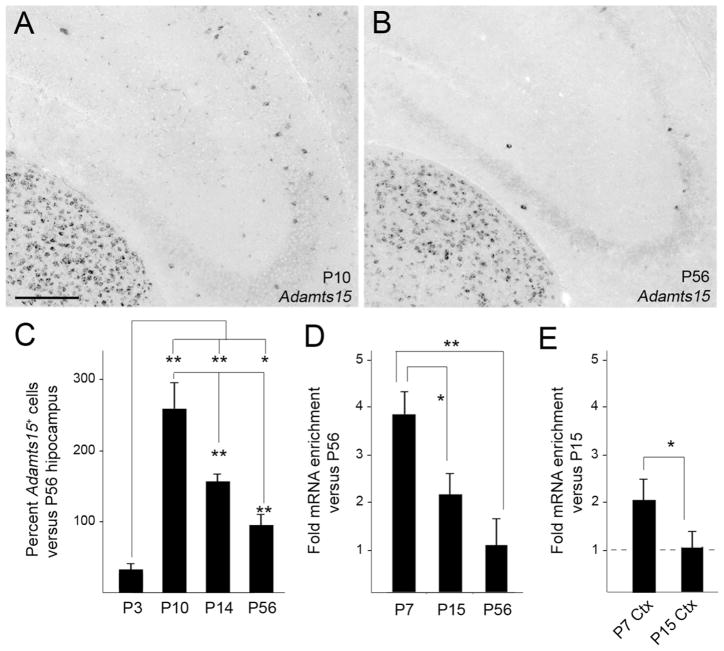
In addition to providing clear evidence that Adamts15 mRNA is generated by Parv+ interneurons, it appeared that more cells were expressing this aggrecanase early in development than in the adult hippocampus. To test whether Adamts15 mRNA expression is developmentally regulated in hippocampus we performed in situ hybridization and qPCR at ages corresponding to the migration of interneurons into the hippocampus (P3), to the peak period of synaptogenesis and circuit formation (P7-P10), to the maturation of hippocampal circuits (P14) and to the establishment of the adult-like hippocampus (P56) (Figure 5A,B and data not shown). In situ hybridizations revealed few if any cells containing Adamts15 mRNA at P3 (Figure 5C). In contrast, the highest number of Adamts15 mRNA containing cells in hippocampus was observed at P10. The number of Adamts15-expressing cells decreased in the P14 hippocampus and remained reduced (compared to the number at P10) into adulthood (Figure 4A–C).
Figure 5.
Developmental expression of Adamts15 mRNA in hippocampus. A,B. Localization of Adamts15 mRNA by in situ hybridization in coronal sections of P10 and P56 mouse hippocampus. C. Percent of Adamts15-expressing cells (labeled by in situ hybridization) compared to numbers in the adult (P56). Differs from indicated samples at P<0.001 (**) or P<0.05 (*) by Tukey-Kramer Difference Between Means Test. Error bars indicate Standard Error of the Mean (SEM). D. Quantitative RT-PCR (qPCR) shows that the highest levels of Adamts15 mRNA in the mouse hippocampus are present at P7. Levels of Adamts15 mRNA were normalized to Gapdh levels in each sample. Differs from indicated samples at P<0.001 (**) or P<0.05 (*) by Tukey-Kramer Difference Between Means Test. n=3 mice, per age. Error bars indicate SEM. E. qPCR shows the developmental pattern of Adamts15 mRNA expression follows a similar trend: Cortical levels of Adamts15 mRNA are higher at P7 than at P15. *Differs from P15 at P<0.05 by Tukey-Kramer Difference Between Means Test. n=3 mice, per age. Error bars indicate SEM. Scale bar in A = 200 μm for A,B.
To quantitate the overall levels of Adamts15 mRNA in the developing hippocampus we used qPCR. Hippocampal RNA was isolated from P7, P15 and P56 hippocampi. Highest levels of Adamts15 mRNA expression were observed at P7, which were nearly 4 fold higher than adult (P56) levels and 2 fold higher than levels at P15 (Figure 5D). Adamts15 expression in neocortex followed a similar trend, with highest levels at the end of the first week of postnatal hippocampal development (Figure 5E).
Taken together, our results reveal that a select subset of telencephalic interneurons expresses Adamts15 mRNA and that its expression by these cells coincides with periods of neural circuit formation and synaptogenesis. Interestingly, we previously observed similar developmental patterns of Adamts15 mRNA expression in dLGN coinciding with visual system circuit formation (Brooks et al. 2013).
Developmental expression of other aggrecan-cleaving ADAMTS proteases in mouse hippocampus
The distinct spatiotemporal expression of ADAMTS15 suggests a unique role for this extracellular protease in hippocampal development. However, since several other ADAMTSs cleave aggrecan, are enriched in brain (Tang 2001, Mayer et al. 2005; Yuan et al. 2002), and are potentially generated by interneurons (Okaty et al. 2009), we sought to determine if other members of the aggrecanase subfamily of ADAMTSs were present in the developing hippocampus. qPCR indicated that mRNA from four other members of this subfamily (i.e. ADAMTS1,4,5, and 8) were present in adult hippocampus. In regards to their developmental expression patterns, Adamts1 and Adamts8 mRNA levels changed little from P7 to P56, whereas Adamts4 and Adamts5 both exhibit dynamic expression profiles during this period. Like Adamts15, Adamts5 mRNA expression was highest at P7 and quickly decreased by the end of the second postnatal week of development (Figure 6). Previous single-cell transcriptome studies indicate that this metalloprotease may be generated by fast-spiking Parv+ cortical interneurons and its expression decreases throughout development (Okaty et al. 2009), although Adamts5 mRNA is not observable in the hippocampus in on-line in situ hybridization databases (http://mouse.brain-map.org/)(RRID:nif-0000-00509). In contrast to Adamts15 and Adamts5, the developmental pattern of Adamts4 expression differed from all other aggrecanases examined here. A dramatic enrichment in Adamts4 mRNA levels was observed at P15, compared with levels in P7 and P56 hippocampi (for example, P15 hippocampal Adamts4 mRNA levels were 12.65+/−1.57 [SEM] fold higher than at P7; n=3, P=0.002 by Tukey-Kramer Difference Between Means) (Figure 6).
Figure 6.
