Summary
The human paracaspase MALT1 plays a central role in NF-κB signaling both as a protease and scaffolding protein. Knocking out MALT1 leads to impaired NF-κB signaling and failure to mount an effective immune response. However, it is unclear to which degree it is the scaffolding function versus the proteolytic activity of MALT1 that is essential. Previous work involving a MALT1 inhibitor with low selectivity suggests that the enzymatic function plays an important role in different cell lines. To help elucidate this proteolytic role of MALT1 we have designed activity-based probes that inhibit its proteolytic activity. The probes selectively label active enzyme and can be used to inhibit MALT1 and trace its activity profile helping to create a better picture of the significance of the proteolytic function of MALT1.
Keywords: acyloxymethyl ketone, B-cell, protease, activity-based probes
Introduction
The human paracaspase MALT1 (mucosa-associated lymphoid tissue lymphoma translocation protein 1) plays a vital role in NF-κB signaling downstream of T and B cell receptors (Lucas et al., 2001; Ruefli-Brasse et al., 2003; Ruland et al., 2003). In addition, other receptors such as Dectin-1 and G-protein coupled receptors have also been described to signal through MALT1 (Wegener and Krappmann, 2007). To signal, MALT1 forms a complex with the CARD-containing proteins CARMA1 (also known as CARD11) as well as its direct binding partner Bcl10 (Gaide et al., 2002; Pomerantz et al., 2002; Uren et al., 2000; Wang et al., 2002). Once the upstream signal is received, assembly of the CARMA1-Bcl10-MALT1 (CBM) complex ensues, allowing other downstream proteins such as TRAF6 and NEMO (NF-κB essential modulator, also known as IKK-γ) to assemble upon it (Oeckinghaus et al., 2007; Sun et al., 2004; Zhou et al., 2004). From this angle, the presumptive protease MALT1 seems to play an important but non-proteolytic role. However, upon its discovery almost 15 years ago (Akagi et al., 1999; Dierlamm et al., 1999; Morgan et al., 1999), its homology to the CD clan of proteases was noted (Uren et al., 2000). In 2008, the first proteolytic substrates A20 and Bcl10 were discovered (Coornaert et al., 2008; Rebeaud et al., 2008), followed by CYLD, RelB, Regnase-1, and Roquin (Hailfinger et al., 2011; Jeltsch et al., 2014; Staal et al., 2011; Uehata et al., 2013). In addition, the NF-κB inducing kinase NIK (Rosebeck et al., 2011) has been reported to be cleaved by an oncogenic fusion of the cellular inhibitor of apoptosis 2 (cIAP2, also known as AIP2) with MALT1 (cIAP2-MALT1), which occurs with varying frequencies in MALT lymphoma of different sites (Isaacson and Du, 2004). The effects of the cleavages have been described elsewhere (Coornaert et al., 2008; Hailfinger et al., 2011; Jeltsch et al., 2014; Rebeaud et al., 2008; Rosebeck et al., 2011; Staal et al., 2011; Uehata et al., 2013), but to summarize, the cleavages of A20 and RelB seem to have mostly fine-tuning effects on NF-κB signaling, whereas the cleavage of Bcl10 is required for increased cellular adhesion to fibronection after T cell activation, and CYLD cleavage plays a role in JNK signaling. Cleavage of the RNase Regnase-1 and the RNA binding protein Roquin disrupts the regulation of several mRNAs, some of which are involved in NF-κB signaling or are NF-κB target genes. The cleavage of NIK relieves it from intramolecular inhibition through its own TRAF3 domain and allows uncontrolled non-canonical NF-κB signaling. The overall importance of MALT1 for T and B cell development has been shown using MALT1 knockout mice, which fail to mount an effective immune response and NF-κB signaling is prevented (Ruefli-Brasse et al., 2003; Ruland et al., 2003).
Importantly, if the proteolytic activity of MALT1 is inhibited but the protein is still present to fulfill its scaffolding function, NF-κB signaling is decreased but not ablated (Cabalzar et al., 2013; Coornaert et al., 2008; Lucas et al., 2001; Rebeaud et al., 2008). Thus, questions remain about the importance of the proteolytic activity of MALT1. Initial results using B cell lymphoma cell lines and the broad-spectrum protease inhibitor Z-VRPR-fluoromethylketone (Z-VRPR-FMK), pointed toward an important role in the activated B cell-like subset of diffuse large B cell lymphoma (ABC-DLBCL) (Ferch et al., 2009; Hailfinger et al., 2009). When ABC-DLBCL cell lines were treated with Z-VRPR-FMK, cell viability declined, providing the first proof that inhibiting MALT1 proteolytic activity could be an important treatment option for this aggressive form of lymphoma. However, further studies are needed to confirm this, especially considering the low selectivity of the inhibitor used, most likely inhibiting not only MALT1 but also a broad spectrum of other proteases, making clear conclusions difficult. A mouse knock in model expressing the catalytic mutant of MALT1 (C464A) will certainly provide enlightening insights.
Here we describe inhibitory active site probes of MALT1 that enable visualization and quantification of MALT1 activity at a certain time point or after a stimulus. These activity-based probes consist of a peptide sequence determining enzyme selectivity, an electrophilic warhead, which irreversibly inhibits the targeted protease, and a label that allows detection and activity monitoring.
Results and Conclusions
Synthesis of activity-based probes directed against MALT1
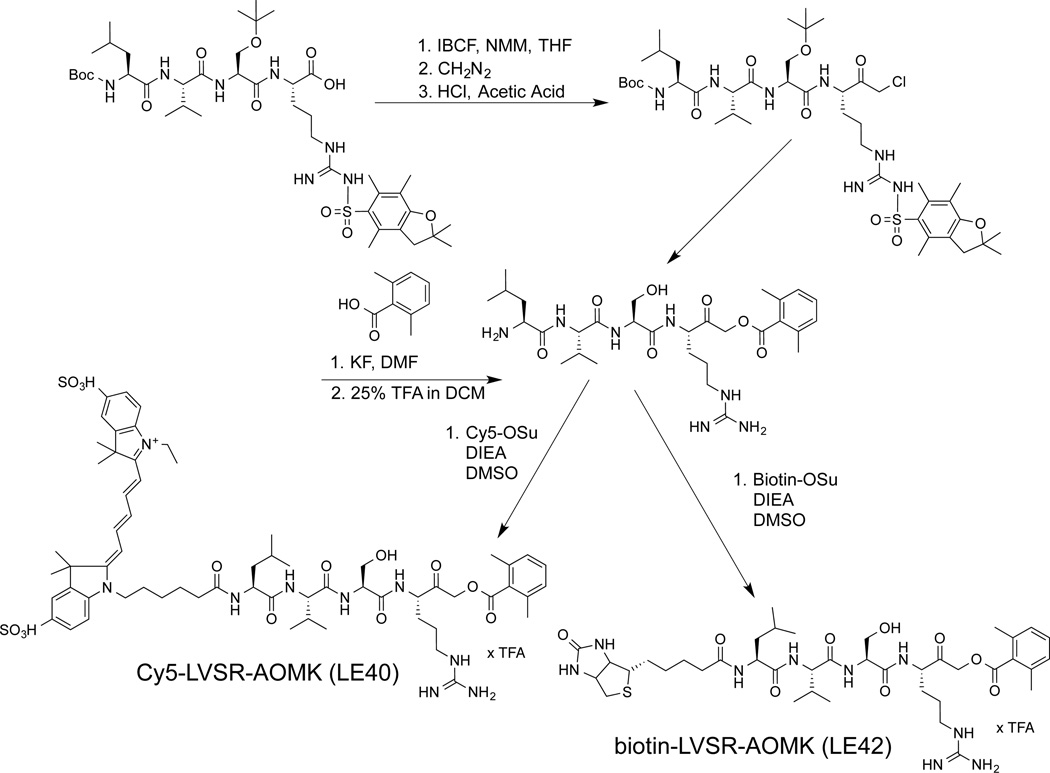
We used the cleavage site specificity determined earlier (LVSR↓) (Hachmann et al., 2012) to design MALT1 activity-based probes. We chose an acyloxymethyl ketone (AOMK) electrophile that preferentially inhibits cysteine proteases and has been used for probe targeting other clan CD proteases (Berger et al., 2006; Edgington et al., 2013; Kato et al., 2005a; Kato et al., 2005b; Powers et al., 2002). We included a Cy5 fluorophore or a biotin label, which allows affinity purification as well as direct imaging of probe modification (Figure 1).
