Abstract
This report describes the volatile organic compounds (VOCs) associated with human cerumen (earwax) and the effects of ethnicity/race and variation on the ATP-binding cassette, sub-family C, member 11 gene (ABCC11). A single nucleotide polymorphism (SNP) in ABCC11 affects the cerumen VOC profiles of individuals from African, Caucasian, and Asian descent. Employing gas chromatography/mass spectrometry (GC/MS) we have identified the nature and relative abundance of cerumen VOCs from 32 male donors. Our results show that cerumen contains a complex mixture of VOCs and that the amounts of these compounds vary across individuals as well as across ethnic/racial groups. In six of the seven compounds whose detected concentrations were found to be statistically different across groups, individuals of African descent (AfD) > Caucasian descent (CaD) > Asians descent (AsD). Our findings also reveal that ABCC11 genotype alone does not predict the type and relative levels of volatiles found in human cerumen, and suggest that other biochemical pathways must be involved. Examination of the composition and diversity of external auditory canal microbiota in a small subset of our subject population revealed that the ear microbiota may not be directly correlated with either ethnic group membership or ABCC11 genotype.
Keywords: cerumen, volatile organic compounds, earwax, GC/MS, genetics, ear microbiota
INTRODUCTION
Odors emitted by the human body are studied for their potential application in a variety of areas including forensics (Curran et al. 2010; DeGreeff et al. 2011; Prada and Furton 2008, 2012), volatile disease biomarkers (Horvath et al. 2008; Kwak et al. 2013; Kwak and Preti 2011; Poling et al. 2010; Preti et al. 1988; Prugnolle et al. 2009), the design of perfumes and deodorants (Behan et al. 1996; Caroprese et al. 2009; Natsch et al. 2005; Wysocki et al. 2009), and the ecology of host-seeking insects (Bernier et al. 2000; Dormont et al. 2013; Logan et al. 2008; Seenivasagan et al. 2014; Tauxe et al. 2013). However, only recently have individuals’ genetic, cultural, and ancestral backgrounds been explored with regard to human odor production (Harker et al. 2014; Martin et al. 2010; Prokop-Prigge et al. 2014). Much of the latter has focused on diversity that exists in a gene central to human odor production, viz., ATP-binding cassette, sub-family C, member 11 (ABCC11). This gene is represented in the human population by two alleles (C and T) resulting in three genotypes: CC (homozygous dominant), CT, and TT (recessive). The protein encoded by ABCC11, a member of the superfamily of ATP-binding cassette (ABC) transporters, facilitates the movement of various molecules across cell membranes. Furthermore, a functional, nonsynonymous single nucleotide polymorphism (SNP; rs17822931) in ABCC11 influences both apocrine and ceruminous gland secretions (Martin et al. 2010; Yoshiura et al. 2006). It has been reported that a SNP in ABCC11, 538 C→T, leads to a G180R substitution in the corresponding protein that results in the loss of ability to secrete metabolites. While the T allele is seen frequently (80–95%) in East Asian populations (Chinese, Japanese, and Korean), it is quite rare (0–3%) among individuals of European and African descent (Yoshiura et al. 2006). As a result, the cerumen (earwax) produced by East Asians typically exhibits a dry, white phenotype and is strikingly different from the wet, yellow cerumen produced by non-Asians. It also has been suggested that a SNP (rs9938025) in the PKD1L3 gene also may influence cerumen type (23andMe 2011). Individuals of Caucasian ancestry who are homozygous AA for SNP rs9938025 were found to have moderately lower odds of a dry cerumen phenotype compared to GG homozygotes, or AG heterozygotes (23andMe 2011). While this gene has not been linked previously to any body odor production (PKD1L3 and the related gene PKD2L3 form an ion channel involved in sour reception (Ishimaru et al. 2006)), we examined the relationship between cerumen VOCs and the (rs9938025) genotype within our subject population.
Several studies have addressed various biological aspects of ABCC11 rs17822931, including the close histological and functional relationship between ceruminous and apocrine sweat glands (Toyoda et al. 2009). This relationship is believed to explain the connection between cerumen type and axillary odor production. TT homozygotes for rs17822931 display few characteristic axillary odorants while the C allele is associated with sufficient production of axillary odor (Harker et al. 2014; Martin et al. 2010; Preti and Leyden 2010). We recently have described the nature and abundance of cerumen odor (Prokop-Prigge et al. 2014), and in the present report examine the effects of ethnicity and the ABCC11 SNP on cerumen volatile profiles. We hypothesized that cerumen volatiles, analogous to axillary odorants, can provide individual-specific information. As skin microbial composition strongly influences the production of human body odors (James et al. 2013a; Verhulst et al. 2010, 2011), we also investigated the influence of the ear microbiota on cerumen VOC production in a small subset of our subject population.
METHODS AND MATERIALS
Collection of Cerumen
Thirty-two male donors aged 21–40 years were enrolled in the study. All volunteers were informed about the aims of this study and provided written consent. The study was approved by the University of Pennsylvania Institutional Review Board (IRB) for Research Involving Human Subjects (Project # 816984). For 7–10 d prior to collection, subjects were instructed to bathe/shower with fragrance-free liquid soap/shampoo (Symrise, Inc. Teterboro, NJ, USA; provided by us) to reduce the influence of exogenous VOCs from consumer products during analysis. The subjects also were instructed not to use cotton-tipped applicators in their ears or apply any cologne or perfumed sprays during the entirety of the study.
Cerumen was collected from both ears of the donors: N = 10 of African descent (AfD), average age 30 ± 2, N = 11 of Caucasian descent (CaD), average age = 32 ± 4; and N = 11 of Asian descent (AsD), average age = 27 ± 2. Cerumen was collected on sterile, 6-in, cotton-tipped, wooden applicators (Fisher Scientific). The cotton applicator was inserted 10–15 mm into the subject’s external auditory canal and gently swabbed. The applicator was removed from the ear and cerumen was transferred to a pre-weighed 4 ml clear glass vial (Supelco Corp. St. Louis, MO, USA) by rotating the cotton tip for 20 sec on the bottom and sides of the vial. Collections were performed on at least three separate occasions on non-consecutive days. The cerumen sample mass was recorded after each collection.