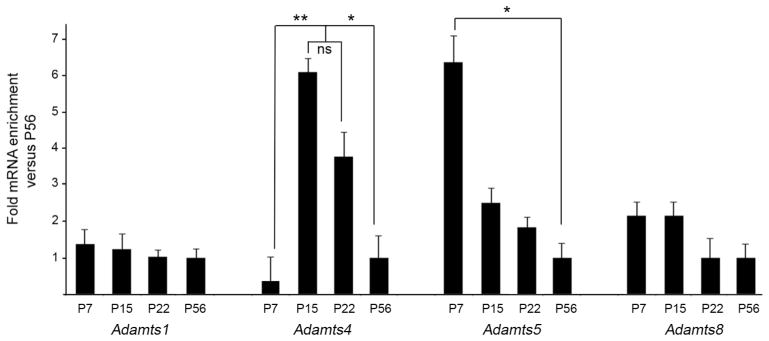
Developmental expression of other aggrecan-cleaving ADAMTSs in mouse hippocampus. Quantitative RT-PCR (qPCR) shows developmental changes in the expression of Adamts1, Adamts4, Adamts5 and Adamts8 mRNA in mouse hippocampus at P7, P15, P22 and P56. Levels of Adamts were normalized to Gapdh levels in each sample. Differs from indicated samples at P<0.001 (**) or P<0.05 (*) by Tukey-Kramer Difference Between Means Test. n=3 mice, per age. Error bars indicate SEM.
Not only were similar levels of Adamts4 mRNA generated in neocortex as in hippocampus (at P7 cortical Adamts4 mRNA levels were 1.14+/−0.09 [SEM] fold different than hippocampal levels; n=3, P=0.9 by Tukey-Kramer Difference Between Means; at P15 cortical Adamts4 mRNA levels were 0.86+/−1.57 [SEM] fold different than hippocampal levels; n=3, P=0.7 by Tukey-Kramer Difference Between Means) but cortical expression levels underwent similar changes throughout development. At P15, cortical Adamts4 mRNA levels were nearly 10 fold higher than levels at P7 (9.44+/−2.49 [SEM] fold higher at P15 versus P7; n=3, P=0.02 by Tukey-Kramer Difference Between Means). Thus, in both hippocampus and neocortex Adamts4 mRNA levels exhibit dramatic upregulation at the end of the second postnatal week of mouse development, a period that coincides with several important aspects of neural development, such as myelination and activity-dependent refinement and maturation of neural circuits.
Developmental changes in Adamts4 expression in non-neuronal cells
We were particularly intrigued by the expression pattern of Adamts4 mRNA, which encodes the most well studied aggrecanase. ADAMTS4 is enriched in brain compared to many other tissues and has the highest affinity and activity for cleaving aggrecan (Tang 2001; Porter et al. 2005). To investigate the cellular expression pattern of Adamts4 mRNA, we generated DIG-labeled Adamts4 riboprobes and performed in situ hybridization in coronal sections of mouse hippocampus. At P15, Adamts4-expressing cells were sparsely distributed throughout the neuronal and synaptic layers of hippocampus (Figure 7A). Similar patterns of sparsely distributed Adamts4-expressing cells were also observed in adjacent regions of dorsal thalamus and neocortex, confirming qPCR data showing similar quantitative levels of Adamts4 mRNA in these regions (see above).
In contrast to the sparse distribution of Adamts4-expressing cells in hippocampal neuropil, an enrichment of these cells was observed in the fimbria and alveus, two fiber tracts that carry axons into and out of the hippocampus (Figure 7A). Similar patterns of Adamts4 mRNA expression were observed in neuron-free regions of the neocortex and thalamus: Adamts4-mRNA appeared enriched in the white matter tract in the deepest cortical layer (adjacent to the alveus), in the corpus callosum, and in the optic tract (Figure 7A,B and data not shown). The presence of Adamts4-expressing cells in regions devoid of neurons suggests glial cells generate this aggrecanase.
Since our analysis of cellular Adamts4 mRNA expression was performed at the peak of Adamts4 expression (P15), we next addressed how these cellular expression patterns changed over time, as overall levels of hippocampal Adamts4 mRNA decreased significantly (Figure 6). In situ hybridization revealed significantly fewer Adamts4-expressing cells in all regions of the hippocampus and neocortex at early ages of postnatal development (i.e. P3 and P7)(Figure 7B,E,H,K) and in adult tissues (7D,G,J,M). Cell counts for all hippocampal regions, and for just the neuron-free regions of hippocampus, revealed the same pattern: ~8 fold more cells generated Adamts4 mRNA at P15 compared to P7 or P60 (Figure 7N–P).
Oligodendrocyte expression of Adamts4 in hippocampus and neocortex
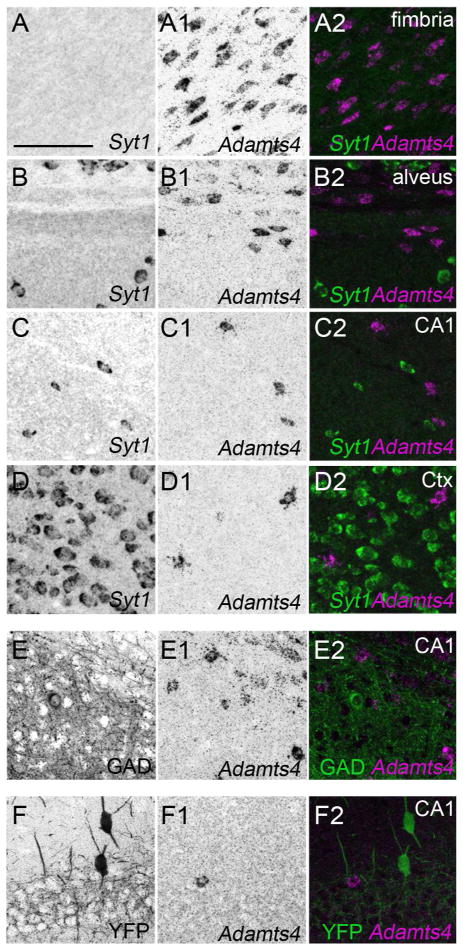
Finally, we used in situ hybridization to determine which cell types generate Adamts4 in mouse hippocampus and neocortex. To test whether neurons in any part of the telencephalon generate Adamts4 mRNA we performed double fluorescent in situ hybridization with DIG-labeled Adamts4 riboprobes and FL-conjugated riboprobes against Syt1. No co-expression of Adamts4 and Syt1 was observed in any part of the P21 mouse hippocampus or neocortex (Figure 8A–D). Likewise, we failed to find colocalization of Adamts4 mRNA in GAD67- or Parv-expressing interneurons (Figure 8E,F). While these expression patterns differ from one previous report in rat (Yuan et al. 2002), they match the pattern of Adamts4 mRNA expression in mouse brain reported in the Allen Brain Atlas (http://mouse.brain-map.org/) (RRID:nif-0000-00509).
