Abstract
Both pluripotent Embryonic Stem Cells (ESCs), established from preimplantation murine blastocysts, and Epiblast Stem cells (EpiSCs), established from postimplantation embryos, can self-renew in culture or differentiate into each of the primary germ layers. While the core transcription factors (TFs) OCT4, SOX2, and NANOG are expressed in both cell types, the gene expression profiles and other features suggest that ESCs and EpiSCs reflect distinct developmental maturation stages of the epiblast in vivo. Accordingly, ‘naïve’ or ‘ground state’ ESCs resemble cells of the ICM, whereas ‘primed’ EpiSCs resemble cells of the postimplantation egg cylinder. To gain insight into the relationship between naïve and primed pluripotent cells, and of each of these pluripotent states to that of non-pluripotent cells, we have used FAIRE-seq to generate a comparative atlas of the accessible chromatin regions within ESCs, EpiSCs, multipotent Neural Stem cells (NSCs) and Mouse Embryonic Fibroblasts (MEFs). We find a distinction between the accessible chromatin patterns of pluripotent and somatic cells that is consistent with the highly related phenotype of ESCs and EpiSCs. However, by defining cell-specific- and shared regions of open chromatin, and integrating these data with published gene expression- and ChIP analyses, we also illustrate unique features of the chromatin of naïve- and primed cells. Functional studies suggest that multiple stage-specific enhancers regulate ESC- or EpiSC- specific gene expression, and implicate auxiliary TFs as important modulators for stage-specific activation by the core TFs. Together these observations provide insights into the chromatin structure dynamics accompanying transitions between these pluripotent states.
Keywords: Pluripotent stem cells, accessible chromatin, transcriptional regulation, gene expression, enhancers
INTRODUCTION
Embryogenesis entails a program of events directing the ordered specification of cells within the pluripotent epiblast to an array of progenitor and differentiated cell fates. Pluripotent cells derived from pre-gastrulation mouse embryos and propagated in culture provide an in vitro model for defining features of the pluripotent state and those that set the stage for lineage-specific differentiation. Two types of murine pluripotent cells have been described: Embryonic Stem Cells (ESCs), established from cells of the Inner Cell Mass (ICM) of preimplantation blastocysts, and Epiblast Stem cells (EpiSCs), established from later, postimplantation embryos [1]. Although both cell types are pluripotent, they exhibit several distinguishing properties. ESCs self-renew in the presence of LIF and BMP, and can differentiate into extraembryonic endoderm (XEN), each of the three somatic lineages, or the germline, and efficiently contribute to chimeras [1, 2]. EpiSCs can also differentiate into each of the embryonic germ layers and germ cells [3–6] but, are not capable of differentiation toward XEN [7], are poorly incorporated into blastocyst chimeras, and their self-renewal requires FGF2 and Activin. While the core TFs OCT4, SOX2, and NANOG are expressed in both pluripotent cell types, ESCs and EpiSCs display distinct gene expression profiles, and many additional TFs that are important for ESC self-renewal are absent in EpiSCs [4, 6]. Thus ESCs and EpiSCs have been posited to represent two distinct states reflecting the developmental maturation stages of the epiblast in vivo: the ‘naïve’ or ‘ground state’ of ESCs resembling cells of the ICM, and the more mature ‘primed’ state of EpiSCs resembling pluripotent cells of the postimplantation egg cylinder just proximal to gastrulation [1]. Although the chromatin structure and molecular networks of ESCs have been intensely studied [8–10], far less is known regarding the molecular components of EpiSCs, and their relationship to those of ESCs.
Cell fate decisions, stage-specific gene expression, and the generation of specialized somatic cell types rely on specific combinations of transcription factors (TFs) that target distinct regulatory DNA elements to establish stage- or lineage-specific gene expression patterns. Since regulatory DNA elements targeted by these TFs generally reside within ‘accessible’ or ‘nucleosome-free regions’ of the genome, changes in TF- and promoter and enhancer activities in different developmental stages are reflected in a reorganization of the patterning of accessible regions. Thus insights into lineage relationships among cell types may be discerned by comparing their genome-wide patterns of accessible chromatin. Here we have used FAIRE-seq [11, 12] to generate a comparative atlas of accessible chromatin regions within ESCs, EpiSCs, NSCs, and MEFs, and to derive a profile of the chromatin dynamics accompanying epiblast maturation and the exit from pluripotency. We show that progression through each of the developmental stages represented by ESCs, EpiSCs, and the somatic cells is accompanied by the loss of a subset of open chromatin sites present in the progenitor stage, the retention of another subset of progenitor sites, and the concomitant acquisition of new sites in the more mature cell. Comparison of ESC- and EpiSC FAIRE-seq data, and their integration with previously reported gene expression and ChIP-seq analyses [13–15], define putative active and poised regulatory regions that are common to both pluripotent cell types, as well as stage-specific accessible regions that are unique to the naïve or primed state. ESC sites that become closed in EpiSCs include promoters and enhancers for genes expressed only in ESCs, and ‘poised’ promoters for genes of an alternate, XEN cell fate. Many ESC-specific FAIRE sites coincide with binding sites for OCT4/SOX2/NANOG TFs in ESCs, suggesting that these TFs are redistributed to distinct target DNA sites in EpiSCs. Functional testing of selected ESC- or EpiSC-specific accessible regions confirmed their ability to act as stage-specific regulatory enhancers. Together these data define shared and distinguishing chromatin features of naive- and primed- cells, and provide insights into the dynamics of chromatin structure that accompany transitions between these pluripotent states.
MATERIALS AND METHODS
Additional detailed Methods are provided in the Supporting Information
Cell culture
E14 Mouse embryonic stem cells were maintained in 2i/LIF medium (Supporting Information, [16]), and E3 Epiblast Stem cells were maintained as in [17]. Neural Stem cells were generated by ESCs differentiation as in [18] The character and integrity of each of these cell states was confirmed by RT-PCR analysis using oligonucleotide primers for the detection of key markers of the naïve or primed pluripotent state, or of NSCs (Supporting Information Fig. S1). Mouse Embryo Fibroblasts were derived from E16.5 embryos.
FAIRE-seq
107 cells of each cell line were crosslinked in 1% Formaldehyde for 10 minutes. Replicate samples were processed for FAIRE and Illumina sequencing [11, 12, 19]. FASTQ files were generated using CASAVA 1.8. (Supporting Information Table S1). FAIRE-seq genomic DNA reads were aligned to the mouse genome (mm9) and peak calling was performed with Qeseq [20] using default parameters.
Comparison of ESC FAIRE peaks with DNaseI data
ESC FAIRE-seq peaks were compared with previously reported ESC DNaseI hypersensitive sites (downloaded from UCSC genome browser (http://genome.ucsc.edu/) [21], Supporting Information Table S2). ROC curves were generated to calculate the correspondence of ESC FAIRE peaks with DNaseI data (Supplemental Methods and Fig. S2). DNaseI treatment of ESCs and EpiSCs was performed as previously [22] and assayed using PCR and oligonucleotide primers complementary to several genomic regions harboring FAIRE peaks.
Genomic Distribution and Shared peak Analyses
Annotations for the mouse genome (mm9) obtained from the UCSC table browser [23] included TSSs, exons, introns and promoter regions (i.e. rup to 10kbp upstream from a TSS where no other gene present). Regions without annotation were labeled ‘Intergenic’. Proximal regions were defined as regions within 2kbp from a TSS. The number of base pairs within a FAIRE peak overlapping a specific feature (i.e. proximal/distal, promoter/exon/intron/intragenic) was counted. Overlap of FAIRE peaks among cell lines was determined by computing the fraction of peaks for each cell line having at least one nucleotide in common with peaks from other cell lines. For Hierarchical clustering analysis, the genome was partitioned into 1kbp bins and a binary profile was calculated for FAIRE peaks of each cell line. Hierarchical clustering, ROC curves, and the Kruskal–Wallis one-way analysis of variance test [24], were done using MATLAB (The MathWorks, Natick, MA).
FAIRE clusters
Genomic regions containing high density clusters of FAIRE peaks were identified for each FAIRE dataset (threshold ≥5) using Galaxy (https://usegalaxy.org/). Cell line-specific clusters were identified by subtracting those that displayed a 50bp or more overlap with a cluster in any other cell line.