Cerumen Volatile Sampling
Following cerumen collection, the sample vial was tightly capped with a white silicone/-TFE septum-containing screw cap and incubated in a 37°C water bath for 30 min. Solid-phase microextraction (SPME) was performed using a 2 cm, 50/30 µm divinylbenzyene/carboxen/polydimethylsiloxane ‘Stableflex’ fiber (Supelco Corp. St. Louis, MO, USA). The fiber was introduced into the vial, and the headspace VOCs were collected for an additional 30 min at 37°C. The SPME fiber then was inserted into the injection port of a GC/MS, and the VOCs were desorbed for 1 min at 230 °C.
GC/MS Analysis of Cerumen Volatiles
A Thermo Scientific ISQ single quadrupole GC/MS with Xcalibur software (ThermoElectron Corp.) was used for separation and analysis of the desorbed VOCs. The GC/MS was equipped with a Stabilwax column, 30 m × 0.32 mm with 1.0 µm film thickness (Restek Corp.). The injection port was set at 230°C. The oven temperature was held at 60°C for 4 min, raised to 230°C at 6°C min−1, and maintained at 230°C for 40 min. Helium carrier gas constantly flowed at 2.5 ml min−1.
The mass spectrometer was operated in electron ionization mode with an electron energy of 70 eV with a scan rate of 2 scans/s over the range of m/z 40–400 and an ion source temperature of 200°C. Identification of structures/compounds was performed using the National Institute of Standards and Technology Library, comparisons with known literature compounds and by comparison to commercially available standards. All standards were purchased from either Sigma-Aldrich or Alfa Aesar at the highest available purity and used as received.
The mass spectra of all peaks ~1% above the baseline and within a retention time range of 5–35 min were investigated. Exogenous components that arose from unwanted sources that can be attributed to liquid soap and cosmetic products (e.g., siloxanes, dodecanol), solvents (e.g., traces of acetone and chlorinated solvents), as well as compounds arising from the cotton-tipped applicators, septa, and column bleed (as identified by analyzing control samples of empty vials in which a clean, sterile cotton tip had been swabbed inside the vial) were excluded from analysis. Compounds consistently detected in a majority of donors were normalized by sample mass and by use of an external standard injected daily (methyl stearate) and then subjected to non-parametric analyses (there were too many violations of assumptions underlying parametric analyses) (IBM SPSS Statistics, v. 20) and multi-response permutation procedure (MRPP; (McCune et al. 2002; Parker and Mason 2009)) in the vegan package in R, a freeware, statistical and graphing software package. Global analysis for total composition differences were conducted first followed by pairwise comparisons across donor groups. Dimensions for nonmetric multidimensional scaling plots were derived in the vegan package in R and plotted in SigmaPlot 12.
Genotyping
Saliva was collected from all subjects, and DNA was isolated following kit manufacturer recommendations (DNA Genotek, Ottawa, Canada). Subjects were genotyped for the SNP 538 C→T in ABCC11 (rs17822931) and for SNP (rs9938025) in PKD1L3. Genotyping was performed using a 5’-exonuclease reaction (TaqMan) from Applied Biosystems (Foster City, CA, USA) and read in a 96-well plate format on a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA).
Collection of Microbiota, DNA Extraction, Amplification, and Sequencing of 16S rRNA Genes
Collection of microbiota samples were performed using sterile, foam-tip, buccal cell collection swabs (Epicenter, Madison, WI, USA) on separate days from cerumen collection for volatile analysis. During sample collection, the applicator was inserted 10–15 mm into the subject’s external auditory canal and swabbed in a circular motion approximately 10 times. The applicator tip was transferred to a sterile 1.5 ml Eppendorf tube and stored at −30°C until ready for further processing.
To isolate DNA, each swab was placed in 300 µl of Yeast Cell Lysis Solution (Epicentre MasterPure Yeast DNA Purification kit), and 0.5 µl of ReadyLyse Lysozyme solution (Epicentre) was added before incubation for 1 h at 37°C with shaking. Samples then were processed with bead beating for 10 min at maximum speed on a vortex mixer with 0.5 mm glass beads (MoBio) followed by a 30 min incubation at 65°C with shaking. Subsequent steps of DNA isolation were performed as previously described (Gardner et al. 2013). PCR of 16S rRNA genes was performed on a 2 µl sample DNA using barcoded forward primer 27F and a barcoded reverse primer 534R using a dual indexing strategy described in (Fadrosh et al. 2014). PCR was performed in duplicate with an Accuprime Taq DNA Polymerase High Fidelity kit (Invitrogen). The cycling conditions were as follows: 94°C for 3 min, then 35 cycles of 94°C for 45 sec, 50°C for 60 sec, and 72°C for 90 sec. Negative (no template and mock swab) controls were treated similarly and did not produce visible PCR products or sequencing reads. Duplicates were combined, and PCR products were purified using the Agencourt AMPure XP kit according the manufacturer's instructions (Beckman Coulter, Pasadena, CA, USA). Fifty ng of each sample were pooled and purified with the MinElute PCR Purification kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Sequencing was performed on an Illumina MiSeq Instrument using 300 bp paired-end chemistry at the University of Pennsylvania Next Generation Sequencing Core. The pre-processing pipeline similar to that described in (Fadrosh et al. 2014) was used to prepare sequences for input into QIIME 1.6.0 (Caporaso et al. 2010) for data analysis. A total of 215,594 paired-end sequencing reads were included in the analysis, with a mean of 35,932 and a median of 38,205 sequences per sample. Potential sequencing artifacts outside of the 502–534 base pair length window were removed using mothur (Schloss et al. 2009). Sequences were clustered into OTUs (operational taxonomic units, a proxy for ‘species’) using the UCLUST method (Edgar 2010) at 97% sequence similarity. Sequences then were taxonomically classified using the RDP classifier (Wang et al. 2007) at a confidence threshold of 0.8. Unclassified sequences were removed. Each 16S amplicon pool was subsampled at an even depth of 16,622 sequences for downstream processing. Alpha diversity metrics (Shannon diversity index and observed species-level OTUs) and beta diversity metrics (weighted and unweighted UniFrac) (Lozupone and Knight 2005) were calculated for each sample. 16S rRNA gene sequences are available in the NCBI Shortread Archive (SRA) under accession number PRJNA259879.