Figure 8.
Hippocampal and cortical neurons do not generate Adamts4 mRNA. A–D. Double fluorescent in situ hybridization with riboprobes directed against Adamts4 and Syt1 show that in all regions of the P21 hippocampus and neocortex cells expressing Adamts4 do not co-express Syt1 mRNA. E. Fluorescent in situ hybridization coupled with immunostaining for GAD67 revealed that GAD67+ interneurons in the P21 hippocampus do not generate Adamts4 mRNA. F. Fluorescent in situ hybridization coupled with immunostaining for YFP in P37 Parv-Cre::Thy1-STOP-YFP15 mice revealed that Parv+ interneurons in the P15 hippocampus do not generate Adamts4 mRNA. Scale bar = 100 μm for all panels.
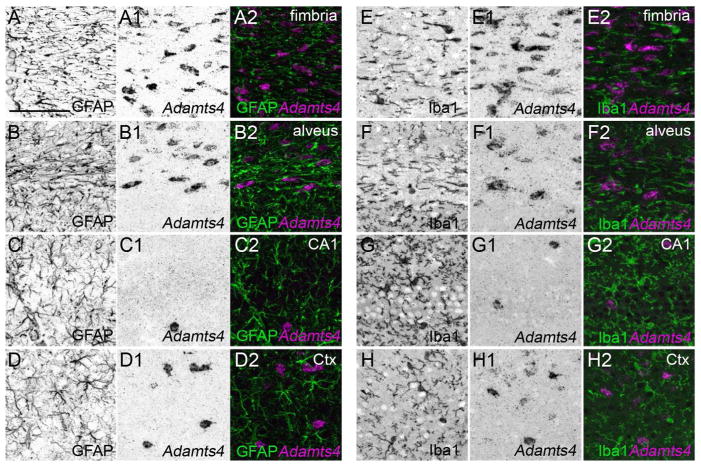
Based upon the lack of neuronal expression of Adamts4 mRNA and its enrichment in regions of telencephalon that lack neurons, we next assessed whether microglia, astrocytes or oligodendrocytes generate mRNA for this aggrecanase. In all regions of hippocampus and neocortex, Adamts4-expressing cells lacked both GFAP and Iba1 (Figure 9), indicating that neither GFAP+ astrocytes nor Iba1+ microglial cells generate Adamts4 mRNA. In contrast, Adamts4-expressing cells were immunoreactive for Olig2, a transcription factor expressed by oligodendrocytes (Nishiyama et al. 2009)(Figure 10), indicating that Adamts4 is generated by the myelinating glial cells of the brain. Taken together with its peak level of expression during the second and third postnatal weeks, these results indicate that ADAMTS4 may play important roles in ECM remodeling during myelination.
Figure 9.
Astrocytes and microglia do not generate Adamts4 mRNA. Fluorescent in situ hybridization coupled with immunostaining for GFAP (A–D) or Iba1 (E–H) revealed that neither Iba1-positive microglia or GFAP-positive astrocytes in the P21 hippocampus (A–C, E–G) or layer V of neocortex (D,H) generate Adamts4 mRNA. Scale bar = 100 μm for all panels.
Figure 10.
Oligodendrocytes generate Adamts4 mRNA. Fluorescent in situ hybridization coupled with immunostaining for Olig2 revealed that oligodendrocytes in the hippocampus (A–C) and neocortex (D) generate Adamts4 mRNA. Arrows highlight cells that co-express Adamts4 mRNA and Olig2 protein. Scale bar = 100 μm for all panels.
Region-specific differences in Adamts4 expression
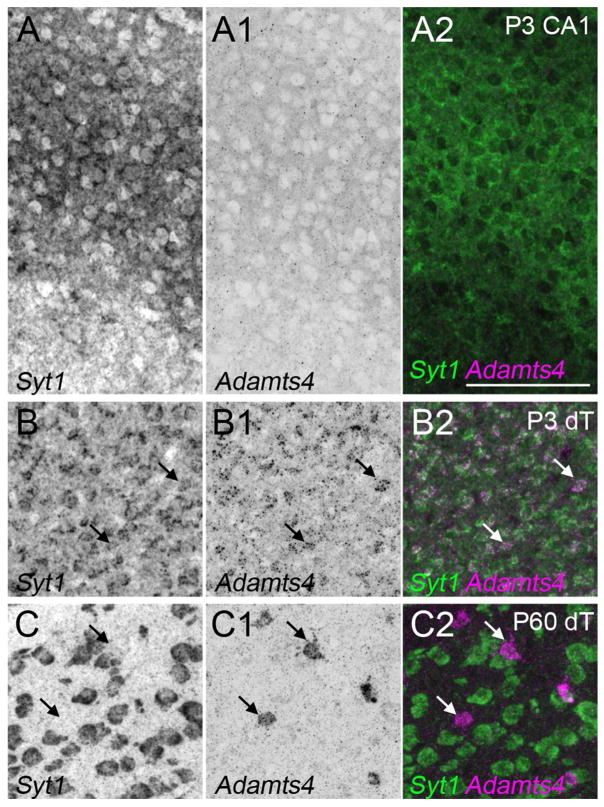
The finding that Adamts4 mRNA was not generated by neurons in the P15 telencephalon was somewhat surprising given our previous observation that neonatal Syt1-expressing thalamic neurons generate Adamts4 (Brooks et al. 2013). For this reason we tested whether hippocampal neurons transiently expressed Adamts4 perinatally. At P3 we failed to detect Adamts4 mRNA in Syt1-expressing hippocampal neurons, despite seeing robust expression by neurons in the dorsal thalamus (dT)(Figure 11). Interestingly, widespread neuronal expression of Adamts4 is transient in dT, and by P60 Adamts4 mRNA appeared to be generated solely by non-neuronal cells in this region (Figure 11). These results suggest that Adamts4 can be expressed by both neurons and glial cells, but that expression is not only developmentally regulated but is also regulated in a nuclei-specific manner.
Figure 11.
Adamts4 mRNA is not generated by neurons at early stages of hippocampal development. Double fluorescent in situ hybridization with riboprobes directed against Adamts4 and Syt1 at P3 and P60. These data show that at early postnatal ages (i.e. P3), neurons within the hippocampus do not generate Adamts4 mRNA. This is in contrast to expression in the dorsal thalamus (dT)(including the dLGN), where Syt1-expressing neurons express Adamts4 at P3. Expression of Adamts4 remains present in dorsal thalamus at later time points, but is no longer generated by neurons. Scale bar = 100 μm for all panels.