Plasmid Construction and luciferase assays
Primers with flanking BglII sites (Supporting Information Table S3) were used to PCR amplify test fragments. PCR products were inserted into the luciferase reporter plasmids as indicated in Figure legends. ESCs or EpiSCs were transfected with reporter plasmids using Lipofectamine. Lysates were prepared and luciferase assays were performed as instructed by the manufacturer (Promega). Lysates were normalized by protein concentration (Bio-Rad Protein Assay Reagent, Bio-Rad). The luciferase activities of test constructs were normalized relative to the a control luciferase construct as indicated in each figure.
RESULTS
A comparative atlas of Open Chromatin in murine ESCs, EpiSCs, NSCs, and MEFs
‘Nucleosome-free regions’ within chromatin consist of displaced- or relatively disordered nucleosome arrays and exhibit increased susceptibility to nucleases such as DNaseI [25]. FAIRE-seq (Formaldehyde-assisted identification of regulatory elements-seq) provides an alternative to nuclease-based methods for determining genome-wide accessible chromatin patterns within cultured cells or tissues [12, 26].
To create a comparative atlas of open chromatin within ESCs, EpiSCs, NSCs, and MEFs, replicate samples of formaldehyde-crosslinked chromatin were subjected to FAIRE [11], sequenced using the Illumina GAIIx, and the resulting reads aligned to the mouse genome. Each set of replicate samples displayed a high degree of correlation (r=0.99), supporting the general reproducibility of the data Supporting Information (Table S1). Comparison of ESC FAIRE peaks with previously reported ESC DNaseI data showed a high degree of correlation in the open regions identified by each method (Supporting Information Fig. S2). FAIRE-seq peaks displaying an enrichment value of 5 or greater over genomic DNA showed 0.86 correlation and an FDR of <0.05 compared with DNaseI data, and therefore represent high confidence peaks in ESCs (Supporting Information Fig. S2). Replicates were combined into a single sample for each cell line that was used for all subsequent analyses. The four cell lines exhibited 80,214 to 204,234 FAIRE peaks at enrichment value of ≥ 5 (Supporting Information Tables S4 and S5). The results of additional experimental assessment of several genomic regions harboring identified FAIRE peaks using partial DNaseI digestion of permeabilized ESCs and EpiSCs further supported the general agreement of accessible regions identified using FAIRE and DNaseI sensitivity (Supporting Information, Fig S3).
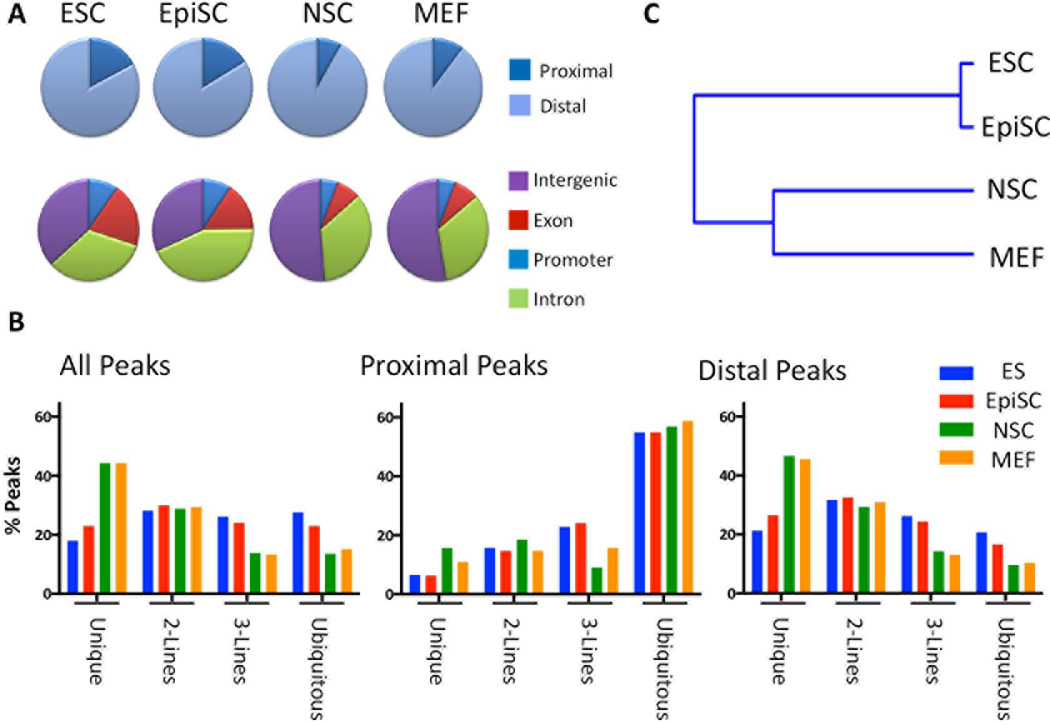
Classification of FAIRE peaks according to their genomic alignment with several annotated features showed that a greater proportion (~ 25%) of the open chromatin within pluripotent ESCs and EpiSCs localized near TSSs than did that within NSCs or MEFs (~15% proximal, Fig. 1A, top row). Determination of the genomic distribution of FAIRE peaks within annotated Exons, Introns, upstream promoters, or no annotated feature (‘Intergenic’) showed ESCs and EpiSCs to exhibit a remarkably similar distribution pattern which was distinct from that shared by NSCs and MEFs (Fig. 1A, bottom row). A greater percentage of the open chromatin in ESCs and EpiSCs was observed at promoters and exons whereas that of NSCs and MEFS was more often observed within intergenic regions. These observations are consistent with the notion that both ground state and primed pluripotent cells exhibit a similar overall chromatin structure that is distinguished by a greater percentage of accessible chromatin at promoter regions.
Figure 1. FAIRE-seq reveals an overall distinction in the chromatin structure of pluripotent- and non-pluripotent cells.
A) Pie charts depicting the genomic distribution of FAIRE peaks in each cell line with respect to annotated transcription start sites (TSSs, Proximal= ± 2 kb from an annotated TSS; Distal ≥ 2kb from a TSS), or that map to genomic regions corresponding to Promoters, Exons, Introns, or Intergenic regions. B) Percentage of FAIRE sites from each cell line (enrichment score ≥ 5) that overlaps sites in one or more of the other cell lines. This analysis was performed comparing all FAIRE sites for each cell line (All peaks, Left panel), and was separately performed for those mapping Proximal to a TSS (Proximal peaks, Center panel), or distal to a TSS (Distal peaks, Right panel). The percentage of peaks that is common to all four cell lines (Ubiquitous), common to three cell lines (3 lines), two cell lines (2 lines), or specific to each cell type (Unique) is shown for each cell type. C) Hierarchical clustering of FAIRE sites among the cell lines shows the close relatedness of the chromatin structure of ESCs and EpiSCs, and their segregation from that of NSCs and MEFs.
The percentage of common or unique accessible regions was determined by identifying overlapping genomic regions among the FAIRE peaks of all cell lines. Between 15–28% of FAIRE peaks in each cell line were common to all four cell types (All Peaks, Ubiquitous, Fig. 1B). On average, approximately 56% of FAIRE peaks mapping proximal to a TSS corresponded to ‘ubiquitously open’ chromatin regions, in contrast to an average of 14% of the Distal peaks (Fig. 1B, Proximal Peaks, or Distal Peaks, Ubiquitous). Conversely, FAIRE peaks that were unique to each of the cell lines typically localized at distal positions (Unique, Fig. 1B).
Compared to NSCs or MEFs, a smaller percentage of ESCs or EpiSCs FAIRE peaks were found to be unique (e.g. approximately 20% of ESC or EpiSC FAIRE peaks are unique to each cell line, compared to 40% of MEFs or NSCs FAIRE peaks, Figure 1A). This would be expected if a significant number of accessible regions are specifically in common in the two pluripotent cell lines. Accordingly, pair-wise hierarchical clustering analysis showed a tight clustering of the FAIRE peak profile within ESCs and EpiSCs that was distinct from those of MEFs and NSC (Fig. 1C). Together these data are consistent with the notion that a significant number of accessible chromatin regions are specifically shared between ESCs and EpiSCs and that both naïve and primed pluripotent cells exhibit an overall chromatin structure that is distinct from that of more differentiated cells.