Figure 1.
Synthesis scheme of the probes Cy5-LVSR-AOMK and biotin-LVSR-AOMK.
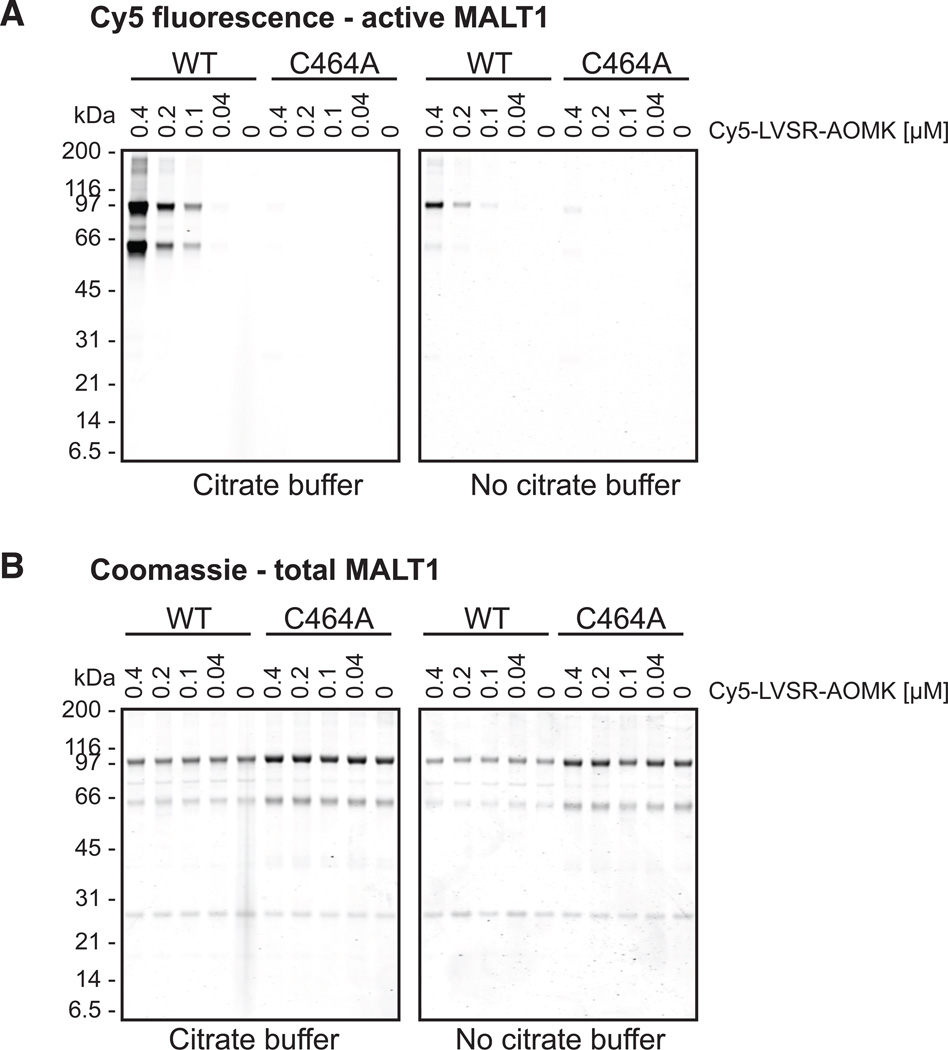
Preferential binding to activated recombinant MALT1
To test whether the inhibitors can distinguish between wild type and catalytic mutant on the one hand and activated and non-activated MALT1 on the other hand, we incubated purified recombinant enzyme with Cy5-LVSR-AOMK in the presence or absence of sodium citrate, which we and others have shown previously to activate MALT1 in vitro (Hachmann et al., 2012; Wiesmann et al., 2012). As shown in Figure 2, the activity-based probe can clearly differentiate between MALT1-WT and the catalytic mutant MALT1-C464A. No labeling is seen for the catalytic mutant whereas Cy5-LVSR-AOMK labels MALT1-WT in a dose-dependent manner, confirming that the probe indeed binds the active site. Labeling is strongly decreased in the absence of sodium citrate, indicating that the probe can distinguish between active and inactive protease. That some binding is seen in non-activating conditions is most likely due to a combination of the high concentration of enzyme present in vitro and substrate- (or in this case inhibitor-) induced generation of an active conformation, as previously reported (Wiesmann et al., 2012; Yu et al., 2011). As can be seen in Figure 2A, two protein species are labeled by Cy5-LVSR-AOMK. The upper species represents full-length MALT1, whereas the lower species was determined via mass spectrometry analysis to most likely be a C-terminal fragment encompassing the catalytic domain as well as the C-terminal extension including the Ig3-domain (Figure S3). The minimal number of peptides observed in mass spectrometry originating from the N-terminal region of MALT1 are likely carryover in the gel from the full-length protein. The intriguing idea that the C-terminal fragment could be an autoproteolytic cleavage product is contradicted by the fact that the catalytic mutant also shows this product. Since no fragments of this size are seen in cell culture experiments and no convincing cleavage sites are found in the sequence preceding the catalytic domain, we conclude that the cleavage is most likely due to an E. coli protease during expression and represents an in vitro artifact leading to two active species.
Figure 2.
Cy5-LVSR-AOMK can distinguish between MALT1-WT and C464A and preferentially binds under activating conditions. MALT1-WT or MALT1-C464A (400 nM) were incubated with Cy5-LVSR-AOMK at different concentrations in buffer with or without citrate for 30 min at 37°C, followed by TCA precipitation and SDS-PAGE. Fluorescence was scanned (A), followed by staining with a coomassie-based protein stain (B). Data are representative of three independent experiments.
To confirm that probe binding indeed leads to the inactivation of the enzyme and to obtain evidence that biotin-LVSR-AOMK also binds and inhibits MALT1, we measured the MALT1 rate of inhibition. Pseudo first order calculations (Table 1) revealed that the rate of inhibition is at least in the range of 103 M−1s−1 for the inhibitors Cy5-LVSR-AOMK and biotin-LVSR-AOMK, slightly exceeding the rate of inhibition of the previously used inhibitor Z-VRPR-FMK.
