Abstract
A novel microfluidic protein digestion system with nanostructured and bioactive inner surface was constructed by an easy biomimetic self-assembly strategy for rapid and effective proteolysis in 2 minutes, which is faster than the conventional overnight digestion methods. It is expected that this work would contribute to rapid online digestion in future high-throughput proteomics.
The ability to reliably identify and accurately quantify any protein targets in the proteome is an essential task in life sciences research and a fundamental challenge in clinical diagnosis.1 Specifically, mass spectrometry (MS)-based proteomics has continued to draw considerable interests, because MS can provide direct and intrinsic information regarding the peptides and screen multiple peptides simultaneously, thereby potentially allowing for the identification and quantification of all proteins contained in a sample.1–3 Notably, the digestion of proteins into peptides is a critical step in proteomics.4, 5 However, conventional in-solution and in-gel protein digestion by enzymes (e.g. trypsin) are compromised due to their restricted conditions, labour-intensive procedure, difficult recovery, and prolonged digestion time, making the digestion one of the bottlenecks in high-throughput proteomics.6, 7 Recently, it has been demonstrated that immobilization of enzymes on nanostructured material surface can enhance enzyme stability and increase enzyme-to-substrate ratio, thereby improving the overall digestion efficiency.8, 9 These advancements greatly encourage researchers to explore trypsin-immobilized nanostructures for high-throughput proteomics.10–12
Microfluidic devices have been demonstrated to be highly efficient tools and platforms in chemical synthesis, practical microanalysis and biomimetic tissue engineering due to the dramatically low sample requirement and enhanced speed, efficiency and sensitivity over conventional strategies.13–15 Besides the miniaturization, the potential for integration with designed nanostructures, various biomolecules or hybrid composites has opened the door to developing functional microfluidic devices for high-density, parallel sample processing, and high-throughput analysis.16–19 For example, enzyme molecules were entrapped in a microfluidic filter device by flow-induced gelation for sensitive biosensing of organophosphorus compounds.20 Furthermore, peptide/Pd nanowires were self-assembled in a microfluidic system for microchemical hydrogenation and Suzuki coupling reactions.21 In addition, via the integration of antibody, catalase and Pt catalyst into a microfluidic chip with uniquely designed channels, a multiplexed volumetric bar-chart chip was developed for visualized quantification of protein and DNA biomarkers.22, 23 Despite the successes in this area, few reports focus on the development of microfluidic devices with nanostructure for the rapid digestion of proteins.24, 25 This is because, besides the poor stability and recyclability of free trypsin molecules, the efficiency of microfluidic digestion system would be greatly challenged by the low density of active trypsin biomolecules due to the limited monolayer modification and small surface area of the smooth microfluidic inner surface as well as the complicated, rigorous and long-time chemical functionalization procedure. Therefore, it is still of great demand and high interest to develop simple and effective methods to prepare microfluidic devices with nanostructured and dense functional surfaces under mild conditions for rapid and efficient digestion of proteins in proteomics.
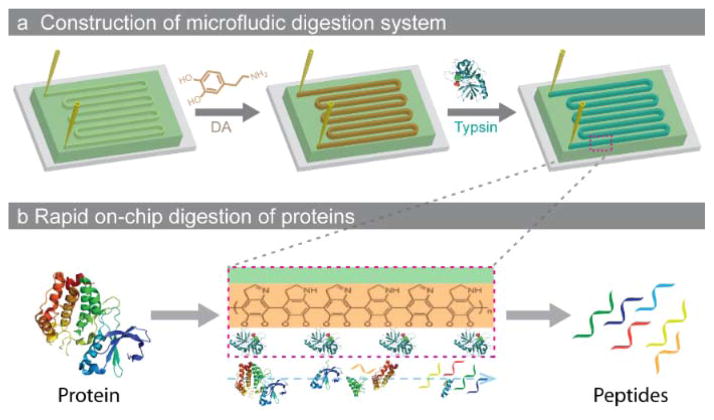
Inspired by the bioadhesion property of marine mussels,26 polydopamine have been suggested for the covalent modification of various inorganic and organic substrates. Owing to its durable stability, chemical versatility, excellent dispersibility and extraordinary biocompatibility, polydopamine is particularly suited for grafting biomolecules. Herein, the simple biomimetic surface chemistry was applied to functionalize the microfluidic channels, and after further covalent attachment of trypsin, the constructed microchip has been applied to rapid protein digestion. As shown in Scheme 1, the microfluidic chip with a meandering channel was first prepared using standard PDMS-based soft lithography with a mold of silicon wafer patterned with photoresist SU-8 (Fig. S1). The dopamine (DA) solution was then injected into the microchannel to form the functional polydopamine (PDA) layer via the self-polymerization process. Further injection of trypsin solution would result in the formation of the enzymatic surface due to the presence of catechol hydroxyl-groups on the PDA layer.26
Scheme 1.