DISCUSSION
Remodeling of the ECM is a critical step in the development of the nervous system as neurons and glia undergo extensive morphogenesis to transform into their adult-like forms. Although brain ECM is rich in lecticans, which comprise a family of chondroitin sulfate proteoglycans that includes aggrecan, brevican, neurocan, and versican, we lack a fundamental understanding of the distribution and developmental expression of lectican-cleaving enzymes in the normal, developing brain. In the present study we examined the expression and developmental regulation of two such lectican-cleaving metalloproteinases, ADAMTS15 and ADAMTS4. Both proteases are expressed in the mammalian brain (Tang 2001; Yuan et al. 2002; Cross et al. 2006; Dancevic et al. 2013) and share an ability to cleave both aggrecan and versican, although with differing affinities and activities (Tortorella et al. 2000; Sandy et al. 2001; Porter et al. 2005; Stupka et al. 2012). ADAMTS4 also harbors an ability to cleave brevican (Mathews et al. 2000), however at present it remains unclear whether this activity is shared by ADAMTS15. Antibodies against ADAMTS-cleaved lecticans have been particularly useful in localizing the activity of ADAMTSs in the normal and injured nervous system (Matthews et al. 2000; Yuan et al. 2002; Mayer et al. 2005; Ajmo et al. 2008); however, since many members of the aggrecanase subfamily of ADAMTSs (i.e. ADAMTS1,4,5,8,9,15) share similar abilities to cleave these proteoglycans the distribution and expression pattern of each ADAMTS in brain remains unclear. In fact, while it appears that both neurons and astrocytes are capable of generating and secreting these enzymes in vitro (Hamel et al. 2005; Hamel et al. 2008), we lack a general understanding of whether these enzymes are generated by neurons, glia, or both in the normal, non-pathological brain. Here, we sought to remedy this by characterizing the cellular and developmental expression patterns of ADAMTS15 and ADAMTS4. These studies led us to uncover several novel features of ADAMTS expression: 1). ADAMTSs are expressed in cell-specific manners in the mammalian brain. For example, in the hippocampus, ADAMTS15 is generated by Parv+ interneurons, while ADAMTS4 is generated by oligodendrocytes. 2). The cellular expressions of single ADAMTS genes are distinct in different brain regions. For example, in the hippocampus and neocortex ADAMTS15 is selectively expressed by inhibitory interneurons, whereas in the dorsal thalamus it is generated by excitatory thalamic relay neurons. Likewise, ADAMTS4 is generated by neurons in the perinatal thalamus, but appears exclusively expressed by oligodendrocytes in the developing hippocampus and neocortex. 3). The expression of many (but not all) ADAMTSs is developmentally regulated in brain. The peak of mRNA expression of both ADAMTS15 and ADAMTS4 coincides with critical aspects of the development of the cell-type generating them. The peak expression of ADAMTS15 by neurons (in all regions examined here and in previous studies [Brooks et al. 2013]) coincides with synaptogenesis and neural circuit formation. Oligodendroglial expression of ADAMTS4 coincides with the peak of myelination, a developmental process that requires substantial oligodendrocyte morphogenesis and ECM remodeling as axons becomes ensheathed by lipid-rich myelin and nodes of Ranvier are assembled (Colognato and Tzvetanova, 2011; Mitew et al. 2013).
Taken together our results suggest important cell-specific roles for ADAMTSs during brain development. Clearly, one important function for ADAMTSs during brain development is the remodeling and cleavage of lectican-rich ECM. The increased expression of ADAMTS15 by interneurons during the critical period and before the emergence of perineuronal nets, suggests its expression may limit lectican-rich matrix deposition and therefore suggests it may play an important role in preventing premature closing of the critical period. This possibility raises several important questions for future experiments: Can increasing ADAMTS15 expression in Parv+ interneurons reopen the critical period? And, is ADAMTS15 expression regulated by neuronal activity? The recent discovery that ADAMTS15 expression is upregulated in the hippocampi of patients suffering from medial temporal lobe epilepsy hints that the expression of this metalloproteinase may indeed be linked to neuronal activity (Venugopal et al. 2012).
It is also possible that ADAMTSs exert their function by cleaving non-lectican components of brain ECM. Although lecticans appear to be the most prominent components of brain ECM, several other ECM molecules are critical for neural development and may be targets of ADAMTSs. Prominent examples include Reelin, a large glycoprotein with important roles in directing neuronal migration, axonal targeting and synapse formation (D’Arcangelo et al. 1995; Borrell et al. 1999; Su et al. 2011) and Thrombospondin and Hevin, two glial-derived matricellular molecules that stimulate synaptogenesis (Christopherson et al. 2005; Eroglu et al. 2009; Kucukdereli et al. 2011). All three of these ECM components are cleaved by ADAMTSs (Dickinson et al. 2003; Weaver et al. 2010; Hisanaga et al. 2012; Krstic et al. 2012; Koie et al. 2014) and are abundant in the developing brain parenchyma. An interesting feature of Reelin, and several other components of brain ECM including Agrin and nonfibrillar Collagens, is that cleavage by extracellular proteases liberates “active” fragments of these ECM molecules which regulate synapse formation (Stephan et al. 2008; Matsumoto-Miyai et al. 2009; Su et al. 2012). It remains unclear whether ADAMTSs liberate such synaptogenic cues from brain ECM.
Lastly, while we have discussed the potential roles of the enzymatic functions of these metalloproteinases in the developing brain, it is important to point out that non-enzymatic functions of ADAMTSs have been suggested from in vitro studies. Specifically, ADAMTS4 and ADAMTS5 have the ability to stimulate neurite extension by activating a MAP ERK 1/2 kinase pathway, an activity that is independent from lectican-cleavage (Hamel et al. 2008). Taken together with results described here, this suggests that the perinatal expression of ADAMTS15 (in all brain regions) and ADAMTS4 (specifically in dorsal thalamus) may contribute a direct role in neural circuit formation, in addition to roles in ECM remodeling. Although it is possible that oligodendrocyte-derived ADAMTS4 in fiber tracts of the postnatal hippocampus and neocortex contributes a direct influence on axonal outgrowth, it seems unlikely based upon its upregulation after the time at which most axonal tracts have already been established. A more likely scenario may be that glial-derived ADAMTS4 is important for directly or indirectly influencing the extension of oligodendrocyte processes while they seek out and myelinate axons.
Acknowledgments
We thank Mr. Aboozar Monavarfeshani and other members of the Fox lab for RNA samples and for helpful discussions regarding this project and manuscript. This work was supported, in part, by the National Institutes of Health (NIH) – National Eye Institute Grants EY021222 (MAF). We thank Dr. G. Valdez (Virginia Tech Carilion Research Institute) for providing Parv-cre and Thy1-STOP-YFP15 mice.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests.