Table 1.
kobs/I values for inhibition of MALT1 by the probes/inhibitors used in this study. Standard errors were derived from the linear regression analyses of the kobs/I plots.
| Inhibitor | kobs/I (M−1s−1) |
|---|---|
| biotin-LVSR-AOMK | 9.1 (±1.6) × 103 |
| Cy5-LVSR-AOMK | 9.3 (±1.6) × 103 |
| Z-VRPR-FMK | 6.0 (±0.7) × 103 |
Detection of MALT1 activity upon ectopic expression in mammalian cells
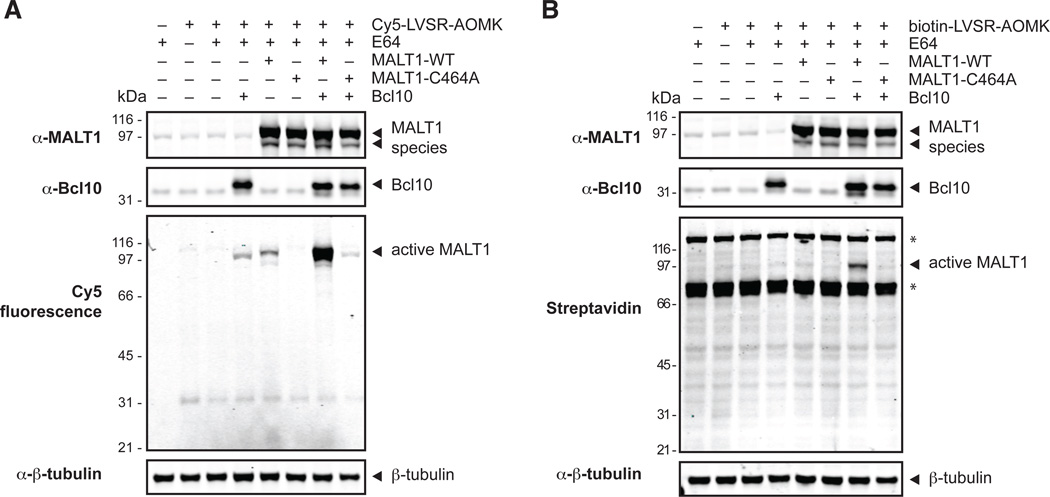
We next determined whether the activity-based probes can be used to detect active versus inactive MALT1 in lysates from a mammalian cell line transfected with the enzyme. MALT1 activity has been reported to be activated upon its co-overexpression with Bcl10 (Coornaert et al., 2008; Rebeaud et al., 2008). We therefore overexpressed MALT1 and Bcl10 in different combinations in human HEK-293A cells, followed by cell lysis in the presence of probe, SDS-PAGE and Western blot analysis. As shown in Figure 3A, overexpression of MALT1-WT in combination with Bcl10 leads to Cy5 fluorescence of a protein species around 100 kDa. Overexpression of MALT1 alone shows fluorescence as well, albeit to a much lesser degree. Overexpression of Bcl10 alone or in the presence of MALT1-C464A likewise leads to the appearance of a faint fluorescently labeled protein. Note that the small size difference is due to the Flag-tag of the overexpression construct. No fluorescence can be seen when MALT1-C464A is expressed alone, again indicating that Cy5-LVSR-AOMK cannot bind the catalytic mutant but requires an intact active site. Furthermore, this data confirms that the probe has generally low cross-reactivity towards other proteins in the cell.
Figure 3.
Cy5-LVSR-AOMK can detect active MALT1 after overexpression. Different combinations of Bcl10, MALT1-WT or C464A, respectively, were overexpressed in HEK-293A cells. Cells were lysed in the presence of Cy5-LVSR-AOMK (A) or biotin-LVSR-AOMK (B), respectively, precleared, and subjected to SDS-PAGE, fluorescence scanning, and Western blot analysis with the indicated antibodies. E-64 was added or omitted in the lysis buffer to control for cathepsin binding of the probe. Note that endogenous and Flag-tagged versions of the proteins migrate slightly differently during the SDS-PAGE. The asterisks mark non-specific bands. Data are representative of three independent experiments.
To inhibit binding of the probe to cathepsins, members of cysteine protease clan CA – some of which have substrate specificities that could overlap with MALT1 – all samples contained the protease inhibitor E-64, which has been shown to potently inhibit cathepsins (Barrett et al., 1982). Under these experimental conditions the probe bound most strongly to MALT1 and only a faint additional species around 31 kDa was seen. To determine whether this cross-reactivity would increase in the absence of E-64, we omitted the inhibitor from one sample. No significant intensity increase of the 31 kDa species could be seen, suggesting that the probe does not substantially cross-react with cathepsins, further validating its high degree of selectivity.
The biotin probe also enables the detection of active MALT1 after overexpression of MALT1-WT and Bcl10 (Figure 3B). No other probe-labeled proteins were detected, most likely due to the lower overall intensity of the biotin probe. Overall, it can be concluded that both probes can label and inhibit active MALT1 obtained from total cell lysates after overexpression in HEK-293A cells.
The blot for MALT1 antigen reveals two species, one at approximately the same size as endogenous MALT1, which can be seen in the non-transfected samples, and one that migrates at approximately 10 kDa lower. The lower species is not detected by the Flag antibody if the Flag-tag is placed at the N- instead of C-terminus (Figure S4). In the WT overexpression samples, this protein species is labeled by the probe suggesting that it represents an active C-terminal fragment. The fragment does not match the size of the species observed in the recombinant E. coli-produced samples (Figure 2). Again, both WT and mutant MALT1 are cleaved and the question remains whether the cleavage is autocatalytic or caused by another protease. It is theoretically possible that endogenous wild type MALT1 performs the cleavage in the mutant samples. However, this seems unlikely because the cleavage occurs to the same extent in all samples. While we did not further investigate this phenomenon, we speculate that MALT1 is cleaved by another protease. Whether this is only the case when MALT1 is overexpressed or whether it also occurs at endogenous levels cannot be definitively concluded due to the low intensity of the endogenous protein.
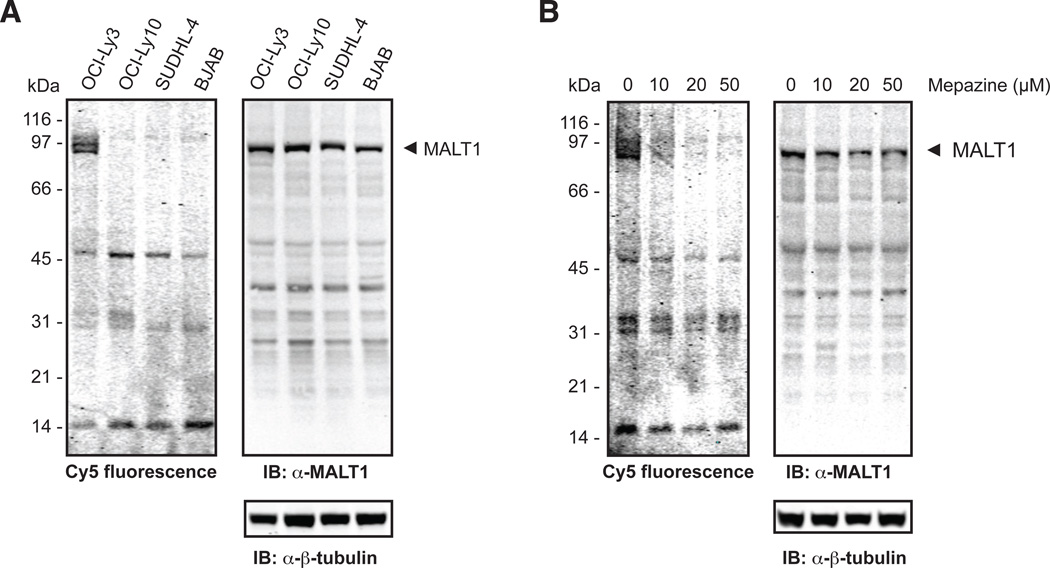
Cy5-LVSR-AOMK can detect endogenous MALT1 activity in OCI-Ly3 ABC-DLBCL cells
Having established that MALT1 activity can be detected in an overexpression system, we wanted to determine whether the more sensitive probe Cy5-LVSR-AOMK can be used to detect MALT1 activity in ABC-DLBCL cell lines. These have previously been reported to exhibit constitutive MALT1 activity, whereas cell lines derived from the germinal center B cell-like DLBCL subtype (GCB-DLBCL) did not (Ferch et al., 2009; Hailfinger et al., 2009). As shown in Figure 4A, the probe was able to visualize MALT1 activity in the ABC-DLBCL cell line OCI-Ly3, but not the ABC-DLBCL cell line OCI-Ly10 nor the GCB-DLBCL cell lines SUDHL-4 and BJAB (note that the faint fluorescent signal visible around 97 kDa in lanes 2–4 does not match in size with the MALT1 species detected by the antibody). While we were surprised to see the difference between OCI-Ly3 and OCI-Ly10 cells, the result nonetheless proved that the probe was sensitive enough to detect endogenous MALT1 activity and directly confirmed the constitutive MALT1 activity of OCI-Ly3 cells shown indirectly in previous publications (Ferch et al., 2009; Hailfinger et al., 2009). We asked why we could not observe activity of MALT1 as claimed previously for OCI-Ly10 cells (Ferch et al., 2009; Hailfinger et al., 2009). Along these lines we observed a decrease in signal intensity generated by active MALT1 in the OCI-Ly3 cell line with increasing time in culture. Upon thawing of a fresh vial the signal was back to its initial intensity (data not shown). This shows that culture conditions can lead to changes that affect MALT1 activity. While we could not solve the discrepancy between our results concerning MALT1 activity for the OCI-Ly10 cell line and previously published data, we speculate that culture conditions and subclones could well cause differences in MALT1 activity. Possibly, lymphoma-derived cells initially under the influence of MALT1 as a driver mutation become insensitive and acquire additional survival characteristics during prolonged passages. This speculation is consistent with our findings, but further clarification is beyond the scope of the current work. However, since our results show that the probe can detect active MALT1 if present at sufficient concentrations, such as in OCI-Ly3 cells or under overexpression conditions, it has the potential to help determine whether MALT1 is active in the cells and conditions investigated in a given study.