Schematic illustration of (a) the construction of microfluidic protein digestion system and (b) the rapid on-chip digestion of proteins.
Fig. 1a and b show the photos and optical microscopy images of the PDMS chip before and after modification with PDA, respectively. Because the unmodified chip is transparent, it is difficult to distinguish the channel and PDMS solid wall. However, after PDA modification, the channel became yellow/brown and could be clearly observed (Fig. 1b inset). The optical microscopy shows that a translucent layer was attached on the surface of the whole channel, which indicates the formation of the PDA layer. SEM characterization was conducted to reveal the structure and composition details of the PDA layer. As shown in Fig. 1c and 1d, the modified inner surface of the channel is dense and coarse, and the PDA layer was assembled by numerous nanoparticles with size range of 50–100 nm. Furthermore, the inner surface topography of PDA modified channel was further characterized by the atomic force microscopy (AFM) at nano-scale. As shown in Fig. S2, before modification, the inner surface of the PDMS channel is relatively smooth. However, apparently, the inner surface of the PDA modified channel exhibits heterogeneous characteristics. The root mean square roughness (RMS) of the inner surface of the PDMS and PDA modified PDMS is calculated to be 4 nm and 169 nm, respectively. It should be noted that, compared with the smooth surface, the coarse surface assembled by the particles would provide much larger active surface and facilitate the interaction between the functional surface and the targets. To further confirm the successes of modification, Raman spectra of the channel’s surface were recorded (Fig. S3a). Compared with that of the unmodified microfluidic chips, new and strong bands (1326 cm−1 and 1589 cm−1) were presented in the PDA functionalized surface, which can be ascribed to the stretching and deformation vibration of catechol. These results demonstrate the successful modification of the PDMS channel with PDA. After further modification with trypsin molecules, as shown in the attenuated total reflection - Fourier transform infrared (ATR-FTIR) spectra (Fig. S3b), new characteristic absorption peaks of trypsin with weak intensities can also be observed in the range of 1400–1800 cm−1 and 3000–3750 cm−1, besides the strong adsorption peaks of the PDMS substrate. The above results demonstrate the successful construction of the microfluidic digestion system.
Fig. 1.
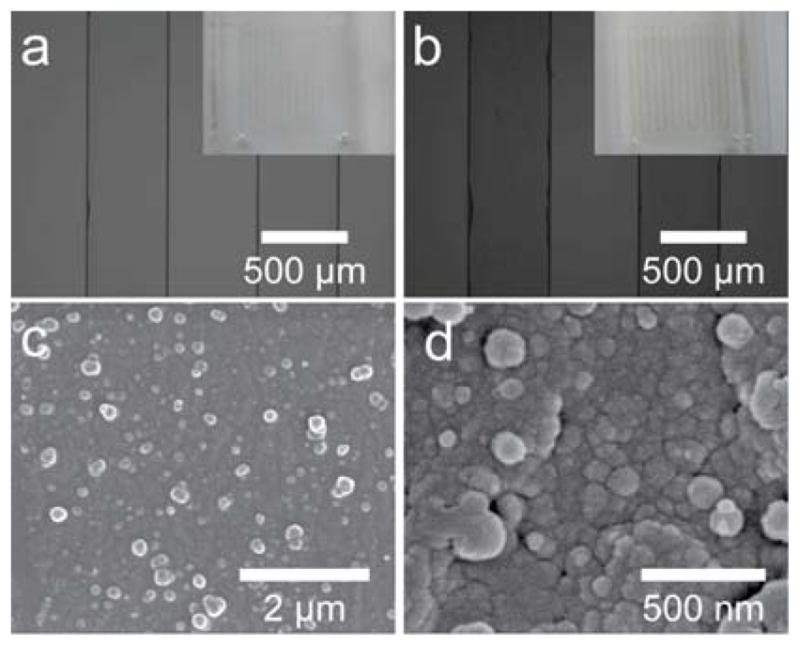
Optical microscope images of the microchannel before (a) and after (b) functionalization; insets are the photos of the microchip, respectively. SEM images with different magnification of the inner surfaces of the modified channels (c and d).