RESULTS
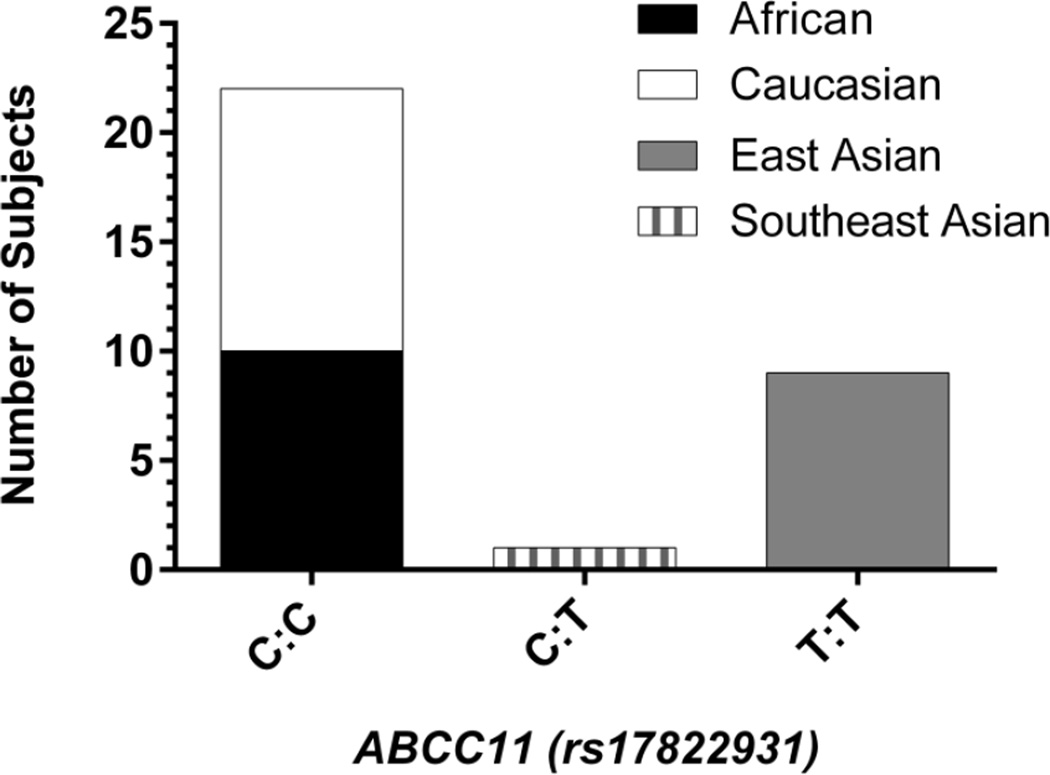
The ABCC11 genotype (rs17822931) of each volunteer was analyzed (Fig. 1). Eleven AsD donors, 10 of East Asian (Chinese, Korean, or Japanese) descent, were identified as TT homozygotes; 1 Southeast Asian donor from the Philippines was characterized as a CT heterozygote. The remaining 21 subjects, 11 CaD and 10 AfD, were CC homozygotes. These findings match the expected allele frequencies for these populations (Yoshiura et al. 2006). For the remaining analyses, East Asian and Southeast Asian groups were collapsed into a group designated simply as “AsD.” We found no correlation among the SNP (rs9938025) in PKD1L3 and either cerumen type (i.e., wet vs dry) or the relative levels of cerumen VOCs identified in our subject population.
Fig. 1.
ABCC11 genotype (rs17822931) of 32 cerumen donors
To investigate whether there were differences in cerumen odor profiles with regard to different ABCC11 genotypes and ethnicity, cerumen was collected from all genotyped donors. The amount collected from each donor displayed high variability (100 µg – 14 mg) with an average of 1.2 ± 0.4 mg across all subjects. There were no significant differences in amount collected based on ethnicity. The cerumen samples from the CaD and AfD donors were noticeably different from samples collected from AsD donors. This was consistent with previously reported findings (Yoshiura et al. 2006). While the samples from CaD and AfD donors were yellow and sticky in nature, cerumen collected from the AsD donors was dry, flaky and colorless.
Several identified compounds were attributed to exogenous sources, i.e., typical organic compounds found in room air, consumer product fragrances, as well as shampoos and soaps as previously reported in (Prokop-Prigge et al. 2014). For the purposes of this study, we focused on the endogenous compounds most commonly detected in all subjects. These VOCs are listed in Table 1. The major odorants found in human cerumen consist of short straight-chain and branched, volatile C2–C6 organic acids. The identified volatiles were qualitatively similar across all three groups of donors yet varied significantly in the relative amounts detected when comparing across donor groups.
Table 1.
Volatile organic compounds (VOCS) FOUND CONSISTENTLY IN THE HEADSPACE OF HUMAN CERUMEN SAMPLES AS DETECTED BY SOLID-PHASE MICROEXTRACTION GC/MS TECHNIQUESa
| RT | Compound | Major ion | Comparison across ethnicities (P-values)b |
|---|---|---|---|
| 11.94 | 6-methyl-5-heptene-2-one | 43 | 0.741 |
| 14.86 | acetic acid | 60 | 0.004 |
| 16.69 | propanoic acid | 74 | 0.054 |
| 17.25 | isobutyric acid | 43 | 0.033 |
| 18.48 | butyric acid | 60 | 0.002 |
| 19.28 | 2-methylbutanoic acid | 74 | 0.006 |
| 19.28 | isovaleric acid | 60 | 0.004 |
| 20.59 | valeric acid | 60 | 0.001 |
| 22.45 | trans-geranylacetone | 151 | 0.485 |
| 22.61 | hexanoic acid | 60 | 0.001 |
| 23.66 | dimethylsulfone | 79 | 0.002 |
| 24.42 | heptanoic acid | 60 | 0.026 |
| 25.34 | phenol | 94 | 0.695 |
| 26.21 | octanoic acid | 60 | 0.958 |
RT = retention time.