Modeling accessible chromatin dynamics during differentiation
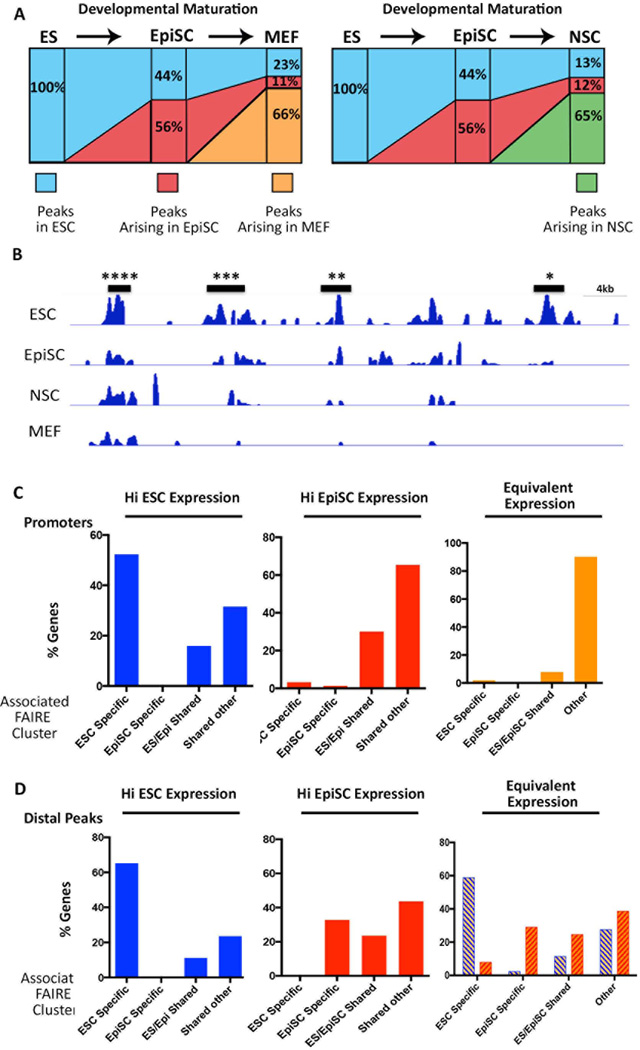
The cell lines were organized according to the developmental stage they represent and the genesis and fate of FAIRE peaks were monitored along this ‘progression’. 168,588 of the 441,855 ESC FAIRE peaks were absent in EpiSCs, and an additional 125,936 ESC sites were not observed in MEFs. In total, 44% of EpiSC FAIRE peaks, and 23% of MEF FAIRE peaks were in common with ESC FAIRE sites (Fig. 2A), suggesting that accessible chromatin regions found in ESCs become progressively lost in the course of differentiation. 56% of EpiSC FAIRE peaks corresponded to newly acquired accessible regions not present in ESCs (Fig. 2A). 11% of MEF FAIRE peaks were shared with these new EpiSC FAIRE peaks, however the majority (66%) of MEF FAIRE peaks represented newly acquired accessible regions. Interestingly, a similar trajectory of FAIRE peak loss and acquisition was observed in the comparison of ESCs and EpiSCs with NSCs (i.e. approximately 25% of NSC FAIRE peaks were in common with ESCs, 12% in common with peaks previously acquired in EpiSCs, and 65% newly acquired accessible regions in NSCs (Fig. 2A). These observations support the notion that progression through differentiation is accompanied by the loss of a fraction of progenitor cell accessible regions, and the concomitant acquisition of new sites in the more mature cell type.
Figure 2. Changes in FAIRE peak distribution model developmental transitions.
A) Model for changes to the chromatin landscape upon differentiation. Each column represents 100% of the FAIRE sites for the cell line indicated on top. The percentage of sites overlapping a FAIRE site in the ‘progenitor’ (cell line to the left) is shown. Sites not overlapping sites in the progenitor were designated as newly acquired accessible regions (Arising). B) FAIRE peaks clusters at putative regulatory DNA elements. A segment of the mouse genome showing clusters of ESC FAIRE peaks that are common to all cell lines (****), shared with 2- (***)- or 1 other line (**), or are ESC-specific (*). C) Determination of the type of FAIRE cluster (ESC-specific, EpiSC-specific, ES/Epi shared, or shared between 2 or more cell types (other)), that is associated with promoters of the top 1000 differentially genes in ESCs and EpiSCs. Each panel depicts the analysis for the gene set indicated at the top, i.e. genes more highly expressed in ESCs (HI ESC Expression), EpiSCs, (HI EpiSC Expression), or Equivalently expressed in the two cell lines. In each case, the number of promoters associated with each type of FAIRE cluster was determined. D) The type of distal FAIRE clusters (>2kb<100 kb from the TSS) that is associated with the same sets of genes analyzed in (C). Interestingly, genes that are equivalently expressed in ESCs and EpiSCs are associated with distinct distal accessible regions in ESCs and EpiSCs (blue- or orange hash marks, respectively).
FAIRE peak clusters reflect regulatory DNA elements defining distinct cell states
Previous studies showed that FAIRE peaks occur in high density clusters at regulatory elements with functional relevance to a particular cell type [12]. Thus to gain insight into elements regulating stage-specific gene expression, we first determined the FAIRE peaks clusters for each cell line (Supporting Information Table S6), and used GREAT analysis (Genomic Regulatory Element Annotation Tool [27] to predict the genes regulated by each of these putative regulatory regions. The ontology and expression patterns of genes predicted to be associated with ESC-, EpiSC-, NSC-, or MEF- clusters were consistent with the phenotype of each cell type (Table 1, Supporting Information Table S7). We next identified FAIRE clusters that were specific to ESCs or EpiSCs by subtracting those overlapping clusters present in any of the other three cell lines (e.g. ESC-specific FAIRE clusters show no overlap with clusters in EpiSCs, NSCs, or MEFS, Fig. 2B and Supporting Information Table S6). GREAT analysis showed that ESC-specific FAIRE clusters (ie putative ESC-specific regulatory regions) associated with genes expressed in either embryonic- or extraembryonic cells of the preimplantation embryo (TS 3–8) and with roles in early development (Table 1, Supporting Information Table S7). Many genes associated with EpiSC-specific FAIRE clusters were also those with predicted roles in embryogenesis but, in contrast to ESC-associated genes, related to slightly later developmental processes such as morphogenesis, regionalization, or commitment (Table 1). However, the most significant gene classes associated with EpiSC-specific clusters consisted of genes expressed at later developmental stages (TS 12–17, Table 1) than that thought to be represented by EpiSCs [4, 6], and those reported to be targets of Polycomb Complex proteins in human ESCs ([28], Table 1, Supporting Information Table S7). These data suggest that many regulatory DNAs within EpiSC-specific open chromatin regulate genes typically expressed in EpiSCs and the late blastocyst, while another subset is associated with genes that are poised for later expression in differentiated cells. Interestingly, analysis of an additional dataset of FAIRE clusters that are common to ESCs and EpiSCs, but neither of the other two cell lines showed that some genes associated with these elements are expressed in the embryo (TS4), but others are typically expressed at later developmental stages and are targeted by the Polycomb Complex proteins in human ESCs ([28], Table 1, Supporting Information Table S7).
TABLE I.