Figure 4.
Cy5-LVSR-AOMK can detect active endogenous MALT1 in OCI-Ly3 cells. (A) The ABC-DLBCL cell lines OCI-Ly3 and OCI-Ly10 and the GCB-DLBCL cell lines SUDHL-4 and BJAB were lysed in the presence of Cy5-LVSR-AOMK and E-64, precleared, and subjected to SDS-PAGE, fluorescence scanning, and Western blot analysis with the indicated antibodies. Note that the MALT1 band detected via Cy5 fluorescence in the OCI-Ly3 sample correlates with the band detected by the MALT1 antibody at ~ 90 kDa (lanes 1). The faint fluorescent bands visible in lanes 2–4 do not correspond to the species detected by the antibody, but migrate at a slightly higher MW. (B) OCI-Ly3 cells were treated with the indicated concentrations of mepazine for 4 h, followed by lysis as in B except that the lysis buffer was supplemented with the appropriate concentrations of mepazine. Data are representative of three independent experiments. IB: immunoblot.
Several additional fluorescent bands (visible at lower molecular weights around 45 kDa, 31 kDa, and 14 kDa) were detected in all DLBCL lines, but their intensities were lower than that of the fluorescent MALT1 species at approximately 90 kDa. To investigate whether these species represent active cleavage products of MALT1 or nonspecific binding of the probe, we pre-incubated OCI-Ly3 cells with increasing concentrations of the reversible MALT1 inhibitor mepazine (Nagel et al., 2012; Schlauderer et al., 2013). This caused a dramatic decrease in the intensity of the fluorescent species at 90 kDa, while the intensity of the lower molecular weight species remained unchanged, suggesting that they represent non-specific probe binding rather than MALT1 cleavage products. There is a formal possibility that the bands represent MALT1 cleavage fragments insensitive to mepazine. However, considering that the cross-reacting species are seen equally in all investigated DLBCL cell lines regardless of MALT1 activation status, it seems unlikely that the species are MALT1 cleavage products. The identity of the cross-reacting species was not investigated further at this point. Overall, Cy5-LVSR-AOMK was successfully used to detect active endogenous MALT1 and the constitutive activity of MALT1 in OCI-Ly3 ABC-DLBCL cells was confirmed in a direct manner.
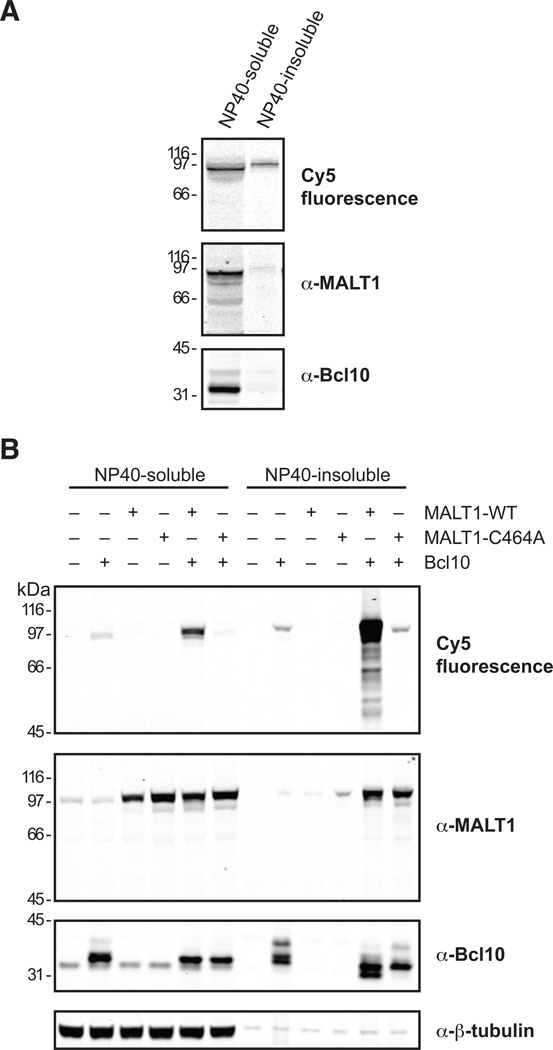
Endogenous active MALT1 is mostly NP40-soluble while overexpression leads to the formation of NP40-insoluble aggregates
After establishing that the activity-based probes can label active MALT1 in cell lysates, we deployed the Cy5 probe to address questions of MALT1 biology. The localization and assembly of components of the CBM complex have been a subject of considerable research with varying results depending on activation status and overexpression versus endogenous proteins (Che et al., 2004; Gaide et al., 2002; Guiet and Vito, 2000; Izumiyama et al., 2003; Qiao et al., 2013; Rossman et al., 2006; Schaefer et al., 2004; Yan et al., 1999). Until now it has not been possible to differentiate between active material and total protein. To determine the subcellular location of MALT1 activation we employed imaging flow cytometry and confocal microscopy. Our results were equivocal (data not shown), which we attribute to the hydrophobic nature of the Cy5 probe, such that even after extensive washing it remained partially bound to cells. This rendered it difficult to distinguish between cell penetration and nonspecific binding to the outside of cells. We conclude that the Cy5 probe shows at best low cell penetration potential, and that further optimization is needed if the probe is to be utilized for the detection and inhibition of MALT1 in intact cells.