For a complete list of compounds identified see ref (Prokop-Prigge et al. 2014); however, the presence of many of these compounds were erratic across all subjects.
Kruskal-Wallis test
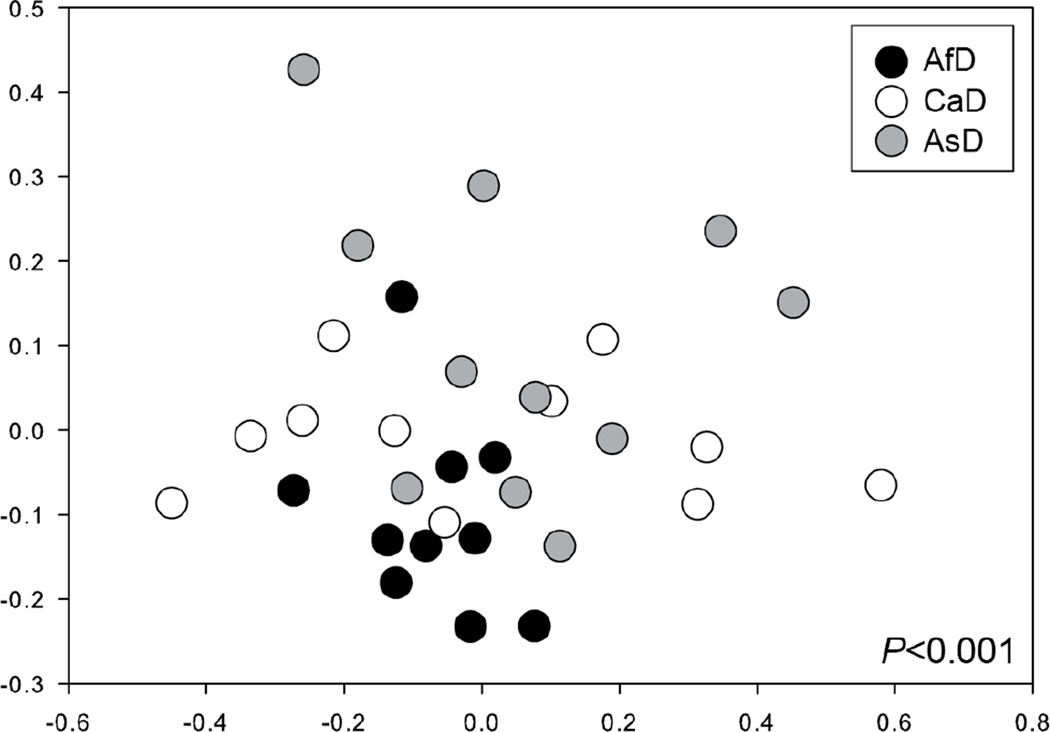
Non-metric multidimensional scaling, a non-parametric analog of principal component analysis, was employed to analyze the data, grouped by ethnicity, across all 14 compounds listed in Table 1. We first tested for global differences in VOC profiles using MRPP. VOC profiles differed significantly across ethnic groups (A=0.107, δ=0.581, P=0.001). We next conducted pairwise comparisons of VOC profiles between ethnic groups and found that AsD VOC profiles were significantly different from AfD VOC profiles (A=0.142, δ=0.570, P=0.001) and CaD VOC profiles (A=0.040, δ=0.599, P=0.033), though the latter difference is less distinct. Further, AfD and CaD VOC profiles were significantly different as well (A=0.069, δ=0.573, P=0.032). We then plotted VOC profiles in non-metric multidimensional space (NMS) to visualize how similar total VOC profiles were among individuals from each donor group (Fig. 2; stress = 15.8). Based on the NMS plot, VOC profiles from AfD subjects were less variable (i.e., points are closer together) than within the other two groups.
Fig. 2.
Non-metric multidimensional scaling plot showing global differences in volatile organic compound (VOC) profiles across individuals of African descent (AfD), Caucasian descent (CaD), and Asian descent (AsD). Data shown for 32 subjects, across all 14 compounds listed in Table 1.
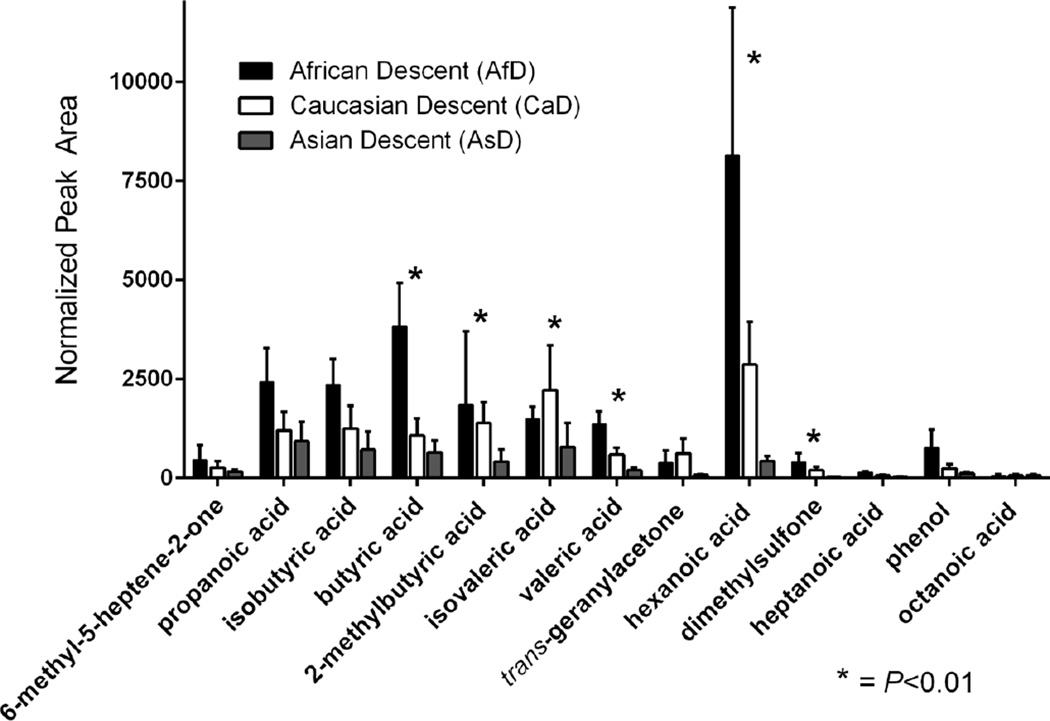
Subsequently, to probe the impact of donor group on the levels of individual compounds, we employed another non-parametric test, the Kruskal-Wallis test. The level of significance was established at P≤0.01 because there were multiple tests. Table 1 also shows the seven compounds that were significantly different across donor groups and the calculated P-values. In six of the seven compounds that were found to be statistically different, AfD > CaD > AsD in the amount of volatiles produced (Fig. 3).