GREAT (Genomic Region Element Annotation Tool) Ontological analysis of genes associated with FAIRE
| Cell line | All ESC clusters |
All EpiSC clusters |
ESC-specific clusters |
EpiSC- specific clusters |
ESC/EpiSC shared Clusters |
|---|---|---|---|---|---|
| Biological Process | negative regulation of gene expression 0 |
embryo development 7.6E-278 |
protein complex subunit organization 2.9E-134 |
embryonic morphogenesis 1.1E-64 |
regulation of cellular metabolic process 2.7E-285 |
| chordate embryonic development 3.6E-319 |
negative regulation of gene expression 2.E-229 |
in utero embryonic development 1.5E-132 |
pattern specification process 4.20E-62 |
regulation of macromolecule metabolic process 2.1E-272 |
|
| in utero embryonic development 8.3E-213 |
embryonic morphogenesis 2.4E-191 |
posttranscriptional regulation of gene expression 1.7E-105 |
regionalization 3.6E-54 |
regulation of gene expression 3.7E-239 |
|
| Mouse Phenotype | prenatal lethality 0 |
abnormal embryonic tissue morphology 5.2E-287 |
prenatal growth retardation 2E-224 |
abnormal neuron differentiation 10E-45 |
prenatal lethality 6.5E-155 |
| embryonic lethality 0 |
embryonic lethality during organogenesis 1.1E-253 |
decreased embryo size 1.04E-215 |
partial neonatal lethality 5.8E-40 |
abnormal embryogenesis/ development 1.9E-143 |
|
| embryonic lethality between implantation and placentation 0 |
abnormal neural tube closure 7.8E-129 |
abnormal placenta morphology 1.85E-146 |
abnormal nervous system tract 7.1E-38 |
abnormal nervous system development 8.8E-117 |
|
| Expression | TS4 inner cell mass 0 |
TS21_embryo; head 1.2E-283 |
TS4_inner cell mass 0 |
TS17_brain 2.4E-91 |
TS17_embryo 2E-298 |
| TS4 extraembryonic component 0 |
TS17_limb 5.59E-272 |
TS4_extraembryonic component 0 |
TS15_embryo; mesenchyme 3.3E-63 |
Theiler_stage_19 1.1E-254 |
|
| Theiler stage 3 0 |
TS3_4–8 cell stage 1.3E-253 |
Theiler stage 3 0 |
TS12_embryo 1.7E-57 |
TS23_metanephros 3.9E-240 |
|
| Perturbation | Genes up-regulated in CD34+ cells isolated from bone marrow of CML 0 |
Genes with copy number losses in primary neuroblastoma tumors. 5.2E-204 |
Genes constituting the BRCA1-PCC network 0 |
'PRC2 targets': trimethylated H3K27 mark in their promoters and are bound by SUZ12 and EED 7.9E-86 |
'H3K27 bound': genes possessing the H3K27me3 mark in their promoters in human embryonic stem cells 8.5E-126 |
| Set 'Nanog targets': genes in human embryonic stem cells. 0 |
Housekeeping genes identified as expressed across 19 normal tissues. 1.4E-120 |
Genes down-regulated in prostate cancer after knockdown of NIPP1 0 |
Genes with copy number gains in primary neuroblastoma tumors. 3.9E-26 |
Eed target genes in human embryonic stem cells 2.2E-106 |
|
| Set 'Sox2 targets': genes in human embryonic stem cells 0 |
Genes up-regulated in lymphoblastoid cells 7.2E-105 |
Set 'Sox2 targets': genes in human embryonic stem cells 7.2E-268 |
Genes bearing H3K27me3 mark or bound by SUZ12 or EED 4.8E-19 |
'PRC2 targets': trimethylated H3K27 mark in their promoters and are bound by SUZ12 and EED 4E-97 |
Together these observations suggest that transition from the naïve to the primed pluripotent state entails inactivation of regulatory elements controlling expression of genes specific to the pre-implantation embryo (ESC specific elements), maintenance of open chromatin at a common set of elements for genes that are expressed in both pluripotent stages or are poised for later activation (ESC/EpiSC shared elements), and acquisition of new accessible regions in EpiSCs that may contain regulatory elements for genes expressed in the late blastocyst, or are poised for activating gene expression after exit from the pluripotent state.
Promoter regions for genes that are differentially expressed between ESCs and EpiSCs exhibit distinct chromatin features
Publically available gene expression profiles of ESCs cultured in 2i/LIF and EpiSCs cultured in FGF2 and ACTIVIN were analyzed using GEO2R to identify genes that are differentially expressed in these two cell lines ([13], Supporting Information Tables S2 and S8). Nearly 5,000 (14%) of the 35,400 microarray probes exhibited a 2-fold or greater difference in gene expression (Supporting Information Table S8). Several, such as Pecam1 and Zfp42, which had previously been shown to be highly expressed in ESCs and the blastocyst ICM were significantly downregulated or extinguished in EpiSCs [4, 6]. The expression of others, such as Fgf5 and Eomes, was dramatically higher in EpiSCs and the postimplantation epiblast compared to ESCs [4, 6] (Supporting Information Table S8). Expression of Sox2 and Oct4 was equivalent in both pluripotent cell types although Nanog expression was slightly downregulated in EpiSCs. These microarray data were validated for a subset of genes using qRT-PCR of mRNA isolated from our ESCs and EpiSCs (Supporting Information Fig. S4).
We then examined the FAIRE clusters associated with the promoters or distal regions of each of the top 1000 differentially expressed genes, or 200 genes displaying equivalent levels of expression in ESCs and EpiSCs (Fig. 2 C and D). The majority of promoters for genes more highly expressed in ESC (Hi ESC expression, Fig. 2C) mapped within ESC-specific FAIRE clusters, suggesting that promoters of ESC-specific genes are accessible only in ESCs. In contrast, most promoters for genes more highly expressed in EpiSCs (Hi EpiSC Expression, Fig. 2C) corresponded to FAIRE clusters common to both EpiSCs and ESCs (and sometimes also MEFS or NSCs), suggesting that the promoters for genes that become activated in EpiSCs are already accessible in ESCs. Notably, promoters for genes with equivalent expression in the two cell lines were generally associated with FAIRE clusters shared among all cell lines (Equivalent Expression, Fig. 2C). In contrast, Distal peaks associated with either differentially expressed- or equivalently expressed genes tended to correspond with cell-specific FAIRE clusters (Fig. 2D).
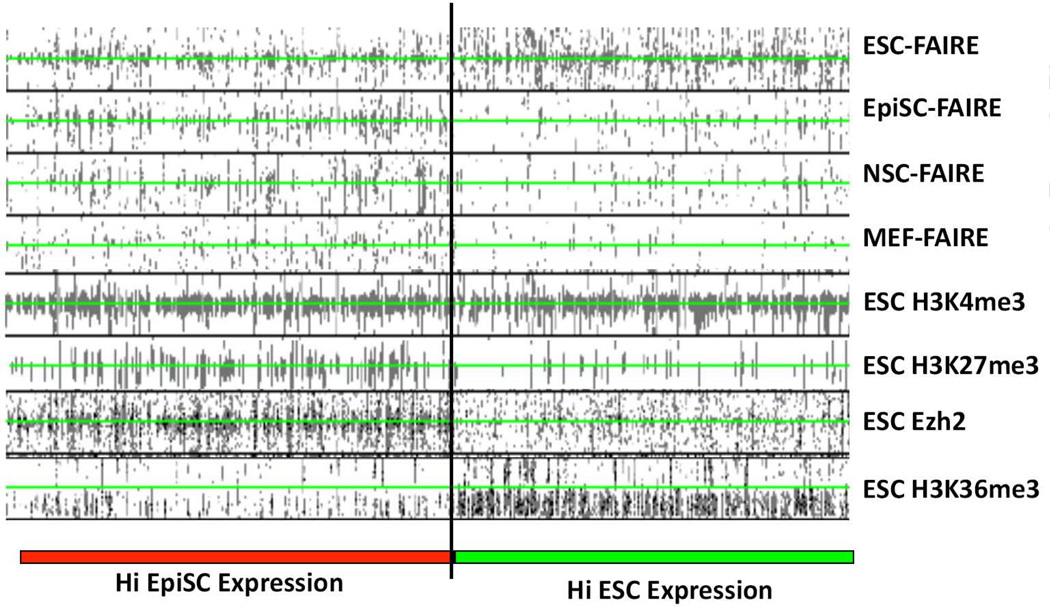
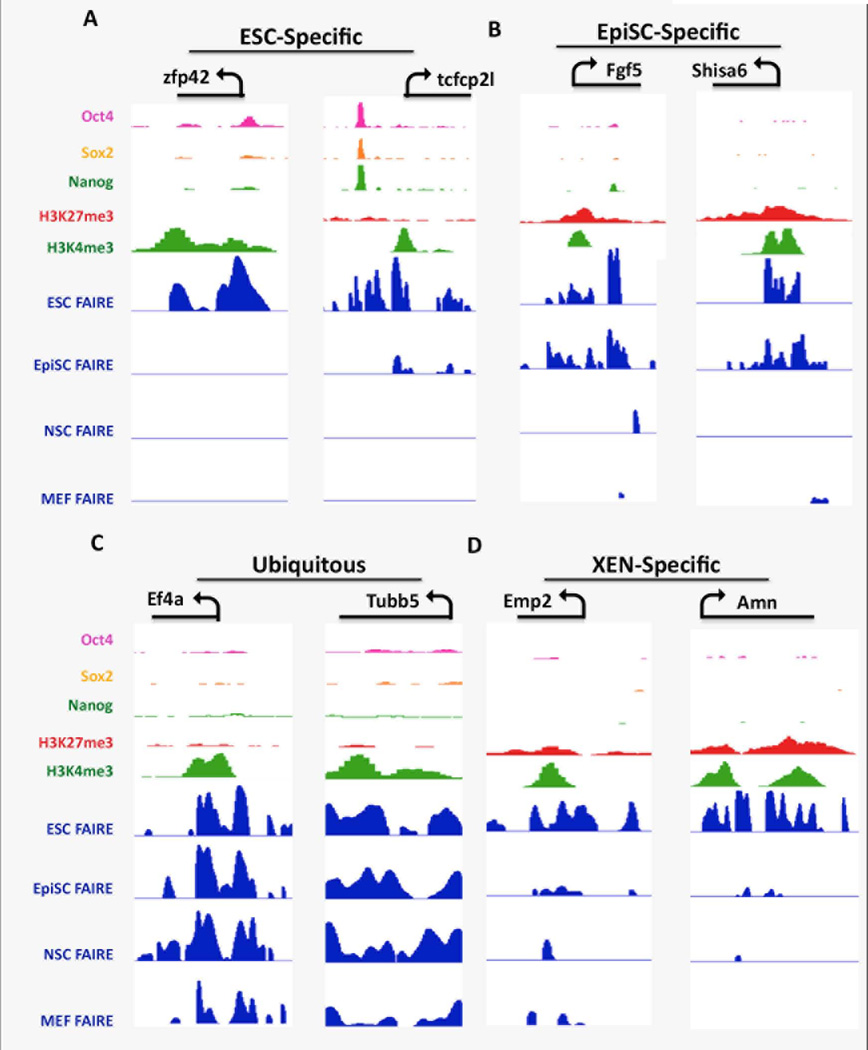