Notwithstanding this limitation, we were able to obtain data on cell compartmentalization comparing endogenous versus overexpressed Bcl10 and MALT1 by employing differential detergent extraction. In the endogenous scenario, OCI-Ly3 cells constitutively activate MALT1 due to an activating mutation in CARMA1 (Lamason et al., 2010; Lenz et al., 2008). In the overexpression scenario, high protein concentrations of Bcl10 and MALT1 in HEK-293A cells activate the protease, most likely by causing constitutive oligomerization. In OCI-Ly3 cells the majority of endogenous MALT1, both active and total protein, as well as Bcl10, was detected in the NP40-soluble fraction (Figure 5A). This allowed us to conclude that the majority of active and total MALT1 is either present in soluble form in OCI-Ly3 cells or in a form that is solubilized under the lysis conditions used. In stark contrast to the OCI-Ly3 cell line, the overwhelming amount of active MALT1 was detected in the NP40-insoluble fraction upon ectopic coexpression of MALT1 and Bcl10 in HEK-293A cells (Figure 5B). Bcl10 was divided relatively evenly between the NP40-soluble and -insoluble fractions, whereas MALT1 protein was only present at substantial amounts in the NP40-insoluble fraction when co-expressed with Bcl10. Importantly, when overexpressed alone, MALT1 remained soluble. Significant amounts of cleaved Bcl10 are seen only in the NP40-insoluble fraction of cells overexpressing both Bcl10 and MALT1-WT, confirming that probe binding correlates with MALT1 proteolytic activity. The fact that the majority of cleaved Bcl10 is detected in the NP40-insoluble fraction together with the vast majority of active MALT1 suggests that Bcl10 associated with active MALT1 is cleaved preferentially.
Figure 5.
Endogenous MALT1 is NP40-soluble, whereas overexpressed active MALT1 is mostly NP40-insoluble. OCI-Ly3 cells (A) or HEK-293A cells transfected as indicated (B) were lysed in the presence of Cy5-LVSR-AOMK, followed by centrifugation and separation of the lysates into NP40-soluble and -insoluble fractions, SDS-PAGE, fluorescence scanning, and Western blot analysis with the indicated antibodies. Data are representative of three independent experiments.
We conclude that upon overexpression Bcl10 forms NP40-insoluble aggregates that contain a large fraction of total Bcl10. MALT1 on the other hand stays soluble when overexpressed alone, but is recruited to the aggregates upon co-overexpression with Bcl10. Whether active MALT1 seen in the NP40-soluble fraction originates from MALT1 that is active outside of the aggregates, or whether the aggregates are partially solubilized during lysis releasing some of the active MALT1, could not be determined from the experimental setup. Due to the formation of NP40-insoluble aggregates we conclude that total cell lysates should be used for analysis when studying the CBM complex to avoid missing crucial information.
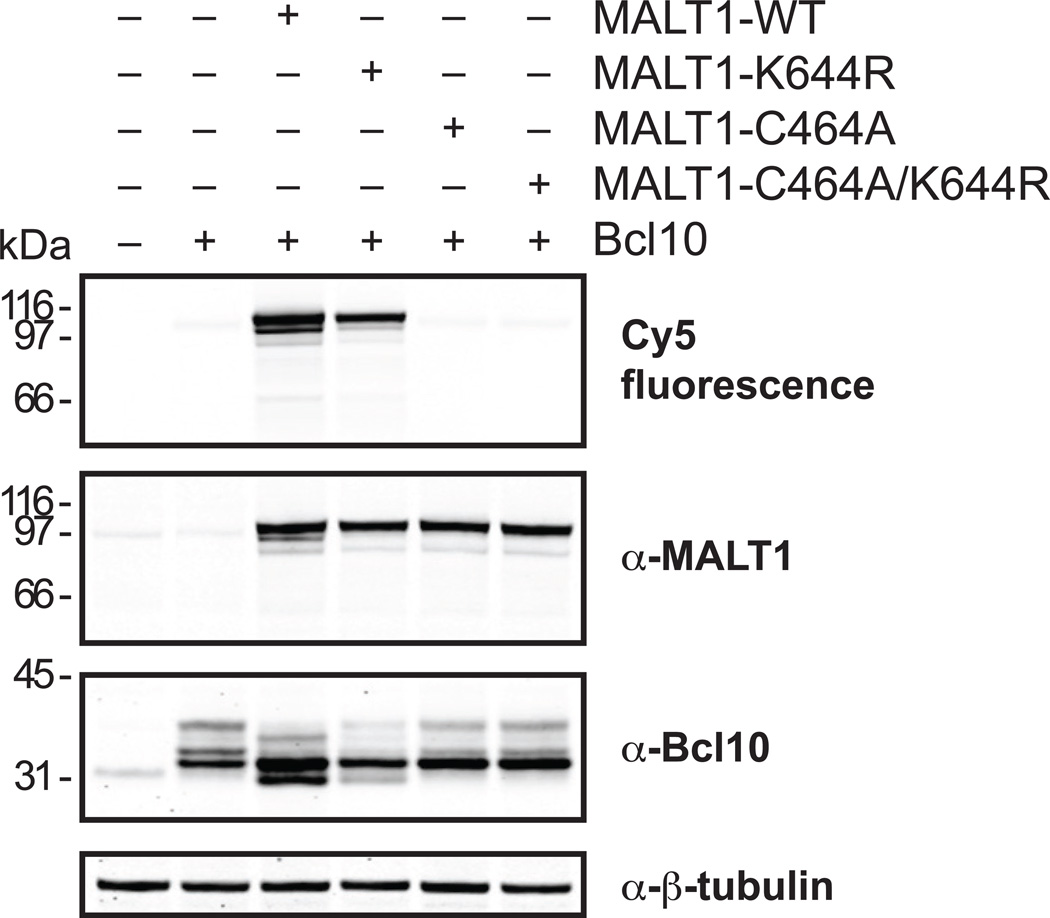
Mutation of MALT1 at K644 decreases but does not eliminate MALT1 activity
As another example of probe application, we investigated the importance of ubiquitination for MALT1 activity. It has been reported that monoubiquitination of MALT1 at K644 is essential for its proteolytic activity (Pelzer et al., 2013). We created the ubiquitin mutant K644R and checked its effect on MALT1 activity using probe binding as a readout. While overexpression of the catalytic mutant C464A lowered Cy5 probe binding to endogenous levels, the K644R mutant still bound the probe. It was substantially decreased compared to MALT1-WT, but ubiquitination at K644 was clearly not a requirement for MALT1 activity per se, which was supported by the decreased but clearly detectable level of Bcl10 cleavage seen in the K644R sample.
Discussion
We have described probes that can distinguish between active and inactive MALT1 both in the recombinant in vitro setting and in cell lysates.
After purification of MALT1 from E. coli, two distinct species are visible upon SDS-PAGE analysis that correspond to full-length MALT1 and a C-terminal fragment, respectively (Figures 2 and S3). Both represent active species of MALT1 and show probe binding. It is interesting to note that the fluorescence intensity of the two protein species is approximately equal in the sodium citrate containing samples even though the upper species is more abundant as shown by the coomassie stain. In the absence of citrate, labeling of the C-terminal fragment is strongly decreased compared to full-length enzyme. We speculate that this could be due to the absence of the N-terminal domains in this fragment, which have been reported to self-oligomerize (Qiu and Dhe-Paganon, 2011). In the presence of sodium citrate these domains might not play as important a role as in its absence.
Comparing the localization of MALT1 and Bcl10 protein in endogenous versus overexpression conditions revealed a clear discrepancy (Figure 5). While overexpressed active MALT1 localized predominantly in the NP40-insoluble fraction, active endogenous MALT1 was detected mostly in the NP40-soluble fraction. These results provide an example of how overexpression may create artificial conditions that do not accurately represent endogenous cellular processes. It remains to be determined whether accumulation in the NP40-insoluble fraction is simply due to higher protein levels upon overexpression, driving the formation of larger complexes that reach a critical size and thus become NP40-insoluble, or whether an entirely different mechanism of activation occurs under the different conditions. In case of the latter it would be interesting to reexamine the recent results pertaining to the filamentous aggregate structure of the CBM complex (Qiao et al., 2013).