Fig. 3.
Comparison of volatile organic compounds (VOCs) from the headspace of cerumen samples from individuals of African (AfD) (N = 10, black), Caucasian (CaD) (N = 11, white), and Asian (AsD) (N = 11, gray) descent averaged over three sample collections. Acetic acid (not shown because of scale) was elevated in subjects of AfD (39,935 ± 12,579 [s.e.]) compared to CaD (11,453 ± 6,055) and AsD (5,317 ± 2,182), and varied significantly across groups (P=0.004). Error bars represent standard error. Asterisks denote compounds that vary significantly (P<0.01) across ethnic groups. Note that within the homozygotic C:C donors, CaD produced significantly more isovaleric acid.
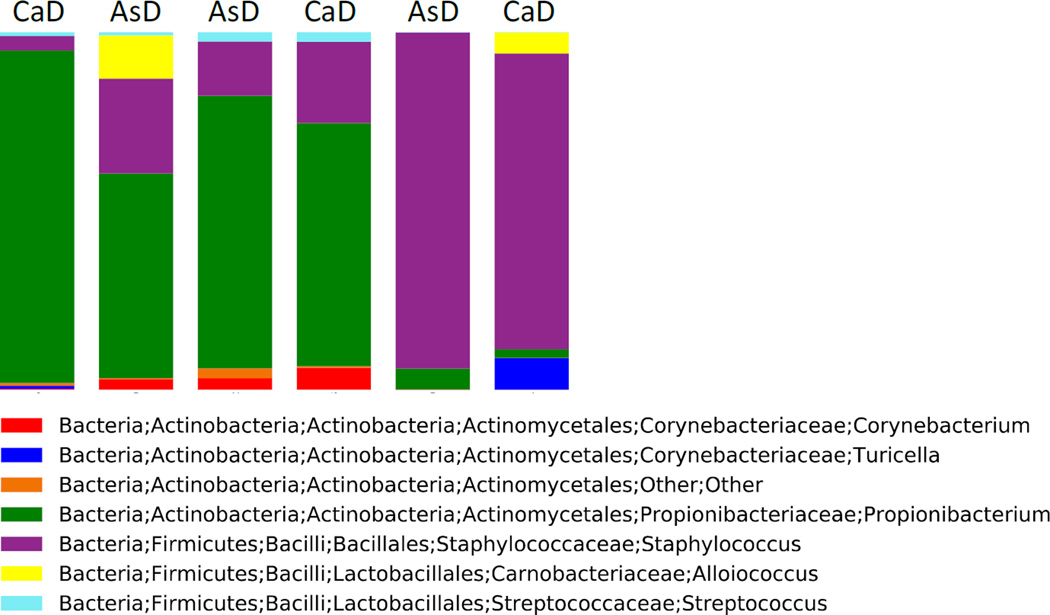
As axillary odor is, in-part, governed by the microbial population residing on the underarm skin’s surface (James et al. 2013a; Leyden et al. 1981; Natsch et al. 2003, 2004, 2005; Taylor et al. 2003), microbiomic analysis was performed to determine if individuals of different ethnicity or ABCC11 genotype exhibit altered microbial profiles of the external ear canal. The diversity and composition of ear bacteria were determined in a small sub-group of subjects (N = 3 AsD and N = 3 CaD). High-throughput 16S ribosomal DNA sequencing showed considerable variability in the small sample size examined (Fig. 4). However, variability was not dependent on donor group or the ABCC11 genotype. No significant differences between CaD and AsD subjects were observed in relative abundance of genus- and phylum-level taxa. No significant differences were observed in alpha diversity, or the within sample bacterial diversity, as calculated by number of species-level OTUs and the Shannon diversity index, which takes into account both richness and evenness. Clustering based on the weighted and unweighted UniFrac were not significant when taking into account donor group, as tested by the ANOSIM test (P=1.0 for both). These findings suggest that factors other than ethnicity and the ABCC11 genotype are responsible for modulating composition and diversity of external auditory canal microbiota.
Fig. 4.
Genus-level relative abundance of bacterial taxa present in three Asian (AsD) subjects and three Caucasian (CaD) subjects.
DISCUSSION
We examined, for the first time, the effects of both genetics and ethnicity/race on human cerumen volatiles. Employing SPME and GC/MS we identified the nature and abundance of cerumen VOCs from 32 male donors. Our results show that cerumen contains a complex mixture of VOCs and that the amounts of these compounds vary across individuals as well as across donor ethnic groups. By examining the composition and diversity of external auditory canal microbiota in a small subset of our subject population, we found that the ear microbiota may not be related to either donor group or ABCC11 genotype.