Examination of the pattern of histone modifications and FAIRE peak density within genomic regions flanking the TSSs of the top 1000 differentially expressed genes in ESCs and EpiSCs (Figure 3) showed that promoter regions of genes that are more highly expressed in ESCs than EpiSCs displayed FAIRE-seq peaks only in ESC chromatin (Fig. 3 and Supporting Information Table S8), and were associated with high levels of H3K36me3 and H3K4me3-modified nucleosomes, that are associated with active gene transcription, in the relative absence of the Polycomb Complex protein Ezh2 or H3K27me3 that are associated with transcriptionally silent genomic regions. The promoter regions of two such genes, Zfp42 and Tcfcp2l1, corresponded to robust FAIRE peaks only in ESC cells, and were enriched for H3K4me3-modified nucleosomes in ESC chromatin (Fig 4A). In addition, high levels of OCT4, SOX2, and NANOG binding were also observed, suggesting that the open chromatin at ESC promoters reflect a high level of binding by TF complexes containing these core factors (Fig 4A and Supporting Information, Fig. S5). No significant FAIRE peaks were found at the promoter regions of these ESC-specific genes in EpiSC, MEF, or NSC chromatin (Figs. 3 and 4A), consistent with the notion that ESC-specific promoters acquire a closed chromatin conformation as the cells transition from the ground state to the primed pluripotent state, and remain closed at later developmental stages. Together these observations support the notion that the closing of chromatin at ESC-specific promoters in EpiSCs reflects, in part, an absence of OCT4, SOX2, and/or NANOG binding at these sites in EpiSCs, despite the fact that these factors are expressed in both pluripotent cell types.
Figure 3. Accessible chromatin patterns and ESC Histone modifications at the promoter regions for differentially expressed genes in ESCs and EpiSCs.
Density plots showing the relationship of the top 1000 differentially expressed genes between ESCs and EpiSCs with FAIRE sites in all four cell lines, and ESC ChIP-seq data for H3K4me3-, H3K27me3-, H3K36me3 modified nucleosomes, or Polycomb Complex component Ezh2 [14, 15]. Genes were ranked according to their expression in ESCs and EpiSCs according to the microarray data of [13] (genes more highly expressed in ESCs, HI ESC Expression, demarcated by green bar; genes more highly expressed in EpiSCs, HI EpiSC Expression, demarcated by red bar). Read densities for FAIRE and ChIP data are shown in black. The TSS for each gene analyzed is centered at the green line of each column. Read densities for each feature indicated at the top of the column is shown for a 5 kbp region flanking each TSS.
Figure 4. Distinct combinations of FAIRE peaks, Histone modifications, and core TF binding in ESC chromatin at the promoter regions for genes that are ubiquitously expressed, or are differentially expressed in ESCs, EpiSCs, or XEN.
IGV view of genomic regions of representative promoters for (A) genes more highly expressed in ESCs (ESC-Specific), (B) genes more highly expressed in EpiSCs (EpiSC-specific) (C) Ubiquitously expressed genes (Ubiquitous), and (D) genes more highly expressed in XEN (XEN-specific). The transcription unit, TSS and direction of transcription is shown on the top of the panel for each representative gene. FAIRE peaks are shown for each of the cell lines as indicated, along with ChIP-seq data derived from ESCs for H3K4me3- and H3K27me3-modified histones and OCT4, SOX2, and NANOG binding [63, 64]. This and all IGV screen shots shown used the following enrichment score scales: FAIRE tracks 0–10, OSN 0–200, H3K27me3 0–5, H3K4me3 0–30).
Promoter regions of genes displaying higher expression in EpiSCs compared to ESCs contained FAIRE peaks in both ESC- and EpiSC chromatin, and, more variably, also in NSCs and/or MEFs, and were associated with both H3K4me3- and H3K27me3-modified nucleosomes, and Ezh2 in ESCs, but not H3K36me3, and are therefore likely to be poised in ESCs (Figure 3, Supporting Information Fig S6). Fgf5 and Shisa6 are both more highly expressed in EpiSCs and promoters for these genes were observed to lie in accessible chromatin in both EpiSCs and ESCs (Fig. 4B). The Fgf5 and Shisa6 promoter regions were highly enriched for both H3K4me3- and H3K27me3-modified histones and are therefore bivalent in ESCs. Interestingly, co-binding of OCT4, SOX2 or NANOG at poised EpiSC promoters within ESC chromatin was rarely observed although peaks of single factors were sometimes noted (Figure 4B, Supporting Information Fig. S5). These observations support the notion that promoters that are destined to become activated as cells transition from the ground state to primed state are likely to be transcriptionally ‘poised’ within accessible chromatin in ESCs.
In contrast to the above observations, broadly expressed genes such as tubulin b5 (tubb5) and the translation initiation factor Eif4a1 displayed robust FAIRE peaks at their promoter regions in all four cell lines (Figure 4C), and an absence of OCT4, SOX2, or NANOG binding in ESC chromatin (Figure 4C and Supporting Information Fig. S8).
Distinctive features of ESC chromatin at promoter regions for genes of extraembryonic lineages
ESCs have the potential to differentiate into cells of the embryonic lineages or extra-embryonic endoderm (XEN) [7, 29]. In current models, a subset of cells of the ICM will mature along the embryonic lineage and contribute to the epiblast, while others will instead give rise to Extraembryonic Endoderm that forms part of the placenta [30]. Although the gene expression profiles of embryonic- and extraembryonic endoderm are largely overlapping, the expression of several genes, such Emp2 and Amn is relatively specific to XEN, and is activated upon differentiation of ESCs into XEN [30–32]. Promoter regions for these genes were found to be enriched for both the H3K4me3 and H4K27me3 marks in ESCs, consistent with the notion that they are in a poised state in ESCs (Fig. 4D, Supporting Information Fig S7). However, in contrast to poised EpiSC promoters, the XEN promoters were associated with FAIRE peaks only in ESC and not EpiSC chromatin (Fig. 4D, Supporting Information Fig S7). These observations suggest that XEN gene promoters are also poised in ESCs and will become activated should the cells differentiate into XEN cells. However if ESCs differentiate along the embryonic pathway, XEN promoters become closed, creating an epigenetic barrier to their reactivation in cells of epiblast lineage.
Differentially expressed genes are associated with multiple stage-specific enhancers
The observations above suggest that the core TFs OCT4, SOX2, and NANOG, although common to both ESCs and EpiSCs, bind to different target sites at differentially active promoters within these two pluripotent cell stages. Cell- and stage-specific patterns of gene transcription are thought to be largely determined by the activities of distal enhancers, and the activity of these elements generally correlates with the acquisition of distally-located accessible chromatin regions or specific combinations of histone modifications [33–36]. Consistent with this, our FAIRE data showed that the majority of accessible chromatin regions that were found to be unique to each of our cell lines were located distal to a TSS (Fig. 1b), and genes expressed at each stage were most often associated with ESC- or EpiSC-specific distal FAIRE clusters (Fig 2D). Thus we functionally assessed selected distal regions for enhancer activity in ESCs and EpiSCs.