Considering the NP40-insoluble nature of overexpressed active MALT1, we reevaluated the results shown in Figure 3 for which NP40-soluble material only had been analyzed. Even though we had to conclude that the amount of active MALT1 was most likely underestimated due to the experimental conditions, the main conclusion – that both Cy5-LVSR-AOMK and biotin-LVSR-AOMK can detect active MALT1 and distinguish between active and inactive MALT1 under overexpression conditions – remains valid.
When we elucidated the significance of MALT1 mutation at K644, which has been reported to alter a ubiquitination site important for its proteolytic activity (Pelzer et al., 2013), we determined that while the mutation clearly decreases MALT1 activity, ubiquitination at this site does not seem to be an absolute requirement (Figure 6). It is important to note that we could not detect a distinct higher molecular weight species representing ubiquitinated MALT1, which could be due to a lack of resolution during SDS-PAGE analysis. Another explanation could be the activity of deubiquitinating enzymes (DUBs) during lysis. The detection of ubiquitinated protein species is challenging due to the highly reversible nature of the modification. For efficient detection the cysteine alkylator N-ethylmaleimide (NEM) is commonly employed to inhibit DUBs, but in our case this was not feasible because NEM also inhibits the cysteine protease MALT1 and probe binding. Therefore we might be slightly underestimating the significance of ubiquitination for proteolytic activity. However, the important point is that in the absence of ubiquitination – whether achieved through the K644R mutation or DUBs – the probe still detects active MALT1. This, in combination with the fact that Bcl10 cleavage is decreased but not eliminated upon expression of the K644R mutant, points to a supporting role for ubiquitination at K644, although possibly not as essential as suggested in a previous report (Pelzer et al., 2013).
Figure 6.
Mutation of MALT1 at K644 decreases its activity. HEK-293A cells were transfected as indicated and lysed in the presence of Cy5-LVSR-AOMK, followed by SDS-PAGE of total cell lysates (no separation into NP40-soluble and insoluble fractions), fluorescence scanning, and Western blot analysis with the indicated antibodies. Note that the antibodies detect both endogenous and Flag-tagged versions of the proteins, which migrate at a slightly different MW. In addition, the MALT1 antibody detects MALT1 cleavage products of unknown significance and the Bcl10 antibody detects multiple higher MW species, which most likely represent posttranslationally modified variants. Data are representative of three independent experiments.
Another interesting observation when comparing the K644R mutant and WT enzyme is that MALT1 seems to be partially cleaved resulting in a slightly smaller fluorescent species than full-length enzyme. Compared to the K644R mutant the cleavage is preferentially seen in the WT enzyme and no cleavage is detected at all in the catalytic mutants, whether they retain their ubiquitin binding site or not. This raises the exciting possibility that the cleavage could potentially be autocatalytic. The difference in size is very small and more readily detected via fluorescence than by immunodetection. Probe fluorescence is therefore a powerful tool that can be used to detect small but potentially important effects involving MALT1 activity.
Significance
We have developed 2 activity-based probes that specifically bind and inhibit active MALT1. The probes can differentiate effectively between active and inactive MALT1 both when using recombinant enzyme, cellular overexpression and endogenous MALT1 in an ABC-DLBCL cell line. We have therefore provided tools that allow the monitoring of MALT1 activity. As examples of probe application we compared the subcellular localization of active MALT1 under endogenous and overexpression conditions as well as the influence of ubiquitin modification of MALT1 at K644 on its activity. We propose that these tools will prove to be useful in further elucidating MALT1 and the importance of its scaffolding versus proteolytic function.
Experimental Procedures
Probe synthesis
See Supplementary information for a detailed description of the probe synthesis.
Protein expression
Recombinant full-length MALT1 was expressed and purified as previously described (Hachmann et al., 2012).
Plasmids
Full-length MALT1 optimized for human expression (WT and catalytic mutant C464A, respectively) and Bcl10 were cloned into pcDNA3.1 containing a C-terminal Flag-tag. The K644R mutant was obtained using site-directed mutagenesis and cloned into the same vector as MALT1-WT and C464A.
Incubation of recombinant MALT1 with Cy5-LVSR-AOMK
Recombinant MALT1 and Cy5-LVSR-AOMK were incubated 30 min at 37°C in MALT1 assay buffer (50 mM HEPES, 100 mM NaCl, 10 mM DTT, 1 mM EDTA, pH 7.5) with or without 0.9 M sodium citrate, followed by TCA precipitation and SDS-PAGE using 4–12% Bolt Bis-Tris gels (Life Technologies). Fluorescence was scanned at 700 nm using an Odyssey infrared scanner (LI-COR Biosciences) and total protein was visualized using coomassie-based InstantBlue stain (expedeon).
Inhibition kinetics
Recombinant full-length MALT1 was incubated with increasing concentrations of inhibitor in MALT1 assay buffer with 0.9 M sodium citrate in the presence of 100 µM of the synthetic tetrapeptide substrate Ac-LRSR-AFC (SM Biochemicals). Kinetics of inhibition were evaluated by monitoring the release of the fluorophore over time using a Gemini Molecular Devices microplate spectrofluorometer. Progress curve data were analyzed by least squares analysis using GraphPad Prism to determine the pseudo-first order rate constant kobs. Derived second order rate constants (kobs/I) were calculated taking into account the factor (1+S/Km), where S is the substrate concentration and Km determined as 37.4 µM (Hachmann et al., 2012).
Cell culture and transfection
HEK-293A cells were cultured in Dulbecco’s Modified Essential Medium supplemented with 10% FBS, L-glutamine and antibiotics. Transfections were performed when the cells had reached a density of ~50% using the NanoJuice transfection reagent (Novagen) according to the manufacturer’s instructions. Cells were cultured for 24 h after transfection. The DLBCL cell lines OCI-Ly3, OCI-Ly10, SUDHL-4, and BJAB were a generous gift from the laboratory of John C. Reed, Sanford-Burnham Medical Research Institute. OCI-Ly3 and OCI-Ly10 were cultured in Iscove’s Modified Dulbecco’s Medium supplemented with 20% human AB serum (Corning), L-glutamine, β-mercaptoethanol, and antibiotics. SUDHL-4, and BJAB were cultured in RPMI 1640 medium supplemented with 10% FBS, L-glutamine, β-mercaptoethanol, and antibiotics.
Cell lysis and inhibitor treatment
24 h after transfection, cells were trypsinized and washed with PBS before lysis with a buffer containing 10 mM Tris, pH 7.4, 75 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, supplemented with 10 mM DTT and protease inhibitors [E-64, MG-132, 3,4-DCI (3,4-dichloroisocoumarin), and leupeptin (all at 10 µM)] unless noted otherwise. For the detection of endogenous MALT1 in DLBCL cells, 2x106 OCI-Ly3 cells or 6x106 OCI-Ly10, SUDHL-4, or BJAB cells, respectively, were washed with PBS and lysed as described above. Where applicable, Cy5-LVSR-AOMK or biotin-LVSR-AOMK, respectively, were added to the lysis buffer at 1 µM. Cells were lysed 15–30 min at 37°C, followed by centrifugation and separation into NP40-soluble and -insoluble fractions. Alternatively, in Figure 6 total cell lysates were obtained by adding SDS sample buffer to the uncleared lysates. Total cell lysates and NP40-insoluble fractions were sonicated to ensure complete solubilization. The lysates were resolved via SDS-PAGE, followed by fluorescence scanning at 700 nm using an Odyssey infrared scanner (LI-COR Biosciences), transfer onto nitrocellulose membrane, and antibody detection. Mepazine hydrochloride was a generous gift from the laboratory of Daniel Krappmann, Helmholtz Zentrum München – German Research Center for Environmental Health, Institute of Molecular Toxicology and Pharmacology.