ROLE OF AUTHORS
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: CL, JMB, MAF. Acquisition of data: CL, JMB, JC, JS. Analysis and interpretation of data: CL, JMB, JC, JS, MAF. Drafting of the manuscript: MAF. Critical revision of the manuscript for important intellectual content: CL, JMB, JC, JS, MAF. Statistical analysis: CL, JMB, JC, JS, MAF. Obtained funding: MAF. Study supervision: MAF.
References
- Abbaszade I, Liu RQ, Yang F, Rosenfeld SA, Ross OH, Link JR, Ellis DM, Tortorella MD, Pratta MA, Hollis JM, Wynn R, Duke JL, George HJ, Hillman MC, Jr, Murphy K, Wiswall BH, Copeland RA, Decicco CP, Bruckner R, Nagase H, Itoh Y, Newton RC, Magolda RL, Trzaskos JM, Burn TC, et al. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. J Biol Chem. 1999;274(33):23443–23450. doi: 10.1074/jbc.274.33.23443. [DOI] [PubMed] [Google Scholar]
- Acosta ML, Bumsted O’Brien KM, Tan SS, Kalloniatis M. Emergence of cellular markers and functional ionotropic glutamate receptors on tangentially dispersed cells in the developing mouse retina. J Comp Neurol. 2008;506(3):506–523. doi: 10.1002/cne.21561. [DOI] [PubMed] [Google Scholar]
- Ajmo JM, Eakin AK, Hamel MG, Gottschall PE. Discordant localization of WFA reactivity and brevican/ADAMTS-derived fragment in rodent brain. BMC Neurosci. 2008;9:14. doi: 10.1186/1471-2202-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura A, Sato H, Coy DH, Schally AV. Radioimmunoassay for GH-release inhibiting hormone. Proc Soc Exp Biol Med. 1975;148(3):784–789. doi: 10.3181/00379727-148-38631. [DOI] [PubMed] [Google Scholar]
- Bateman JF, Boot-Handford RP, Lamande SR. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat Rev Genet. 2009;10(3):173–183. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- Borrell V, Del Rio JA, Alcantara S, Derer M, Martinez A, D’Arcangelo G, Nakajima K, Mikoshiba K, Derer P, Curran T, Soriano E. Reelin regulates the development and synaptogenesis of the layer-specific entorhino-hippocampal connections. J Neurosci. 1999;19(4):1345–1358. doi: 10.1523/JNEUROSCI.19-04-01345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JM, Su J, Levy C, Wang JS, Seabrook TA, Guido W, Fox MA. A molecular mechanism regulating the timing of corticogeniculate innervation. Cell Rep. 2013;5(3):573–581. doi: 10.1016/j.celrep.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carulli D, Pizzorusso T, Kwok JC, Putignano E, Poli A, Forostyak S, Andrews MR, Deepa SS, Glant TT, Fawcett JW. Animals lacking link protein have attenuated perineuronal nets and persistent plasticity. Brain. 2010;133(Pt 8):2331–2347. doi: 10.1093/brain/awq145. [DOI] [PubMed] [Google Scholar]
- Celio MR, Spreafico R, De Biasi S, Vitellaro-Zuccarello L. Perineuronal nets: past and present. Trends Neurosci. 1998;21(12):510–515. doi: 10.1016/s0166-2236(98)01298-3. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120(3):421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Colognato H, Tzvetanova ID. Glia unglued: how signals from the extracellular matrix regulate the development of myelinating glia. Dev Neurobiol. 2011;71(11):924–955. doi: 10.1002/dneu.20966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross AK, Haddock G, Stock CJ, Allan S, Surr J, Bunning RA, Buttle DJ, Woodroofe MN. ADAMTS-1 and -4 are up-regulated following transient middle cerebral artery occlusion in the rat and their expression is modulated by TNF in cultured astrocytes. Brain Res. 2006;1088(1):19–30. doi: 10.1016/j.brainres.2006.02.136. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374(6524):719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- Dancevic CM, Fraser FW, Smith AD, Stupka N, Ward AC, McCulloch DR. Biosynthesis and expression of a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats-15: a novel versican-cleaving proteoglycanase. J Biol Chem. 2013;288(52):37267–37276. doi: 10.1074/jbc.M112.418624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danglot L, Triller A, Marty S. The development of hippocampal interneurons in rodents. Hippocampus. 2006;16(12):1032–1060. doi: 10.1002/hipo.20225. [DOI] [PubMed] [Google Scholar]
- del Rio JA, de Lecea L, Ferrer I, Soriano E. The development of parvalbumin-immunoreactivity in the neocortex of the mouse. Brain Res Dev Brain Res. 1994;81(2):247–259. doi: 10.1016/0165-3806(94)90311-5. [DOI] [PubMed] [Google Scholar]
- Dickinson SC, Vankemmelbeke MN, Buttle DJ, Rosenberg K, Heinegard D, Hollander AP. Cleavage of cartilage oligomeric matrix protein (thrombospondin-5) by matrix metalloproteinases and a disintegrin and metalloproteinase with thrombospondin motifs. Matrix Biol. 2003;22(3):267–278. doi: 10.1016/s0945-053x(03)00034-9. [DOI] [PubMed] [Google Scholar]
- Dityatev A, Frischknecht R, Seidenbecher CI. Extracellular matrix and synaptic functions. Results Probl Cell Differ. 2006;43:69–97. doi: 10.1007/400_025. [DOI] [PubMed] [Google Scholar]
- Dityatev A, Seidenbecher CI, Schachner M. Compartmentalization from the outside: the extracellular matrix and functional microdomains in the brain. Trends Neurosci. 2010;33(11):503–512. doi: 10.1016/j.tins.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Egles C, Claudepierre T, Manglapus MK, Champliaud MF, Brunken WJ, Hunter DD. Laminins containing the beta2 chain modulate the precise organization of CNS synapses. Mol Cell Neurosci. 2007;34(3):288–298. doi: 10.1016/j.mcn.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Eroglu C, Allen NJ, Susman MW, O’Rourke NA, Park CY, Ozkan E, Chakraborty C, Mulinyawe SB, Annis DS, Huberman AD, Green EM, Lawler J, Dolmetsch R, Garcia KC, Smith SJ, Luo ZD, Rosenthal A, Mosher DF, Barres BA. Gabapentin receptor alpha2delta-1 is a neuronal thrombospondin receptor responsible for excitatory CNS synaptogenesis. Cell. 2009;139(2):380–392. doi: 10.1016/j.cell.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MA, Sanes JR. Synaptotagmin I and II are present in distinct subsets of central synapses. J Comp Neurol. 2007;503(2):280–296. doi: 10.1002/cne.21381. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Muller U. Extracellular matrix functions during neuronal migration and lamination in the mammalian central nervous system. Dev Neurobiol. 2011;71(11):889–900. doi: 10.1002/dneu.20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123(Pt 24):4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund TF, Buzsaki G. Interneurons of the hippocampus. Hippocampus. 1996;6(4):347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Frischknecht R, Gundelfinger ED. The brain’s extracellular matrix and its role in synaptic plasticity. Adv Exp Med Biol. 2012;970:153–171. doi: 10.1007/978-3-7091-0932-8_7. [DOI] [PubMed] [Google Scholar]