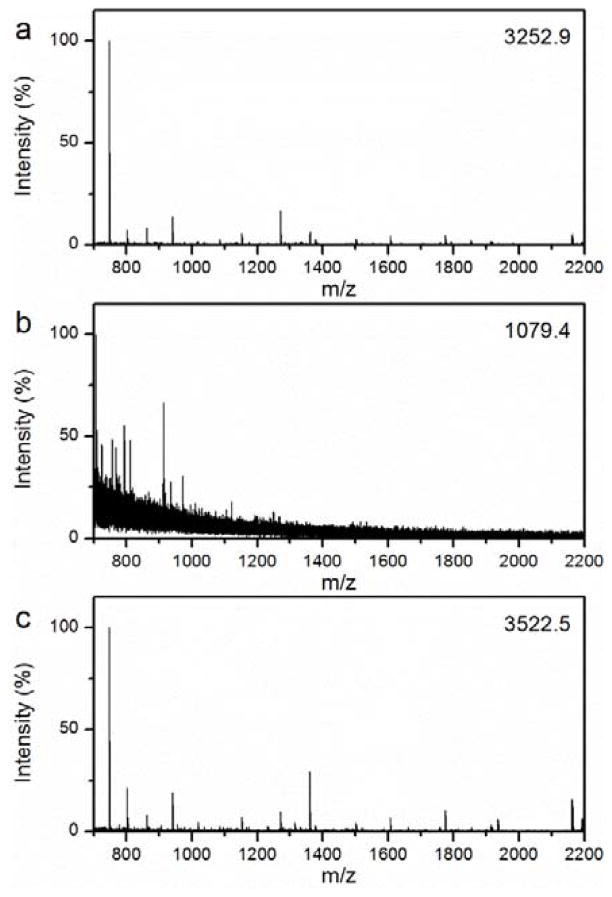
As shown in Scheme 1b, protein digestion can be easily realized in the microfluidic device. The protein solution was infused into the microfluidic digestion system by a syringe pump. The device was heated to 37 °C by a hot plate. Benefiting from the unique nanostructure and specific trypsin activity on the inner surface of the microfluidic channel, the protein molecules were rapidly digested as they passed though the microfluidic device. With matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), the resulting peptide solution could be detected and identified directly and rapidly. The feasibility and performance of the microfluidic digestion system was first evaluated using a model protein myoglobin which is a protein that is resistant to tryptic digestion.27 For comparison, the digestion of myoglobin solution using the most popular protocol, 12 hours in-solution digestion, was also conducted. As expected, the proteins can be effectively digested (Fig. 2a). However, besides the difficult recovery and reuse of free trypsin, the longtime requirement is still a bottleneck for the high throughput process and identification of proteins in proteomics research. Fig. 2b presents the MS of myoglobin digested by the in-solution method for 30 minutes. Only a few peaks with low intensities and signal-to-noise (S/N) ratios can be observed, because such a short time is not enough for all the protein molecules to interact with the limited trypsin molecules. For the digestion process via the microfluidic device, with a flow rate of 2 μL min−1, the protein solution passes through the channel very fast (less than 2 min). To our surprise, as shown in Fig. 2c, high quality MS could be obtained, and many peptide signals with relatively strong intensities and S/N ratios were detected. Furthermore, with the help of a database search, the protein can be effectively identified with a high number of matched peptides (15) and sequence coverage (86%), which is even better than that of the 12 hour in-solution digestion. Table S2 lists detailed information regarding the identified peptides. Notably, the influence of flow rate was investigated from both the theoretical and experimental perspectives, as discussed in the supporting information (Fig. S4 and Table S1).