Antibodies
Anti-MALT1 antibody was from Santa Cruz (B-12), anti-Bcl10 was from Cell Signaling (4237), anti-Flag was from Sigma-Aldrich (clone M2), anti-β-tubulin was from Abcam (ab6046). IRDye 800CW Streptavidin (LI-COR Biosciences) was used for biotin detection.
Supplementary Material
Acknowledgments
We thank Daniel Krappmann and John C. Reed for supplying reagents. We also thank Khatereh Motamedchaboki (Sanford-Burnham Medical Research Institute, Proteomics Facility), supported by NIH grant P30 CA030199, for help with mass spectrometry. This research was supported by NIH grant R01-GM09040 to G.S.S. and R01 EB005011 and R01 HL11630703 to M.B. M.D. received support from the Foundation for Polish Science and a statutory activity subsidy from the Polish Ministry of Science and Higher Education for the Faculty of Chemistry at Wroclaw University of Technology.
Abbreviations used
- ABC-DLBCL
activated B cell-like diffuse large B cell lymphoma
- Ac
acetyl
- AOMK
acyloxymethyl ketone
- AIP2
apoptosis inhibitor 2
- CARD
caspase recruitment domain
- CARMA1
CARD-containing MAGUK (membrane-associated guanylate kinase) 1
- cIAP2
cellular inhibitor of apoptosis protein 2
- 3,4-DCI
3,4-dichloroisocoumarin
- DD
death domain
- DTT
dithiothreitol
- DUB
deubiquitinating enzyme
- FMK
fluoromethylketone
- GCB-DLBCL
germinal center B cell-like diffuse large B cell lymphoma
- HEK
human embryonic kidney
- IAP
inhibitor of apoptosis protein
- Ig
immunoglobulin-like domain
- IKK
IκB kinase
- MALT1
mucosa-associated lymphoid tissue lymphoma translocation protein 1
- NEM
N-ethylmaleimide
- NEMO
NF-κB essential modulator
- NF-κB
nuclear factor κB
- NIK
NF-κB-inducing kinase
- TCA
trichloroacetic acid
- TRAF6
TNF receptor-associated factor 6
- WT
wild-type
- Z
benzyloxycarbonyl
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
J.H. designed and performed the experiments. M.B. and M.D. designed, L.E.E.-M. and M.P. designed and synthesized, and L.E.S. synthesized the probes. J.H, M.B. and G.S.S. conceived the study and wrote the paper.
References
- Akagi T, Motegi M, Tamura A, Suzuki R, Hosokawa Y, Suzuki H, Ota H, Nakamura S, Morishima Y, Taniwaki M, et al. A novel gene, MALT1 at 18q21, is involved in t(11;18) (q21;q21) found in low-grade B-cell lymphoma of mucosa-associated lymphoid tissue. Oncogene. 1999;18:5785–5794. doi: 10.1038/sj.onc.1203018. [DOI] [PubMed] [Google Scholar]
- Barrett AJ, Kembhavi AA, Brown MA, Kirschke H, Knight CG, Tamai M, Hanada K. L-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem J. 1982;201:189–198. doi: 10.1042/bj2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AB, Sexton KB, Bogyo M. Commonly used caspase inhibitors designed based on substrate specificity profiles lack selectivity. Cell Res. 2006;16:961–963. doi: 10.1038/sj.cr.7310112. [DOI] [PubMed] [Google Scholar]
- Cabalzar K, Pelzer C, Wolf A, Lenz G, Iwaszkiewicz J, Zoete V, Hailfinger S, Thome M. Monoubiquitination and Activity of the Paracaspase MALT1 Requires Glutamate 549 in the Dimerization Interface. PloS one. 2013;8:e72051. doi: 10.1371/journal.pone.0072051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che T, You Y, Wang D, Tanner MJ, Dixit VM, Lin X. MALT1/paracaspase is a signaling component downstream of CARMA1 and mediates T cell receptor-induced NF-kappaB activation. J Biol Chem. 2004;279:15870–15876. doi: 10.1074/jbc.M310599200. [DOI] [PubMed] [Google Scholar]
- Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21)p6ssociated with mucosa-associated lymphoid tissue lymphomas. Blood. 1999;93:3601–3609. [PubMed] [Google Scholar]
- Edgington LE, Verdoes M, Ortega A, Withana NP, Lee J, Syed S, Bachmann MH, Blum G, Bogyo M. Functional imaging of legumain in cancer using a new quenched activity-based probe. J Am Chem Soc. 2013;135:174–182. doi: 10.1021/ja307083b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferch U, Kloo B, Gewies A, Pfander V, Duwel M, Peschel C, Krappmann D, Ruland J. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2009;206:2313–2320. doi: 10.1084/jem.20091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaide O, Favier B, Legler DF, Bonnet D, Brissoni B, Valitutti S, Bron C, Tschopp J, Thome M. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat Immunol. 2002;3:836–843. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- Guiet C, Vito P. Caspase recruitment domain (CARD)-dependent cytoplasmic filaments mediate bcl10-induced NF-kappaB activation. J Cell Biol. 2000;148:1131–1140. doi: 10.1083/jcb.148.6.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmann J, Snipas SJ, van Raam BJ, Cancino EM, Houlihan EJ, Poreba M, Kasperkiewicz P, Drag M, Salvesen GS. Mechanism and specificity of the human paracaspase MALT1. Biochem J. 2012;443:287–295. doi: 10.1042/BJ20120035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, Penas EM, Dierlamm J, Chan WC, Staudt LM, et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2009;106:19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailfinger S, Nogai H, Pelzer C, Jaworski M, Cabalzar K, Charton JE, Guzzardi M, Decaillet C, Grau M, Dorken B, et al. Malt1-dependent RelB cleavage promotes canonical NF-kappaB activation in lymphocytes and lymphoma cell lines. Proc Natl Acad Sci U S A. 2011;108:14596–14601. doi: 10.1073/pnas.1105020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson PG, Du MQ. MALT lymphoma: from morphology to molecules. Nat Rev Cancer. 2004;4:644–653. doi: 10.1038/nrc1409. [DOI] [PubMed] [Google Scholar]
- Izumiyama K, Nakagawa M, Yonezumi M, Kasugai Y, Suzuki R, Suzuki H, Tsuzuki S, Hosokawa Y, Asaka M, Seto M. Stability and subcellular localization of API2-MALT1 chimeric protein involved in t(11;18) (q21;q21) MALT lymphoma. Oncogene. 2003;22:8085–8092. doi: 10.1038/sj.onc.1207002. [DOI] [PubMed] [Google Scholar]
- Jeltsch KM, Hu D, Brenner S, Zoller J, Heinz GA, Nagel D, Vogel KU, Rehage N, Warth SC, Edelmann SL, et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote TH17 differentiation. Nat Immunol. 2014;15:1079–1089. doi: 10.1038/ni.3008. [DOI] [PubMed] [Google Scholar]
- Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KA, Salvesen GS, Bogyo M. Activity-based probes that target diverse cysteine protease families. Nat Chem Biol. 2005a;1:33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- Kato D, Verhelst SH, Sexton KB, Bogyo M. A general solid phase method for the preparation of diverse azapeptide probes directed against cysteine proteases. Organic letters. 2005b;7:5649–5652. doi: 10.1021/ol052275v. [DOI] [PubMed] [Google Scholar]