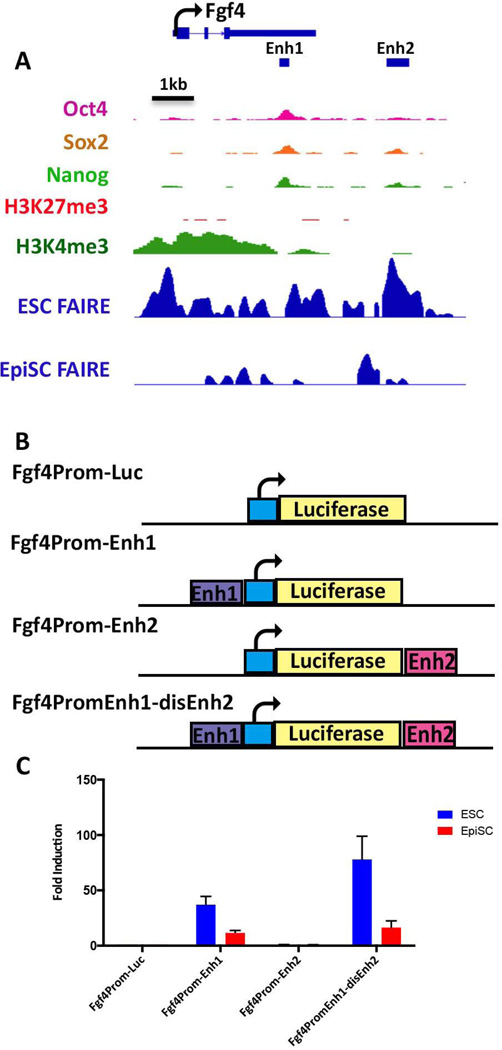
Fgf4 is only expressed in ESCs and, accordingly, a broad region of accessible chromatin within and upstream of the Fgf4 coding sequences, was observed in ESCs but not in the other cell lines (Figure 5A). An enhancer located within the 3’ UTR of the Fgf4 gene is one of the best characterized regulatory elements governing gene expression in ESCs and the blastocyst ICM, and has been shown to be a target of OCT4 and SOX2, and to display activity in ESCs but not in any somatic cells tested [37–39]. To assess whether the FGF4 enhancer is functional in EpiSCs, activation of luciferase reporter gene expression was tested for a plasmid in which this enhancer had been inserted proximal to a minimal Fgf4 promoter element (Fgf4Prom-Enh1, Fig. 5B). As expected, robust activation of luciferase expression was observed in transfected ESCs. In contrast, enhancer activity was significantly lower in EpiSCs than that observed in ESCs (Fig 5C).
Figure 5. FAIRE-seq reveals an additional ESC enhancer associated with the Fgf4 gene.
(A) IGV view of the Fgf4 locus. The TSS and direction of transcriptional elongation are depicted by the arrow on top of the panel, FAIRE peaks derived from ESCs and EpiSCs are shown along with previously reported ChIP data for the indicated histone modifications and OCT4, SOX2, and NANOG binding in ESCs, [63, 64]. Location of the previously characterized Fgf4 enhancer (Enh1) is shown by the black bar, as well as a unique ESC-specific FAIRE peak, Enh2, located 2 kb downstream of the Fgf4 transcription unit. (B) Schematic depiction of luciferase reporter plasmids. The basal plasmid Fgf4prom-luc [65] consists of a minimal region of the Fgf4 promoter and has little activity on its own. Test plasmids contain either Enh1 or Enh2 inserted a shown within Fgf4prom-Luc, or both enhancers contained within the same plasmid in the case of Fgf4PromEnh1-disEnh2(C) Lysates derived from ESCs or EpiSCs transfected with each of the reporter plasmids indicated on the x-axis were assessed for luciferase activity. Activities are expressed as fold activation relative to that of Fgf4prom-Luc.
Upon inspection of the FAIRE peaks at the Fgf4 locus, we noted an additional region, approximately 2 kb downstream of the Fgf4 transcription unit, which was accessible in ESCs but largely absent in EpiSC chromatin (Enh2, Fig. 5A). A luciferase plasmid containing Enh2 at a distal site relative to the minimal FGF4 promoter (Fgf4Prom-Enh2, Fig. 5B) displayed detectable, but modest (i.e.2–3-fold) reporter gene activation. However, plasmid Fgf4PromEnh1-disEnh2, containing Enh1 at the proximal position, and Enh2 at the distal position, displayed an ~80-fold activation of luciferase expression in ESCs, suggesting that these elements can interact to promote gene expression in ESCs (Fig. 5C). These data are consistent with the notion that some distal accessible regions that are present in ESCs but absent in EpiSCs harbor enhancers that are preferentially active in ESCs and include those activated by the core TFs OCT4 and SOX2.
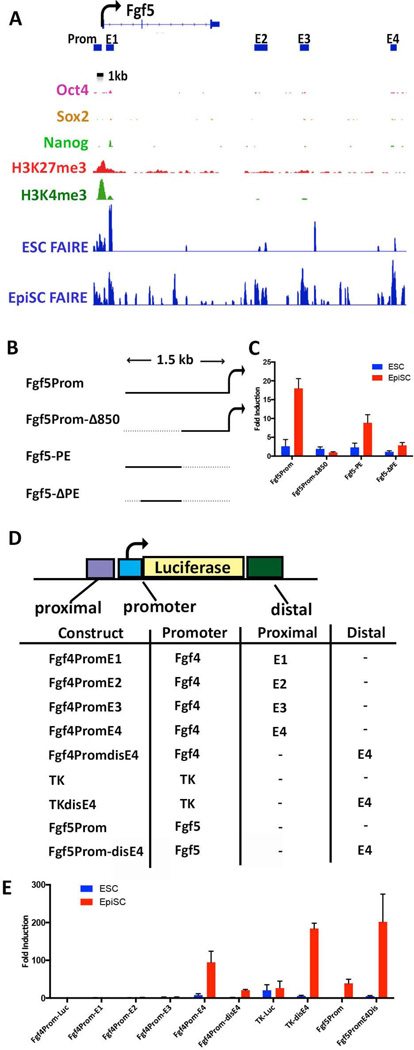
As noted above, Fgf5 is expressed in EpiSCs but not ESCs and the Fgf5 promoter was observed to be accessible in both cell types (although the accessible region is somewhat wider in EpiSCs, Figure 6A). Interestingly, a 1.5 kb DNA fragment spanning the entirety of the accessible chromatin of the Fgf5 promoter region activated luciferase expression 40 fold in EpiSCs but was not active in ESCs (Fgf5Prom, Fig. 6B&C). Deletion of the upstream 850 bp eliminated this activity, suggesting the existence of an EpiSC-specific proximal enhancer (PE) (Fgf5Prom-Δ850, Fig.6 B&C). Accordingly these DNA sequences activated reporter gene expression 8 fold from the minimal Fgf4 promoter in transfected EpiSCs (Fgf5-PE, Fig. 6 B&C). Deletion of the 5’-most 18bp from this fragment eliminated reporter activation, confirming the presence of a proximal enhancer upstream of the Fgf5 promoter (Fgf5Prom-PE, Fig. 6B&C).
Figure 6. EpiSC-specific enhancers within distal FAIRE sites at the Fgf5 locus.
(A) IGV view of the Fgf5 locus. The TSS and direction of transcriptional elongation are depicted by the arrow on top of the panel. ESC and EpiSC FAIRE peaks are shown along with previously reported ESC ChIP data for the indicated histone modifications and OCT4, SOX2, and NANOG binding [63, 64]. Bars under the Fgf5 gene correspond to the Fgf5 promoter region (Prom) and the locations of distal EpiSC-specific FAIRE peaks (regions E1, E2, E3, E4) that were cloned into luciferase reporter plasmids depicted in (B&D). (B) Fgf5 promoter constructs. Fgf5Prom contains the 1.5 bp DNA region spanning the Fgf5 TSS; the 5’-most 850bp of this fragment are deleted in Fgf5Prom-Δ850. Fgf5-PE contains the 850bp region deleted in Fgf5Prom-Δ850 inserted upstream the minimal promoter in fgf4prom. Fgf5-ΔPE is similar to Fgf5-PE but contains an 18bp deletion of the 5’-most sequences of the 850 DNA fragment. (C) Luciferase activity of the indicated reporter plasmids in ESCs and EpiSCs. Fold activation=relative to that of Fgf4prom-luc. (D) Fgf5 Distal regions. Promoter= the Fgf4 minimal promoter, the Herpes Simplex virus TK promoter [65], or the 1.5kb Fgf5 promoter region as indicated in the table. Fgf4prom-E1-E4 contain regions E1, E2, E3, or E4 upstream of the minimal promoter within Fgf4prom-luc. TKdisE4 and Fgf5Prom-disE4 harbor region E4 distal to the promoters. (E) Luciferase activity of the indicated reporter plasmids in ESCs and EpiSCs. Fold activation=relative to that of Fgf4prom-luc.