- Gogolla N, Caroni P, Luthi A, Herry C. Perineuronal nets protect fear memories from erasure. Science. 2009;325(5945):1258–1261. doi: 10.1126/science.1174146. [DOI] [PubMed] [Google Scholar]
- Hamel MG, Mayer J, Gottschall PE. Altered production and proteolytic processing of brevican by transforming growth factor beta in cultured astrocytes. J Neurochem. 2005;93(6):1533–1541. doi: 10.1111/j.1471-4159.2005.03144.x. [DOI] [PubMed] [Google Scholar]
- Hamel MG, Ajmo JM, Leonardo CC, Zuo F, Sandy JD, Gottschall PE. Multimodal signaling by the ADAMTSs (a disintegrin and metalloproteinase with thrombospondin motifs) promotes neurite extension. Exp Neurol. 2008;210(2):428–440. doi: 10.1016/j.expneurol.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig W, Derouiche A, Welt K, Brauer K, Grosche J, Mader M, Reichenbach A, Bruckner G. Cortical neurons immunoreactive for the potassium channel Kv3.1b subunit are predominantly surrounded by perineuronal nets presumed as a buffering system for cations. Brain Res. 1999;842(1):15–29. doi: 10.1016/s0006-8993(99)01784-9. [DOI] [PubMed] [Google Scholar]
- Hedstrom KL, Xu X, Ogawa Y, Frischknecht R, Seidenbecher CI, Shrager P, Rasband MN. Neurofascin assembles a specialized extracellular matrix at the axon initial segment. J Cell Biol. 2007;178(5):875–886. doi: 10.1083/jcb.200705119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisanaga A, Morishita S, Suzuki K, Sasaki K, Koie M, Kohno T, Hattori M. A disintegrin and metalloproteinase with thrombospondin motifs 4 (ADAMTS-4) cleaves Reelin in an isoform-dependent manner. FEBS Lett. 2012;586(19):3349–3353. doi: 10.1016/j.febslet.2012.07.017. [DOI] [PubMed] [Google Scholar]
- Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326(5957):1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Ibata I, Ito D, Ohsawa K, Kohsaka S. A novel gene iba1 in the major histocompatibility complex class III region encoding an EF hand protein expressed in a monocytic lineage. Biochem Biophys Res Commun. 1996;224(3):855–862. doi: 10.1006/bbrc.1996.1112. [DOI] [PubMed] [Google Scholar]
- Jiang M, Swann JW. A role for L-type calcium channels in the maturation of parvalbumin-containing hippocampal interneurons. Neuroscience. 2005;135(3):839–850. doi: 10.1016/j.neuroscience.2005.06.073. [DOI] [PubMed] [Google Scholar]
- Kamphuis W, Mamber C, Moeton M, Kooijman L, Sluijs JA, Jansen AH, Verveer M, de Groot LR, Smith VD, Rangarajan S, Rodriguez JJ, Orre M, Hol EM. GFAP isoforms in adult mouse brain with a focus on neurogenic astrocytes and reactive astrogliosis in mouse models of Alzheimer disease. PLoS One. 2012;7(8):e42823. doi: 10.1371/journal.pone.0042823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karetko M, Skangiel-Kramska J. Diverse functions of perineuronal nets. Acta Neurobiol Exp (Wars) 2009;69(4):564–577. doi: 10.55782/ane-2009-1766. [DOI] [PubMed] [Google Scholar]
- Liu S, Wang J, Zhu D, Fu Y, Lukowiak K, Lu YM. Generation of functional inhibitory neurons in the adult rat hippocampus. J Neurosci. 2003;23(3):732–736. doi: 10.1523/JNEUROSCI.23-03-00732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerever A, Schnack J, Vellinga D, Ichikawa N, Moon C, Arikawa-Hirasawa E, Efird JT, Mercier F. Novel extracellular matrix structures in the neural stem cell niche capture the neurogenic factor fibroblast growth factor 2 from the extracellular milieu. Stem Cells. 2007;25(9):2146–2157. doi: 10.1634/stemcells.2007-0082. [DOI] [PubMed] [Google Scholar]
- Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321(5885):53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koie M, Okumura K, Hisanaga A, Kamei T, Sasaki K, Deng M, Baba A, Kohno T, Hattori M. Cleavage within Reelin repeat 3 regulates the duration and range of the signaling activity of Reelin protein. J Biol Chem. 2014;289(18):12922–12930. doi: 10.1074/jbc.M113.536326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krstic D, Rodriguez M, Knuesel I. Regulated proteolytic processing of Reelin through interplay of tissue plasminogen activator (tPA), ADAMTS-4, ADAMTS-5, and their modulators. PLoS One. 2012;7(10):e47793. doi: 10.1371/journal.pone.0047793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukdereli H, Allen NJ, Lee AT, Feng A, Ozlu MI, Conatser LM, Chakraborty C, Workman G, Weaver M, Sage EH, Barres BA, Eroglu C. Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC. Proc Natl Acad Sci U S A. 2011;108(32):E440–449. doi: 10.1073/pnas.1104977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labelle-Dumais C, Dilworth DJ, Harrington EP, de Leau M, Lyons D, Kabaeva Z, Manzini MC, Dobyns WB, Walsh CA, Michele DE, Gould DB. COL4A1 mutations cause ocular dysgenesis, neuronal localization defects, and myopathy in mice and Walker-Warburg syndrome in humans. PLoS Genet. 2011;7(5):e1002062. doi: 10.1371/journal.pgen.1002062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu VS, Macdonald B, Kalluri R. Structure and function of basement membranes. Exp Biol Med (Maywood) 2007;232(9):1121–1129. doi: 10.3181/0703-MR-72. [DOI] [PubMed] [Google Scholar]