Fig. 2.
MALDI-TOF mass spectra of myoglobin digested by (a) 12 hour in-solution digestion, (b) 30 min in-solution digestion, and (c) rapid on-chip digestion (< 2 min). The number in the upright corner is the intensity of the highest peak.
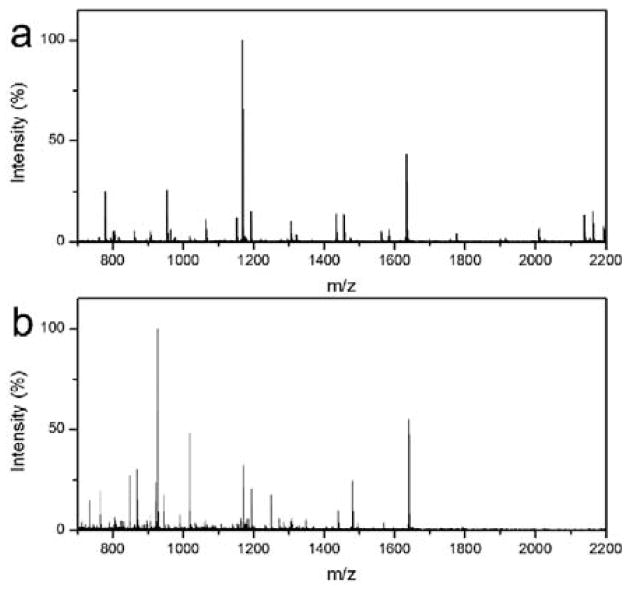
The high performance of the microfluidic digestion system can be attributed to the following reasons. First, the stability of the enzyme structures and interactions between protein subunits can be enhanced through immobilization on a solid support.28–30 In addition, the nanoparticles assembled coarse nanostructure would provide more functional surface for enzyme attachment, while enzyme activity would not decrease dramatically with the mild modification using the PDA based biomimetic method.26, 31 The local density of enzyme molecules is much higher than that of the in-solution counterpart, thereby improving the chances for the interaction between the proteins and trypsin and contributing to the high digestion efficiency. Notably, because the trypsin molecules were covalently grafted on the inner surface of the microfluidic channel assembled with polydopamine particles, the microfluidic digestion system can be regenerated by simply flushing the channel with elution buffer. As discussed and shown in the supporting information (Fig. S5 and S6), the microfluidic system has excellent recyclability. To further investigate the performance of the microfluidic digestion system for various proteins, cytochrome C (cyt-C; Mw=12 kDa) and bovine serum album (BSA; Mw=66 kDa) were demonstrated for the rapid on-chip digestion. Fig. 3a presents the typical mass spectrum of cyt-C digested by the microfluidic digestion system. High quality MS were obtained, and many peptide peaks with relatively strong intensities and S/N ratios were observed. Further identification of these peptides was realized by database search using Mascot. 18 peptides were matched and identified for cyt-C, and the sequence coverage is high at 92% (Table S3). As for the BSA with high molecular weight, high quality MS were also obtained (Fig. 3b), and 28 peptides with sequence coverage of 36% were identified after a database search by Mascot (Table S4). It indicates that the microfluidic digestion system is still effective for proteins with relatively large molecular weight. However, conventional in-solution digestion cannot realize effective digestion in a short time. As shown in Fig. S7, the MS of both the cyt-C and the BSA digested using the 30 minutes conventional digestion present poor quality. The above results demonstrate the advantages of the microfluidic digestion system.