We found that human cerumen contains many of the same VOCs present in human axillary odor (i.e., ketones and short-chain fatty acids < C6), albeit not some of the odorants that characterize the axillae (e.g., (E)-3-methyl-2-hexenoic acid or 3-hydroxy-3-methyl-hexanoic acid). However, the compounds detected in axillary secretions are often technique dependent (Curran et al. 2005, 2010; Dormont et al. 2013; Martin et al. 2014; Mebazaa et al. 2011; Natsch et al. 2004; Penn et al. 2007; Preti et al. 2006; Riazanskaia et al. 2008; Zeng et al. 1991, 1992, 1996). A review of these studies demonstrates that the characteristic axillary odorants are not generally seen using headspace sampling (e.g., thermal desorption or SPME) but are typically detected when solvent extraction techniques are employed. For cerumen, we initially showed that cerumen’s odor appears to stem from the short chained organic acids found using headspace analysis (Prokop-Prigge et al. 2014).
Individuals of AfD and CaD (all C/C homozygotes for rs17822931), on average, emit significantly greater amounts of cerumen VOCs as compared to individuals of AsD (10 T/T homozygotes, one C/T heterozygote). These results are in agreement with previously published reports indicating that a functional C allele of ABCC11 is essential for the formation of human axillary odor (Martin et al. 2010), and the SNP in the ABCC11 gene results in lower levels of axillary odor precursors in T/T homozygotes compared to those carrying the C allele (Harker et al. 2014). Therefore, we hypothesize that cerumen can be used as a surrogate model for axillary odor production. Cerumen can be more quickly and readily sampled than axillary secretions and has the added advantage of bio-accumulation over time. The external ear canal is also less frequently exposed to potential exogenous compounds (e.g., lotions, detergents, deodorants) typically applied to the underarm region.
The findings reveal that ABCC11 alone is not solely responsible for the volatiles found in human cerumen. Despite the lack of a functional ATP-driven efflux pump believed to be responsible for the loss of odor production, the findings demonstrate that a TT genotype, corresponding to a dry cerumen phenotype, results in the production of lower, yet measurable, amounts of cerumen odorants. Other biochemical pathways must be involved in odor production or the secretion of odorous precursors to the skin’s surface. Given that all CaD and AfD subjects in this study were homozygous CC for the ABCC11 SNP, dramatic differences in the levels of cerumen volatiles were still detected. AfD donors produced greater amounts of VOCs than CaD for six out of the seven VOCs that varied significantly across donor group. It is interesting to note, however, that CaD had significantly more isovaleric acid than both AfD and AsD, suggesting that a significant difference in the metabolism or amount of the amino acid leucine, a precursor of isovaleric acid (James et al. 2013b), is present in cerumen of CaD. The results also show no effect of PKD1L3 SNP (rs9938025) on either cerumen type or VOC profile; 23andMe (2011) previously suggested variation in the SNP was associated with cerumen type. In addition, we report wide variation in the microbial residents inhabiting the ear canal in both AsD and CaD. Despite the wet or dry phenotype, the same microbial residents appear to thrive in both, suggesting that other environmental factors influence the type and number of bacterial inhabitants.
Human cerumen, an easily obtained bodily secretion, is an overlooked source of personal and diagnostic information. Previous research from our lab as well as others has shown that underarm odors can convey a great deal of information about an individual, including personal identity, gender, and sexual orientation (Kuhn and Natsch 2009; Martins et al. 2005; Penn et al. 2007). An individual’s metabolic processes, and hence the compounds emitted into the environment, are influenced by genetics, diet, stress, immune status, and the individual’s microbiome. Therefore, an individual’s odor profile is likely to be unique and undoubtedly contains a wealth of both physiological and behavioral information, as reported for many other animals. Our data suggest that human cerumen VOC profiles are quantitatively unique to the individual and suggest that cerumen may act as a site-specific human “odorprint.”
ACKNOWLEGEMENTS
The authors thank Jason Eades and Amanda Tyldsley for technical support. KAP acknowledges support from NIH-NIDCD Postdoctoral Training Grant 5T32DC0014. The genotyping was performed at the Monell Genotyping and DNA/RNA Analysis Core, which is supported, in part, by funding from the NIH-NIDCD Core Grant P30DC01173.
REFERENCES
- 23andMe. [Accessed 11 June 2014];Wet and dry earwax isn't all about one gene. 2011 https://www.23andme.com/about/factoid/cilantro/. Accessed.