Furthermore, several distal accessible regions were observed downstream of the Fgf5 TSS in EpiSCs that were not observed in any of the other cell types (Fig. 6A, E1-E4). To assess whether these represent newly acquired enhancers active in EpiSCs, regions E1-E4 were each inserted proximal to the minimal fgf4 promoter tested for an ability to activate reporter gene transcription in transfected ESCs and EpiSCs (Fig. 6D&E). E4, but none of the others, displayed EpiSC-specific reporter gene activation. Insertion of E4 at a distal position downstream of the TK promoter or Fgf5 promoter resulted in induction of luciferase activity in EpiSCs, whereas no activity was observed for either reporter plasmid in ESCs (Fig.6E). These data show that some, but not all, of newly acquired distal accessible regions in EpiSCs harbor EpiSC-specific enhancers that correlate with the induction of differentially expressed genes in EpiSCs. Together these data support the notion that differentially active enhancers play a prominent role in determining stage-specific gene expression in naïve and primed pluripotent cells.
Discussion
The ordered transitions in gene expression patterns and the enumeration of differentiated cell phenotypes during development are accompanied by progressive and dynamic reorganization of cellular chromatin resulting in changes in the genome-wide patterns of accessible chromatin regions. While insights into lineage relationships among cell types are most often sought by comparing gene expression profiles, comparison of accessible chromatin patterns provides an important additional perspective on key regulatory features underlying distinct cell states. This point is of particular relevance for pluripotent cells whose hallmark feature, i.e. their wide differentiation potential, is defined less by the genes that are overtly expressed than by the vast number of poised, relatively inactive transcriptional regulatory elements residing within accessible chromatin. Here we have used FAIRE-seq technology to create a comparative atlas of the accessible chromatin regions with two major goals in mind. The first was to derive a profile of chromatin dynamics accompanying epiblast maturation and the exit from pluripotency. Our analyses show a clear segregation in the overall pattern of accessible sites in pluripotent cells and somatic lineages that underscores the highly related chromatin structure of ESCs and EpiSCs. Nearly half of the FAIRE peaks detected in EpiSCs were observed to be in common with those of ESCs, and approximately half of these are exclusively shared by ESCs and EpiSCs (Fig 2A). Progression through the developmental stages represented by ESCs, EpiSCs, and the somatic cells show that each ‘differentiation’ step is accompanied by the loss of a subset of open chromatin sites present in the progenitor stage, the retention of another subset of progenitor sites, and the concomitant acquisition of new sites in the more mature cell, consistent with analogous previous studies examining differentiation-related changes in accessible chromatin patterns or histone modifications [12, 40–44].
Our second goal was to gain insight into features distinguishing-, or common to- the naïve and primed pluripotent states. It is important to note that the ESC chromatin utilized in this study derives from cells that have been cultured in 2i/LIF medium rather than the traditional serum/LIF conditions in order to more clearly delineate the ground- and primed- pluripotent states. While 2i and serum-grown cultures display comparable expression levels of key genes associated with pluripotency such as Oct4, Sox2, Nanog, Tbx3, and Fgf4, ESC populations grown in 2i are generally more homogeneous and show effective silencing of lineage-specific genes such as Brachyury and Eomes that are often detectably expressed in ESCs maintained in serum [9]. Accordingly, ESCs maintained in 2i have been reported to more closely resemble cells of the early embryonic blastocyst [9]. The utilization of FAIRE data from NSC and MEFs as controls permits us to better identify those features of ESC and EpiSC chromatin that are specifically shared by these pluripotent cells as well as those that distinguish the ground and primed states. Integration of these FAIRE data with gene expression- and ChIP-seq analyses further permitted us to correlate these chromatin features with transcriptional activity in these two pluripotent cells.
We identify three classes of ESC sites that are ‘extinguished’ in EpiSCs: the inactivation of promoters for gene expression that is specific to ESCs, inactivation of enhancers for genes expressed in ESCs, and the closing of ‘poised’ elements at the promoters for genes of an alternate, XEN cell fate. Although ESCs are functionally pluripotent, the initial decision for these cells upon exiting self-renewal is to differentiate toward the extraembryonic- or epiblast lineage. Thus differentiating ESCs will either give rise to XEN, or will pass through a stage equivalent to the egg cylinder epiblast, roughly approximated by EpiSCs, prior to generation of the definitive somatic lineages [45]. Accordingly, we find that promoter regions for genes specific to XEN or epiblast fates are in accessible chromatin regions and are poised in ESCs (Fig. 4, [46]). These data are consistent with the notion that transition of naïve pluripotent cells towards the primed epiblast fate entails the loss of accessible regions at promoters for genes of the XEN lineage as well as at the promoters for ESC-specific genes such as Zfp42, Prdm14, and Klf factors (Fig. 4 and data not shown). Accordingly, the ‘extinguishing’ of many of these accessible regions correlates with increased levels of DNA methylation at these loci in EpiSCs as reported in a recent MethylCap-Seq analysis [47]. Thus these events would be predicted to create an epigenetic barrier that prevents reversion to the ground state or selection toward a XEN fate, and to reinforce lineage commitment to the primed epiblast fate. Interestingly, promoters for epiblast genes have been reported to be methylated and stably repressed in cells adopting the XEN fate [46] and it is likely that the accessibility at these promoters is extinguished in XEN cells. Thus coordinated and reciprocal alterations to the chromatin accessibility at critical lineage-specific promoters contribute to the canalization of developmental pathways and stabilization of cell fate decisions.
Accessible regions that are common to both ESCs and EpiSCs but neither of the somatic cell types are also of several subtypes: sites that harbor elements for active gene expression in both cell types, or elements that are poised in ESCs but become active for gene expression in EpiSCs or later somatic cell types. We note that many of the shared accessible regions associated with actively transcribed genes include those for key transcription factors such as SOX2 and NANOG that have well established roles in stem cell maintenance, and for genes providing basic metabolic functions that may play a role maintaining robust self-renewal (Table 1). As would be expected, FAIRE clusters that are exclusively shared between ESCs and EpiSCs correlate with binding sites for OCT4, SOX2, and NANOG identified by ChIP-seq in ESCs (Supporting Information Table S9). Additional common accessible regions are associated with promoters for late epiblast genes such as Fgf5 and Shisa6 that are poised in ESCs, and promoters for genes typically expressed in differentiated somatic cell lineages later in embryogenesis, and are likely to be poised in both ESCs and EpiSCs (Table 1).
The functional analyses of cell specific open chromatin further supports the notion that the activity of ESC- and EpiSC- gene promoters is largely controlled by stage-specific enhancers localized within cell-specific accessible regions distal to the TSS. The Fgf4 and Fgf5 genes are each associated with multiple enhancers that may reinforce and fine-tune stage-specific gene expression. Enhancers PE and E4, identified within accessible chromatin upstream of the TSS and downstream of the gene body, respectively, of the Fgf5 gene were each shown to be able to independently activate the reporter gene, but also to act together to elicit EpiSC-specific reporter gene expression. Interestingly, the accessibility of the downstream Fgf5 E4 enhancer region arises de novo in EpiSCs and is not open or associated with chromatin mark combinations previously postulated to mark the positions of poised or latent enhancers in ESCs that might foreshadow the presence of a potential EpiSC enhancer [35, 48, 49] (although regions E1-4 each correspond with minor peaks of H3K4me1 enrichment in ESCs, data not shown). The molecular mechanisms mediating the acquired accessibility and activity of these and other distal sites will provide an interesting avenue for delineating key factors of primed cells. Distal regions E1-E3 did not display enhancer activity in EpiSCs, and their functional significance is presently unclear.