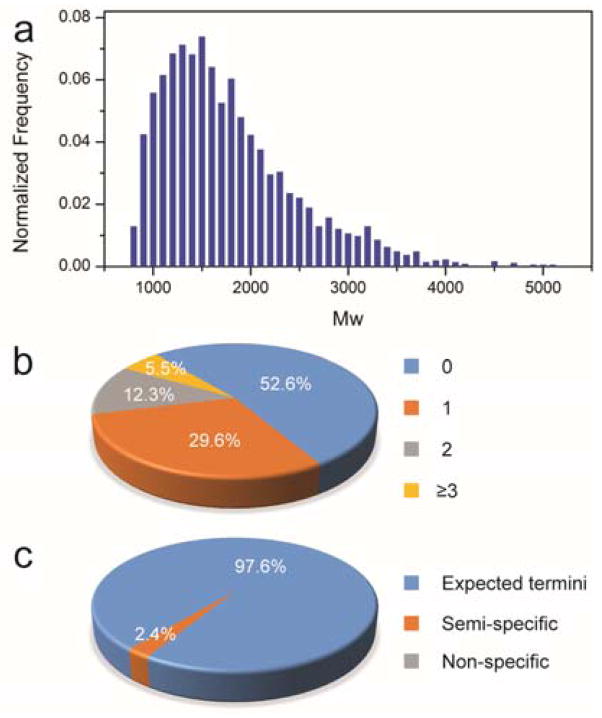
To explore the potential application in practical proteomics, a complex sample of proteins extracted from the cell lysate was digested by the microfluidic digestion system for evaluation. The digest was collected and analyzed by LC-MS for protein identification. The raw data of MS were then submitted to search engine with defining parameters for protein identification. After a database search, a total of 3478 peptides from 737 proteins with high confidence (p<0.05) were identified. The distribution of molecular weights (Mw) for the identified peptides is shown in Fig. 4a, and Mw of the identified peptides are mainly concentrated between 1000 and 2500, which implies the proteins were well digested. Fig. 4b and c represent statistical information of missed cleavages and expected digestion according to the sequence of identified peptides. Apparently, 94.5% of peptides were present with less than two miss-cleavages, although the sample solution passed through the microfluidic channel in two minutes, manifesting the effectiveness of the microfluidic digestion system. As shown in Fig. 4c, most of the peptides are identified with expected termini, and only 2.4% of them were semi-specific peptides which may arise from the over-cleavage. Above results demonstrate that the microfluidic digestion system can be applied for digestion of complex protein mixture extracted from cell lysate.
Fig. 4.
(a) The MW distribution of the identified peptides digested by the microwave-assisted tryptic digestion system using the microfluidic digestion system; distributions of the identified peptides according to the missed cleavages (b) and expected digestion (c).
Conclusions
A novel microfluidic digestion system was constructed via an easy biomimetic covalent self-assembly strategy. The inner microchannel surface with unique nanostructure and high trypsin activity endows the microfluidic device with the capability of rapid and effective on-line digestion of proteins. Model proteins and cell lysate proteins were used to evaluate the effectiveness of the microfluidic digestion system. The results show that the microfluidic digestion system can realize the rapid and effective digestion of proteins in 2 minutes as the solution flows through the microfluidic device. Furthermore, the microfluidic digestion system can be regenerated and reused. Thus, it is expected that this work will contribute to rapid on-line digestion in future high throughput proteomics. Notably, this strategy could also extend to the immobilization of other enzyme molecules for the construction of enzyme-based bio-microreactors, due to their similar properties. Therefore, this work could also present opportunities for the development of novel microfluidic platforms for various rapid on-line enzymatic reactions.
Supplementary Material
Fig. 3.
MALDI-TOF mass spectra of the Cyt-c (a) and BSA (b) digested using the microfluidic digestion system.
Acknowledgments
We appreciate assistance from the Penn State Materials Research Institute and Proteomics & Mass Spectrometry Core for facilities. This work was supported by the National Cancer Institute of the National Institutes of Health under Award Number DP2CA174508.