- Behan JM, Macmaster AP, Perring KD, Tuck KM. Insight into how skin changes perfume. Int J Cosmetic Sci. 1996;18:237–246. doi: 10.1111/j.1467-2494.1996.tb00154.x. [DOI] [PubMed] [Google Scholar]
- Bernier UR, Kline DL, Barnard DR, Schreck CE, Yost RA. Analysis of human skin emanations by gas chromatography/mass spectrometry. 2. Identification of volatile compounds that are candidate attractants for the yellow fever mosquito (Aedes aegypti) Anal Chem. 2000;72:747–756. doi: 10.1021/ac990963k. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroprese A, Gabbanini S, Beltramini C, Lucchi E, Valgimigli L. HS-SPME-GC-MS analysis of body odor to test the efficacy of foot deodorant formulations. Skin Res Technol. 2009;15:503–510. doi: 10.1111/j.1600-0846.2009.00399.x. [DOI] [PubMed] [Google Scholar]
- Curran AM, Prada PA, Furton KG. The differentiation of the volatile organic signatures of individuals through SPME-GC/MS of characteristic human scent compounds. J Forensic Sci. 2010;55:50–57. doi: 10.1111/j.1556-4029.2009.01236.x. [DOI] [PubMed] [Google Scholar]
- Curran AM, Rabin SI, Prada PA, Furton KG. Comparison of the volatile organic compounds present in human odor using SPME-GC/MS. J Chem Ecol. 2005;31:1607–1619. doi: 10.1007/s10886-005-5801-4. [DOI] [PubMed] [Google Scholar]
- DeGreeff LE, Curran AM, Furton KG. Evaluation of selected sorbent materials for the collection of volatile organic compounds related to human scent using non-contact sampling mode. Forensic Sci Int. 2011;209:133–142. doi: 10.1016/j.forsciint.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Dormont L, Bessiere JM, Cohuet A. Human skin volatiles: A review. J Chem Ecol. 2013;39:569–578. doi: 10.1007/s10886-013-0286-z. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes. 2013;62:923–930. doi: 10.2337/db12-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker M, Carvell AM, Marti VP, Riazanskaia S, Kelso H, Taylor D, Grimshaw S, Arnold DS, Zillmer R, Shaw J, et al. Functional characterisation of a SNP in the ABCC11 allele - effects on axillary skin metabolism, odour generation and associated behaviours. J Dermatol Sci. 2014;73:23–30. doi: 10.1016/j.jdermsci.2013.08.016. [DOI] [PubMed] [Google Scholar]
- Horvath G, Jarverud GA, Jarverud S, Horvath I. Human ovarian carcinomas detected by specific odor. Integr Cancer Ther. 2008;7:76–80. doi: 10.1177/1534735408319058. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Inada H, Kubota M, Zhuang H, Tominaga M, Matsunami H. Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc Natl Acad Sci U S A. 2006;103:12569–12574. doi: 10.1073/pnas.0602702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AG, Austin CJ, Cox DS, Taylor D, Calvert R. Microbiological and biochemical origins of human axillary odour. Fems Microbiol Ecol. 2013a;83:527–540. doi: 10.1111/1574-6941.12054. [DOI] [PubMed] [Google Scholar]
- James AG, Cox D, Worrall K. Microbiological and biochemical origins of human foot malodour. Flavour Frag J. 2013b;28:231–237. [Google Scholar]
- Kuhn F, Natsch A. Body odour of monozygotic human twins: a common pattern of odorant carboxylic acids released by a bacterial aminoacylase from axilla secretions contributing to an inherited body odour type. J R Soc Interface. 2009;6:377–392. doi: 10.1098/rsif.2008.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J, Gallagher M, Ozdener MH, Wysocki CJ, Goldsmith BR, Isamah A, Faranda A, Fakharzadeh SS, Herlyn M, Johnson AT, et al. Volatile biomarkers from human melanoma cells. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;931:90–96. doi: 10.1016/j.jchromb.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Kwak J, Preti G. Volatile disease biomarkers in breath: a critique. Curr Pharm Biotechnol. 2011;12:1067–1074. doi: 10.2174/138920111795909050. [DOI] [PubMed] [Google Scholar]
- Leyden JJ, McGinley KJ, Holzle E, Labows JN, Kligman AM. The microbiology of the human axilla and its relationship to axillary odor. J Invest Dermatol. 1981;77:413–416. doi: 10.1111/1523-1747.ep12494624. [DOI] [PubMed] [Google Scholar]
- Logan JG, Birkett MA, Clark SJ, Powers S, Seal NJ, Wadhams LJ, Mordue AJ, Pickett JA. Identification of human-derived volatile chemicals that interfere with attraction of Aedes aegypti mosquitoes. J Chem Ecol. 2008;34:308–322. doi: 10.1007/s10886-008-9436-0. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Saathoff M, Kuhn F, Max H, Terstegen L, Natsch A. A functional ABCC11 allele is essential in the biochemical formation of human axillary odor. J Invest Dermatol. 2010;130:529–540. doi: 10.1038/jid.2009.254. [DOI] [PubMed] [Google Scholar]
- Martin HJ, Reynolds JC, Riazanskaia S, Thomas CL. High throughput volatile fatty acid skin metabolite profiling by thermal desorption secondary electrospray ionisation mass spectrometry. Analyst. 2014;139:4279–4286. doi: 10.1039/c4an00134f. [DOI] [PubMed] [Google Scholar]
- Martins Y, Preti G, Crabtree CR, Runyan T, Vainius AA, Wysocki CJ. Preference for human body odors is influenced by gender and sexual orientation. Psychol Sci. 2005;16:694–701. doi: 10.1111/j.1467-9280.2005.01598.x. [DOI] [PubMed] [Google Scholar]
- McCune B, Grace JB, Urban DL. Analysis of ecological communitites. Gleneden Beach, OR: MjM Software Design; 2002. [Google Scholar]
- Mebazaa R, Rega B, Camel V. Analysis of human male armpit sweat after fenugreek ingestion: Characterisation of odour active compounds by gas chromatography coupled to mass spectrometry and olfactometry. Food Chem. 2011;128:227–235. doi: 10.1016/j.foodchem.2011.02.063. [DOI] [PubMed] [Google Scholar]
- Natsch A, Gfeller H, Gygax P, Schmid J. Isolation of a bacterial enzyme releasing axillary malodor and its use as a screening target for novel deodorant formulations. Int J Cosmetic Sci. 2005;27:115–122. doi: 10.1111/j.1467-2494.2004.00255.x. [DOI] [PubMed] [Google Scholar]
- Natsch A, Gfeller H, Gygax P, Schmid J, Acuna G. A specific bacterial aminoacylase cleaves odorant precursors secreted in the human axilla. J Biol Chem. 2003;278:5718–5727. doi: 10.1074/jbc.M210142200. [DOI] [PubMed] [Google Scholar]