- Libby RT, Lavallee CR, Balkema GW, Brunken WJ, Hunter DD. Disruption of laminin beta2 chain production causes alterations in morphology and function in the CNS. J Neurosci. 1999;19(21):9399–9411. doi: 10.1523/JNEUROSCI.19-21-09399.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Matsumoto-Miyai K, Sokolowska E, Zurlinden A, Gee CE, Luscher D, Hettwer S, Wolfel J, Ladner AP, Ster J, Gerber U, Rulicke T, Kunz B, Sonderegger P. Coincident pre- and postsynaptic activation induces dendritic filopodia via neurotrypsin-dependent agrin cleavage. Cell. 2009;136(6):1161–1171. doi: 10.1016/j.cell.2009.02.034. [DOI] [PubMed] [Google Scholar]
- Matthews RT, Gary SC, Zerillo C, Pratta M, Solomon K, Arner EC, Hockfield S. Brain-enriched hyaluronan binding (BEHAB)/brevican cleavage in a glioma cell line is mediated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family member. J Biol Chem. 2000;275(30):22695–22703. doi: 10.1074/jbc.M909764199. [DOI] [PubMed] [Google Scholar]
- Matthews RT, Kelly GM, Zerillo CA, Gray G, Tiemeyer M, Hockfield S. Aggrecan glycoforms contribute to the molecular heterogeneity of perineuronal nets. J Neurosci. 2002;22(17):7536–7547. doi: 10.1523/JNEUROSCI.22-17-07536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J, Hamel MG, Gottschall PE. Evidence for proteolytic cleavage of brevican by the ADAMTSs in the dentate gyrus after excitotoxic lesion of the mouse entorhinal cortex. BMC Neurosci. 2005;6:52. doi: 10.1186/1471-2202-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain CJ, Fisahn A. Interneurons unbound. Nat Rev Neurosci. 2001;2(1):11–23. doi: 10.1038/35049047. [DOI] [PubMed] [Google Scholar]
- McRae PA, Baranov E, Sarode S, Brooks-Kayal AR, Porter BE. Aggrecan expression, a component of the inhibitory interneuron perineuronal net, is altered following an early-life seizure. Neurobiol Dis. 2010;39(3):439–448. doi: 10.1016/j.nbd.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier F, Kitasako JT, Hatton GI. Anatomy of the brain neurogenic zones revisited: fractones and the fibroblast/macrophage network. J Comp Neurol. 2002;451(2):170–188. doi: 10.1002/cne.10342. [DOI] [PubMed] [Google Scholar]
- Mitew S, Hay CM, Peckham H, Xiao J, Koenning M, Emery B. Mechanisms regulating the development of oligodendrocytes and central nervous system myelin. Neuroscience. 2013 doi: 10.1016/j.neuroscience.2013.11.029. [DOI] [PubMed] [Google Scholar]
- Nicholson C, Sykova E. Extracellular space structure revealed by diffusion analysis. Trends Neurosci. 1998;21(5):207–215. doi: 10.1016/s0166-2236(98)01261-2. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci. 2009;10(1):9–22. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Hagihara K, Suzuki M, Yamaguchi Y. Brevican in the developing hippocampal fimbria: differential expression in myelinating oligodendrocytes and adult astrocytes suggests a dual role for brevican in central nervous system fiber tract development. J Comp Neurol. 2001;432(3):285–295. doi: 10.1002/cne.1103. [DOI] [PubMed] [Google Scholar]
- Okaty BW, Miller MN, Sugino K, Hempel CM, Nelson SB. Transcriptional and electrophysiological maturation of neocortical fast-spiking GABAergic interneurons. J Neurosci. 2009;29(21):7040–7052. doi: 10.1523/JNEUROSCI.0105-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva AA, Jr, Jiang M, Lam T, Smith KL, Swann JW. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci. 2000;20(9):3354–3368. doi: 10.1523/JNEUROSCI.20-09-03354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oohashi T, Hirakawa S, Bekku Y, Rauch U, Zimmermann DR, Su WD, Ohtsuka A, Murakami T, Ninomiya Y. Bral1, a brain-specific link protein, colocalizing with the versican V2 isoform at the nodes of Ranvier in developing and adult mouse central nervous systems. Mol Cell Neurosci. 2002;19(1):43–57. doi: 10.1006/mcne.2001.1061. [DOI] [PubMed] [Google Scholar]
- Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005;386(Pt 1):15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt LF, Prokop A. Introduction: the role of extracellular matrix in nervous system development and maintenance. Dev Neurobiol. 2011;71(11):883–888. doi: 10.1002/dneu.20975. [DOI] [PubMed] [Google Scholar]
- Ricard-Blum S, Salza R. Matricryptins and matrikines: biologically active fragments of the extracellular matrix. Exp Dermatol. 2014;23(7):457–463. doi: 10.1111/exd.12435. [DOI] [PubMed] [Google Scholar]
- Rozario T, DeSimone DW. The extracellular matrix in development and morphogenesis: a dynamic view. Dev Biol. 2010;341(1):126–140. doi: 10.1016/j.ydbio.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandy JD, Westling J, Kenagy RD, Iruela-Arispe ML, Verscharen C, Rodriguez-Mazaneque JC, Zimmermann DR, Lemire JM, Fischer JW, Wight TN, Clowes AW. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441-Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS-4. J Biol Chem. 2001;276(16):13372–13378. doi: 10.1074/jbc.M009737200. [DOI] [PubMed] [Google Scholar]
- Sanes JR. Extracellular matrix molecules that influence neural development. Annu Rev Neurosci. 1989;12:491–516. doi: 10.1146/annurev.ne.12.030189.002423. [DOI] [PubMed] [Google Scholar]
- Sloviter RS. Calcium-binding protein (calbindin-D28k) and parvalbumin immunocytochemistry: localization in the rat hippocampus with specific reference to the selective vulnerability of hippocampal neurons to seizure activity. J Comp Neurol. 1989;280(2):183–196. doi: 10.1002/cne.902800203. [DOI] [PubMed] [Google Scholar]
- Spicer AP, Joo A, Bowling RA., Jr A hyaluronan binding link protein gene family whose members are physically linked adjacent to chondroitin sulfate proteoglycan core protein genes: the missing links. J Biol Chem. 2003;278(23):21083–21091. doi: 10.1074/jbc.M213100200. [DOI] [PubMed] [Google Scholar]