Footnotes
Electronic supplementary information (ESI) available: Experimental details and additional data are provided. See DOI: 10.1039/c000000x/
Notes and references
- 1.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yates JR., III Nat Methods. 2011;8:633–637. [Google Scholar]
- 3.Cheng G, Zhou MD, Zheng SY. ACS Appl Mater Interfaces. 2014;6:12719–12728. doi: 10.1021/am502712a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 5.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 6.Hartmann M, Jung D. J Mater Chem. 2010;20:844–857. [Google Scholar]
- 7.Slysz GW, Schriemer DC. Anal Chem. 2005;77:1572–1579. doi: 10.1021/ac048698c. [DOI] [PubMed] [Google Scholar]
- 8.Xue T, Jiang S, Qu Y, Su Q, Cheng R, Dubin S, Chiu CY, Kaner R, Huang Y, Duan X. Angew Chem Int Ed. 2012;51:3822–3825. doi: 10.1002/anie.201108400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng G, Zheng SY. Sci Rep. 2014;4:6947. doi: 10.1038/srep06947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng Y, Deng C, Qi D, Liu C, Liu J, Zhang X, Zhao D. Adv Mater. 2009;21:1377–1382. [Google Scholar]
- 11.Lin S, Yun D, Qi D, Deng C, Li Y, Zhang X. J Proteome Res. 2008;7:1297–1307. doi: 10.1021/pr700586j. [DOI] [PubMed] [Google Scholar]
- 12.Cheng G, Chen P, Wang ZG, Sui XJ, Zhang JL, Ni JZ. Anal Chim Acta. 2014;812:65–73. doi: 10.1016/j.aca.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 13.He M, Herr AE. Nat Protoc. 2010;5:1844–1856. doi: 10.1038/nprot.2010.142. [DOI] [PubMed] [Google Scholar]
- 14.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 15.Jun Y, Kang E, Chae S, Lee SH. Lab on a Chip. 2014;14:2145–2160. doi: 10.1039/c3lc51414e. [DOI] [PubMed] [Google Scholar]
- 16.Craighead H. Nature. 2006;442:387–393. doi: 10.1038/nature05061. [DOI] [PubMed] [Google Scholar]
- 17.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 18.Weinrich D, Jonkheijm P, Niemeyer CM, Waldmann H. Angew Chem Int Ed. 2009;48:7744–7751. doi: 10.1002/anie.200901480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirsaidov U, Mokkapati VRSS, Bhattacharya D, Andersen H, Bosman M, Ozyilmaz B, Matsudaira P. Lab on a Chip. 2013;13:2874–2878. doi: 10.1039/c3lc50304f. [DOI] [PubMed] [Google Scholar]
- 20.Lu D, Shao G, Du D, Wang J, Wang L, Wang W, Lin Y. Lab on a Chip. 2011;11:381–384. doi: 10.1039/c0lc00337a. [DOI] [PubMed] [Google Scholar]
- 21.Ryoo HI, Lee JS, Park CB, Kim DP. Lab on a Chip. 2011;11:378–380. doi: 10.1039/c0lc00448k. [DOI] [PubMed] [Google Scholar]
- 22.Song Y, Wang Y, Qin L. J Am Chem Soc. 2013;135:16785–16788. doi: 10.1021/ja4085397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song Y, Zhang Y, Bernard PE, Reuben JM, Ueno NT, Arlinghaus RB, Zu Y, Qin L. Nat Commun. 2012;3:1283–1283. doi: 10.1038/ncomms2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji J, Nie L, Qiao L, Li Y, Guo L, Liu B, Yang P, Girault HH. Lab on a Chip. 2012;12:2625–2629. doi: 10.1039/c2lc40206h. [DOI] [PubMed] [Google Scholar]
- 25.Bao H, Chen Q, Zhang L, Chen G. Analyst. 2011;136:5190–5196. doi: 10.1039/c1an15690j. [DOI] [PubMed] [Google Scholar]
- 26.Lee H, Dellatore SM, Miller WM, Messersmith PB. Science. 2007;318:426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo W, Bi H, Qiao L, Wan J, Qian K, Girault HH, Liu B. Mol Biosyst. 2011;7:2890–2898. doi: 10.1039/c1mb05140g. [DOI] [PubMed] [Google Scholar]
- 28.Hudson S, Cooney J, Magner E. Angew Chem Int Ed. 2008;47:8582–8594. doi: 10.1002/anie.200705238. [DOI] [PubMed] [Google Scholar]
- 29.Hartmann M. Chem Mater. 2005;17:4577–4593. [Google Scholar]
- 30.Wang G, Li X, Mo L, Song Z, Chen W, Deng Y, Zhao H, Qin E, Qin C, Tang R. Angew Chem Int Ed. 2012;51:10576–10579. doi: 10.1002/anie.201206154. [DOI] [PubMed] [Google Scholar]
- 31.Lee H, Rho J, Messersmith PB. Adv Mater. 2009;21:431–434. doi: 10.1002/adma.200801222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.