- Natsch A, Schmid J, Flachsmann F. Identification of odoriferous sulfanylalkanols in human axilla secretions and their formation through cleavage of cysteine precursors by a C-S lyase isolated from axilla bacteria. Chem Biodivers. 2004;1:1058–1072. doi: 10.1002/cbdv.200490079. [DOI] [PubMed] [Google Scholar]
- Parker MR, Mason RT. Low temperature dormancy affects the quantity and quality of the female sexual attractiveness pheromone in red-sided garter snakes. J Chem Ecol. 2009;35:1234–1241. doi: 10.1007/s10886-009-9699-0. [DOI] [PubMed] [Google Scholar]
- Penn DJ, Oberzaucher E, Grammer K, Fischer G, Soini HA, Wiesler D, Novotny MV, Dixon SJ, Xu Y, Brereton RG. Individual and gender fingerprints in human body odour. J R Soc Interface. 2007;4:331–340. doi: 10.1098/rsif.2006.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poling A, Weetjens BJ, Cox C, Mgode G, Jubitana M, Kazwala R, Mfinanga GS, Huis In 't Veld D. Using giant African pouched rats to detect tuberculosis in human sputum samples: 2009 findings. Am J Trop Med Hyg. 2010;83:1308–1310. doi: 10.4269/ajtmh.2010.10-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada PA, Furton KG. Human scent detection; a review of its developments and forensic appliactions. Rev Cienc Foren. 2008;1:81–87. [Google Scholar]
- Prada PA, Furton KG. Recent advances in solid phase microextraction for forensic applications. In: Pawliszyn J, editor. Comprehensive sampling and sample preparation. Analytical techniques for scientists. Oxford: Elsevier; 2012. [Google Scholar]
- Preti G, Labows JN, Kostelc JG, Aldinger S, Daniele R. Analysis of lung air from patients with bronchogenic carcinoma and controls using gas chromatography-mass spectrometry. J Chromatogr. 1988;432:1–11. doi: 10.1016/s0378-4347(00)80627-1. [DOI] [PubMed] [Google Scholar]
- Preti G, Leyden JJ. Genetic influences on human body odor: from genes to the axillae. J Invest Dermatol. 2010;130:344–346. doi: 10.1038/jid.2009.396. [DOI] [PubMed] [Google Scholar]
- Preti G, Willse A, Labows JN, Leyden JJ, Wahl J, Kwak J. On the definition and measurement of human scent: comments on Curran et al. J Chem Ecol. 2006;32:1613–1616. doi: 10.1007/s10886-006-9095-y. author reply 1617-1623. [DOI] [PubMed] [Google Scholar]
- Prokop-Prigge KA, Thaler E, Wysocki CJ, Preti G. Identification of volatile organic compounds in human cerumen. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;953–954:48–52. doi: 10.1016/j.jchromb.2014.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, Lefevre T, Renaud F, Moller AP, Misse D, Thomas F. Infection and body odours: evolutionary and medical perspectives. Infect Genet Evol. 2009;9:1006–1009. doi: 10.1016/j.meegid.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Riazanskaia S, Blackburn G, Harker M, Taylor D, Thomas CL. The analytical utility of thermally desorbed polydimethylsilicone membranes for in-vivo sampling of volatile organic compounds in and on human skin. Analyst. 2008;133:1020–1027. doi: 10.1039/b802515k. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seenivasagan T, Guha L, Parashar BD, Agrawal OP, Sukumaran D. Olfaction in Asian tiger mosquito Aedes albopictus: flight orientation response to certain saturated carboxylic acids in human skin emanations. Parasitol Res. 2014;113:1927–1932. doi: 10.1007/s00436-014-3840-x. [DOI] [PubMed] [Google Scholar]
- Tauxe GM, MacWilliam D, Boyle SM, Guda T, Ray A. Targeting a dual detector of skin and CO2 to modify mosquito host seeking. Cell. 2013;155:1365–1379. doi: 10.1016/j.cell.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D, Daulby A, Grimshaw S, James G, Mercer J, Vaziri S. Characterization of the microflora of the human axilla. Int J Cosmet Sci. 2003;25:137–145. doi: 10.1046/j.1467-2494.2003.00181.x. [DOI] [PubMed] [Google Scholar]
- Toyoda Y, Sakurai A, Mitani Y, Nakashima M, Yoshiura K, Nakagawa H, Sakai Y, Ota I, Lezhava A, Hayashizaki Y, et al. Earwax, osmidrosis, and breast cancer: why does one SNP (538G>A) in the human ABC transporter ABCC11 gene determine earwax type? FASEB J. 2009;23:2001–2013. doi: 10.1096/fj.09-129098. [DOI] [PubMed] [Google Scholar]
- Verhulst NO, Qiu YT, Beijleveld H, Maliepaard C, Knights D, Schulz S, Berg-Lyons D, Lauber CL, Verduijn W, Haasnoot GW, et al. Composition of human skin microbiota affects attractiveness to malaria mosquitoes. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0028991. DOI: 10.1371/journal.pone.0028991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst NO, Takken W, Dicke M, Schraa G, Smallegange RC. Chemical ecology of interactions between human skin microbiota and mosquitoes. Fems Microbiol Ecol. 2010;74:1–9. doi: 10.1111/j.1574-6941.2010.00908.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki CJ, Louie J, Leyden JJ, Blank D, Gill M, Smith L, McDermott K, Preti G. Cross-adaptation of a model human stress-related odour with fragrance chemicals and ethyl esters of axillary odorants: gender-specific effects. Flavour Frag J. 2009;24:209–218. [Google Scholar]
- Yoshiura K, Kinoshita A, Ishida T, Ninokata A, Ishikawa T, Kaname T, Bannai M, Tokunaga K, Sonoda S, Komaki R, et al. A SNP in the ABCC11 gene is the determinant of human earwax type. Nat Genet. 2006;38:324–330. doi: 10.1038/ng1733. [DOI] [PubMed] [Google Scholar]
- Zeng C, Spielman AI, Vowels BR, Leyden JJ, Biemann K, Preti G. A human axillary odorant is carried by apolipoprotein D. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6626–6630. doi: 10.1073/pnas.93.13.6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng XN, Leyden JJ, Brand JG, Spielman AI, McGinley KJ, Preti G. An investigation of human apocrine gland secetion for axillary odor precursors. J Chem Ecol. 1992;18:1039–1055. doi: 10.1007/BF00980061. [DOI] [PubMed] [Google Scholar]
- Zeng XN, Preti G, Leyden JJ, Lawley HJ, Sawano K, Nohara I. Analysis of the characteristic odors from the male axillae. J Chem Ecol. 1991;17:1469–1492. doi: 10.1007/BF00983777. [DOI] [PubMed] [Google Scholar]