- Stephan A, Mateos JM, Kozlov SV, Cinelli P, Kistler AD, Hettwer S, Rulicke T, Streit P, Kunz B, Sonderegger P. Neurotrypsin cleaves agrin locally at the synapse. Faseb J. 2008;22(6):1861–1873. doi: 10.1096/fj.07-100008. [DOI] [PubMed] [Google Scholar]
- Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupka N, Kintakas C, White JD, Fraser FW, Hanciu M, Aramaki-Hattori N, Martin S, Coles C, Collier F, Ward AC, Apte SS, McCulloch DR. Versican processing by a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats proteinases-5 and -15 facilitates myoblast fusion. J Biol Chem. 2012;288(3):1907–1917. doi: 10.1074/jbc.M112.429647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Gorse K, Ramirez F, Fox MA. Collagen XIX is expressed by interneurons and contributes to the formation of hippocampal synapses. J Comp Neurol. 2010;518(2):229–253. doi: 10.1002/cne.22228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Haner CV, Imbery TE, Brooks JM, Morhardt DR, Gorse K, Guido W, Fox MA. Reelin is required for class-specific retinogeniculate targeting. J Neurosci. 2011;31(2):575–586. doi: 10.1523/JNEUROSCI.4227-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Klemm MA, Josephson AM, Fox MA. Contributions of VLDLR and LRP8 in the establishment of retinogeniculate projections. Neural Dev. 2013;8:11. doi: 10.1186/1749-8104-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susuki K, Chang KJ, Zollinger DR, Liu Y, Ogawa Y, Eshed-Eisenbach Y, Dours-Zimmermann MT, Oses-Prieto JA, Burlingame AL, Seidenbecher CI, Zimmermann DR, Oohashi T, Peles E, Rasband MN. Three mechanisms assemble central nervous system nodes of Ranvier. Neuron. 2013;78(3):469–482. doi: 10.1016/j.neuron.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL. ADAMTS: a novel family of extracellular matrix proteases. Int J Biochem Cell Biol. 2001;33(1):33–44. doi: 10.1016/s1357-2725(00)00061-3. [DOI] [PubMed] [Google Scholar]
- Tani E, Ametani T. Extracellular distribution of ruthenium red-positive substance in the cerebral cortex. J Ultrastruct Res. 1971;34(1):1–14. doi: 10.1016/s0022-5320(71)90002-5. [DOI] [PubMed] [Google Scholar]
- Tortorella MD, Burn TC, Pratta MA, Abbaszade I, Hollis JM, Liu R, Rosenfeld SA, Copeland RA, Decicco CP, Wynn R, Rockwell A, Yang F, Duke JL, Solomon K, George H, Bruckner R, Nagase H, Itoh Y, Ellis DM, Ross H, Wiswall BH, Murphy K, Hillman MC, Jr, Hollis GF, Newton RC, Magolda RL, Trzaskos JM, Arner EC. Purification and cloning of aggrecanase-1: a member of the ADAMTS family of proteins. Science. 1999;284(5420):1664–1666. doi: 10.1126/science.284.5420.1664. [DOI] [PubMed] [Google Scholar]
- Tortorella MD, Pratta M, Liu RQ, Austin J, Ross OH, Abbaszade I, Burn T, Arner E. Sites of aggrecan cleavage by recombinant human aggrecanase-1 (ADAMTS-4) J Biol Chem. 2000;275(24):18566–18573. doi: 10.1074/jbc.M909383199. [DOI] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29(1):52, 54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- Tsien RY. Very long-term memories may be stored in the pattern of holes in the perineuronal net. Proc Natl Acad Sci U S A. 2013;110(30):12456–12461. doi: 10.1073/pnas.1310158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varea E, Nacher J, Blasco-Ibanez JM, Gomez-Climent MA, Castillo-Gomez E, Crespo C, Martinez-Guijarro FJ. PSA-NCAM expression in the rat medial prefrontal cortex. Neuroscience. 2005;136(2):435–443. doi: 10.1016/j.neuroscience.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Vasudevan A, Ho MS, Weiergraber M, Nischt R, Schneider T, Lie A, Smyth N, Kohling R. Basement membrane protein nidogen-1 shapes hippocampal synaptic plasticity and excitability. Hippocampus. 2010;20(5):608–620. doi: 10.1002/hipo.20660. [DOI] [PubMed] [Google Scholar]
- Venugopal AK, Sameer Kumar GS, Mahadevan A, Selvan LD, Marimuthu A, Dikshit JB, Tata P, Ramachandra Y, Chaerkady R, Sinha S, Chandramouli B, Arivazhagan A, Satishchandra P, Shankar S, Pandey A. Transcriptomic Profiling of Medial Temporal Lobe Epilepsy. J Proteomics Bioinform. 2012;5(2) doi: 10.4172/jpb.1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver MS, Workman G, Cardo-Vila M, Arap W, Pasqualini R, Sage EH. Processing of the matricellular protein hevin in mouse brain is dependent on ADAMTS4. J Biol Chem. 2010;285(8):5868–5877. doi: 10.1074/jbc.M109.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werb Z, Chin JR. Extracellular matrix remodeling during morphogenesis. Ann N Y Acad Sci. 1998;857:110–118. doi: 10.1111/j.1749-6632.1998.tb10111.x. [DOI] [PubMed] [Google Scholar]
- Wlodarczyk J, Mukhina I, Kaczmarek L, Dityatev A. Extracellular matrix molecules, their receptors, and secreted proteases in synaptic plasticity. Dev Neurobiol. 2011;71(11):1040–1053. doi: 10.1002/dneu.20958. [DOI] [PubMed] [Google Scholar]
- Xiao J, Ferner AH, Wong AW, Denham M, Kilpatrick TJ, Murray SS. Extracellular signal-regulated kinase 1/2 signaling promotes oligodendrocyte myelination in vitro. J Neurochem. 2012;122(6):1167–1180. doi: 10.1111/j.1471-4159.2012.07871.x. [DOI] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24(11):2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J Comp Neurol. 2006;499(1):144–160. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- Yamada J, Jinno S. Spatio-temporal differences in perineuronal net expression in the mouse hippocampus, with reference to parvalbumin. Neuroscience. 2013;253:368–379. doi: 10.1016/j.neuroscience.2013.08.061. [DOI] [PubMed] [Google Scholar]
- Yuan W, Matthews RT, Sandy JD, Gottschall PE. Association between protease-specific proteolytic cleavage of brevican and synaptic loss in the dentate gyrus of kainate-treated rats. Neuroscience. 2002;114(4):1091–1101. doi: 10.1016/s0306-4522(02)00347-0. [DOI] [PubMed] [Google Scholar]
- Zimmermann DR, Dours-Zimmermann MT. Extracellular matrix of the central nervous system: from neglect to challenge. Histochem Cell Biol. 2008;130(4):635–653. doi: 10.1007/s00418-008-0485-9. [DOI] [PubMed] [Google Scholar]