- Lamason RL, McCully RR, Lew SM, Pomerantz JL. Oncogenic CARD11 mutations induce hyperactive signaling by disrupting autoinhibition by the PKC-responsive inhibitory domain. Biochemistry. 2010;49:8240–8250. doi: 10.1021/bi101052d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- Lucas PC, Yonezumi M, Inohara N, McAllister-Lucas LM, Abazeed ME, Chen FF, Yamaoka S, Seto M, Nunez G. Bcl10 and MALT1, independent targets of chromosomal translocation in MALT lymphoma, cooperate in a novel NF-kappa B signaling pathway. J Biol Chem. 2001;276:19012–19019. doi: 10.1074/jbc.M009984200. [DOI] [PubMed] [Google Scholar]
- Morgan JA, Yin Y, Borowsky AD, Kuo F, Nourmand N, Koontz JI, Reynolds C, Soreng L, Griffin CA, Graeme-Cook F, et al. Breakpoints of the t(11;18)(q21;q21) in mucosa-associated lymphoid tissue (MALT) lymphoma lie within or near the previously undescribed gene MALT1 in chromosome 18. Cancer Res. 1999;59:6205–6213. [PubMed] [Google Scholar]
- Nagel D, Spranger S, Vincendeau M, Grau M, Raffegerst S, Kloo B, Hlahla D, Neuenschwander M, Peter von Kries J, Hadian K, et al. Pharmacologic Inhibition of MALT1 Protease by Phenothiazines as a Therapeutic Approach for the Treatment of Aggressive ABC-DLBCL. Cancer Cell. 2012;22:825–837. doi: 10.1016/j.ccr.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Oeckinghaus A, Wegener E, Welteke V, Ferch U, Arslan SC, Ruland J, Scheidereit C, Krappmann D. Malt1 ubiquitination triggers NF-kappaB signaling upon T-cell activation. EMBO J. 2007;26:4634–4645. doi: 10.1038/sj.emboj.7601897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer C, Cabalzar K, Wolf A, Gonzalez M, Lenz G, Thome M. The protease activity of the paracaspase MALT1 is controlled by monoubiquitination. Nat Immunol. 2013 doi: 10.1038/ni.2540. [DOI] [PubMed] [Google Scholar]
- Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. EMBO J. 2002;21:5184–5194. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chemical reviews. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- Qiao Q, Yang C, Zheng C, Fontan L, David L, Yu X, Bracken C, Rosen M, Melnick A, Egelman EH, et al. Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly. Mol Cell. 2013;51:766–779. doi: 10.1016/j.molcel.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Dhe-Paganon S. Oligomeric Structure of the MALT1 Tandem Ig-Like Domains. PloS one. 2011;6:e23220. doi: 10.1371/journal.pone.0023220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeaud F, Hailfinger S, Posevitz-Fejfar A, Tapernoux M, Moser R, Rueda D, Gaide O, Guzzardi M, Iancu EM, Rufer N, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol. 2008;9:272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- Rosebeck S, Madden L, Jin X, Gu S, Apel IJ, Appert A, Hamoudi RA, Noels H, Sagaert X, Van Loo P, et al. Cleavage of NIK by the API2-MALT1 fusion oncoprotein leads to noncanonical NF-kappaB activation. Science. 2011;331:468–472. doi: 10.1126/science.1198946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Stoicheva NG, Langel FD, Patterson GH, Lippincott-Schwartz J, Schaefer BC. POLKADOTS are foci of functional interactions in T-Cell receptor-mediated signaling to NF-kappaB. Molecular biology of the cell. 2006;17:2166–2176. doi: 10.1091/mbc.E05-10-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302:1581–1584. doi: 10.1126/science.1090769. [DOI] [PubMed] [Google Scholar]
- Ruland J, Duncan GS, Wakeham A, Mak TW. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity. 2003;19:749–758. doi: 10.1016/s1074-7613(03)00293-0. [DOI] [PubMed] [Google Scholar]
- Schaefer BC, Kappler JW, Kupfer A, Marrack P. Complex and dynamic redistribution of NF-kappaB signaling intermediates in response to T cell receptor stimulation. Proc Natl Acad Sci U S A. 2004;101:1004–1009. doi: 10.1073/pnas.0307858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlauderer F, Lammens K, Nagel D, Vincendeau M, Eitelhuber AC, Verhelst SH, Kling D, Chrusciel A, Ruland J, Krappmann D, et al. Structural analysis of phenothiazine derivatives as allosteric inhibitors of the MALT1 paracaspase. Angewandte Chemie. 2013;52:10384–10387. doi: 10.1002/anie.201304290. [DOI] [PubMed] [Google Scholar]
- Staal J, Driege Y, Bekaert T, Demeyer A, Muyllaert D, Van Damme P, Gevaert K, Beyaert R. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J. 2011;30:1742–1752. doi: 10.1038/emboj.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, Satoh T, Mino T, Suzuki Y, Standley DM, et al. Malt1-Induced Cleavage of Regnase-1 in CD4 Helper T Cells Regulates Immune Activation. Cell. 2013;153:1036–1049. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- Uren AG, O'Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- Wang D, You Y, Case SM, McAllister-Lucas LM, Wang L, DiStefano PS, Nunez G, Bertin J, Lin X. A requirement for CARMA1 in TCR-induced NF-kappa B activation. Nat Immunol. 2002;3:830–835. doi: 10.1038/ni824. [DOI] [PubMed] [Google Scholar]
- Wegener E, Krappmann D. CARD-Bcl10-Malt1 signalosomes: missing link to NF-kappaB. Science's STKE : signal transduction knowledge environment. 2007;2007:e21. doi: 10.1126/stke.3842007pe21. [DOI] [PubMed] [Google Scholar]
- Wiesmann C, Leder L, Blank J, Bernardi A, Melkko S, Decock A, D'Arcy A, Villard F, Erbel P, Hughes N, et al. Structural Determinants of MALT1 Protease Activity. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Yan M, Lee J, Schilbach S, Goddard A, Dixit V. mE10, a novel caspase recruitment domain-containing proapoptotic molecule. J Biol Chem. 1999;274:10287–10292. doi: 10.1074/jbc.274.15.10287. [DOI] [PubMed] [Google Scholar]
- Yu JW, Jeffrey PD, Ha JY, Yang X, Shi Y. Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region. Proc Natl Acad Sci U S A. 2011;108:21004–21009. doi: 10.1073/pnas.1111708108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wertz I, O'Rourke K, Ultsch M, Seshagiri S, Eby M, Xiao W, Dixit VM. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
Supplemental References
- Edgington LE, van Raam BJ, Verdoes M, Wierschem C, Salvesen GS, Bogyo M. An optimized activity-based probe for the study of caspase-6 activation. Chem Biol. 2012;19:340–352. doi: 10.1016/j.chembiol.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington LE, Verdoes M, Ortega A, Withana NP, Lee J, Syed S, Bachmann MH, Blum G, Bogyo M. Functional imaging of legumain in cancer using a new quenched activity-based probe. J Am Chem Soc. 2013;135:174–182. doi: 10.1021/ja307083b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti C, Motamedchaboki K, Magrini A, Palmieri G, Mattei M, Bernardini S, Rosato N, Bottini N, Bottini M. Surface polyethylene glycol conformation influences the protein corona of polyethylene glycol-modified single-walled carbon nanotubes: potential implications on biological performance. ACS nano. 2013;7:1974–1989. doi: 10.1021/nn400409h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.