FAIRE analyses also facilitated the identification of a second ESC-specific enhancer, Fgf4-Enh2, that may function together with our originally characterized enhancer (Enh1) to activate Fgf4 gene transcription in ESCs. Although we noted some reporter gene activation by these Fgf4 enhancers in EpiSCs (Fig 6C), this is likely due to the reported metastable nature of E3 EpiSCs that causes a fluctuating subpopulation of 5% of E3 EpiSCs to display ESC-like transcriptional properties [50]. Enh2, located within a largely ESC-specific accessible region 2 kb downstream of the Fgf4 transcription unit, acts synergistically with Enh1 in ESCs but not in EpiSCs (Fig. 5), and is consistent with the possibility that full function of these enhancers requires an ESC TF(s) that is absent in EpiSCs. ESCs and EpiSCs both express the core TFs OCT4, SOX2, and NANOG but the expression of many auxiliary TFs, such as KLF proteins, ESSRB, PRDM14, and ZFP42, is lacking in EpiSCs. Interestingly, binding and motif analyses have shown KLF- and PRDM14 TFs to co-bind many target DNAs bound by OCT4 and SOX2 in ESCs [51–53]. We had previously shown that Enh1 function depends on binding and activation by the OCT4/SOX2 complex, but that Enh1function also requires a TF binding a GC-rich motif downstream of the OCT4/SOX2 site [38, 39]. Both sequence analysis and ChIP data suggests that this latter motif may bind a KLF factor [54]. If an ESC-specific auxiliary TF(s) such as KLF participates in the selection of core TF target sites, this notion could explain the loss of accessible sites in EpiSCs, and presumably OCT4/SOX2 binding, at regulatory elements for ESC genes (Supporting Information Fig. S5), and is consistent with the demonstrated ability of auxiliary TFs KLF4 or PRDM14 to facilitate reprogramming of EpiSCs to ground state pluripotency [55, 56]. Binding of the core TFs would be predicted to be redirected in EpiSCs by comparable EpiSC-specific auxiliary TFs. Accordingly, during the preparation of our manuscript, Buecker et al [57] published ChIP-seq and gene expression analyses of ‘Epi-like stem cells’, (EpiLSCs, i.e. cells induced by the treatment of ESCs with FGF2 and Activin for 48 hours to display a gene expression profile similar to that of established EpiSC lines [57, 58]), that support many of the conclusions we present here. In particular, EpiLSC-specific gene activation is shown to correlate with the redirection of OCT4 binding to distinct target DNA sites by OTX2, a TF whose expression is upregulated in EpiSCs and that, together with POU3F1 and ZIC proteins, has been suggested to play a role in the maintenance of EpiSC self-renewal [59, 60].
The observations presented here closely parallel previous genome-wide mapping studies of accessible chromatin regions and//or histone modifications across multiple cell types[12, 34, 40–44]. For example, these studies showed genomic regions near TSS’s to be generally accessible across cell types whereas cell specific accessible sites are mainly observed within distal regions[34]. However, more recent data also shows the existence of a subset of promoters displaying an increased susceptibility to DNaseI cleavage and transcriptional activity that is coordinated by the activation of cell-specific enhancers, in agreement with our observations [41]. Furthermore, a comparative study of the DNase hypersensitive sites (DHS) across 49 human cell types, including several within the hematopoietic lineage and cardiomyocytes derived from the directed differentiation of ESCs, illustrates the coordinate extinction, maintenance, and de novo activation of distinct DHS’s accompanying lineage selection and cell fate decisions [40]. As also suggested by our studies, the selective activation and extinction of accessible regions largely parallels the expression or absence, respectively, of cell-specific TFs targeting these sites, suggesting that TFs are the main determinants of chromatin accessibility [41]. Thus, together with our analyses, these considerations support a model in which the combined activities of OCT4, SOX2, and NANOG serve as core factors for the epiblast lineage while stage-specific auxiliary TFs such as KLFs or OTX2 direct the activity of these core TFs to distinct target regulatory DNAs for expression of genes that are specific to naive or primed cells [61, 62]. An analogous role has been proposed for stage-specific co-regulators that act cooperatively with core TFs GATA1 and TAL1 to achieve temporal and differential activation of genes within the erythroid lineages [43]. The stage-specific accessible regions identified here therefore provide an important resource for identifying the mechanisms and TF combinations governing naïve- and primed- pluripotent states.
SUMMARY
Through comparison of the genome-wide patterns of accessible chromatin in two pluripotent cell types and non-pluripotent cells we identify features that distinguish the naïve and primed pluripotent states, and that illustrate some of the progressive changes in chromatin structure that accompany increased degrees of differentiation and are necessary to ordered lineage determination. The stage-specific and shared accessible regions identified in this study provide an important resource for further identifying specific TF combinations defining naïve and primed pluripotent states.
Supplementary Material
AKNOWLEDGEMENTS
We thank Boris Greber (Max Planck Institute, Muenster, Germany) for his gift of the E3 Epiblast Stem cells used in this study, Zuojian Tang and Stuart Brown (CHIBI, NYU School of Medicine) for processing the Illumina sequencing files, Elisa Venturini and the Genome Technology Center for preparation of libraries and Illumina sequencing, Yatong Wang for technical assistance, and Upal Basu Roy for comments on the manuscript. This work was supported by an Empire State Stem Cell Board grant through the New York State Department of Health (NYSTEM Contract #CO24322) to L.D., and a postdoctoral fellowship from NIH NCI institutional training grant 1T32CA160002 awarded to M.M.
Footnotes
Author contribution:
Matthew Murtha: Collection and assembly of data, data analysis and interpretation, manuscript preparation
Francesco Strino: Bioinformatic support and analyses
Zeynep Tokcaer-Keskin: Collection and assembly of data
N. Sumru Bayin: Collection and assembly of data
Doaa Shalabi: Collection and assembly of data
Xiangmei Xi: Collection and assembly of data
Yuval Kluger: Bioinformatic support and analyses
Lisa Dailey: Conception and design, data interpretation, Financial support, manuscript writing and final approval of manuscript.
Data Access: All DNA sequencing files and FAIRE-seq data has been deposited to the Gene Expression Omnibus (GEO) database under accession # GSE 58520.
DISCLOSURE OF POTENTIAL CONFLICT OF INTEREST
The authors indicate no potential conflict of interest
REFERENCES
- 1.Nichols J, Smith A. Pluripotency in the embryo and in culture. Cold Spring Harb Perspect Biol. 2012;4(8):a008128. doi: 10.1101/cshperspect.a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimosato D, Shiki M, Niwa H. Extra-embryonic endoderm cells derived from ES cells induced by GATA Factors acquire the character of XEN cells. Bmc Developmental Biology. 2007;7 doi: 10.1186/1471-213X-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayashi K, Surani MA. Self-renewing epiblast stem cells exhibit continual delineation of germ cells with epigenetic reprogramming in vitro. Development. 2009;136(21):3549–3556. doi: 10.1242/dev.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, Osorno R, Tsakiridis A, et al. In Vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep. 2(6):1571–1578. doi: 10.1016/j.celrep.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 7.Cho LT, Wamaitha SE, Tsai IJ, et al. Conversion from mouse embryonic to extra-embryonic endoderm stem cells reveals distinct differentiation capacities of pluripotent stem cell states. Development. 139(16):2866–2877. doi: 10.1242/dev.078519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers I, Tomlinson SR. The transcriptional foundation of pluripotency. Development. 2009;136(14):2311–2322. doi: 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marks H, Stunnenberg HG. Transcription regulation and chromatin structure in the pluripotent ground state. Biochim Biophys Acta. 2014;1839(3):129–137. doi: 10.1016/j.bbagrm.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nat Cell Biol. 2011;13(5):490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- 11.Giresi PG, Kim J, McDaniell RM, et al. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17(6):877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song L, Zhang Z, Grasfeder LL, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21(10):1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Wu J, Ye S, et al. Modulation of beta-catenin function maintains mouse epiblast stem cell and human embryonic stem cell self-renewal. Nat Commun. 2013;4:2403. doi: 10.1038/ncomms3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marks H, Kalkan T, Menafra R, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149(3):590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453(7194):519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greber B, Wu G, Bernemann C, et al. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6(3):215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Conti L, Pollard SM, Gorba T, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3(9):e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48(3):233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Micsinai M, Parisi F, Strino F, et al. Picking ChIP-seq peak detectors for analyzing chromatin modification experiments. Nucleic Acids Res. 2012;40(9):e70. doi: 10.1093/nar/gks048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Consortium EP. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9(4):e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yaragatti M, Basilico C, Dailey L. Identification of active transcriptional regulatory modules by the functional assay of DNA from nucleosome-free regions. Genome Res. 2008;18(6):930–938. doi: 10.1101/gr.073460.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karolchik D, Hinrichs AS, Furey TS, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32(Database issue):D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruskal WH, Wallis WA. Use of Ranks in One-Criterion Variance Analysis. Journal of the American Statistical Association. 1952;47(260):583–621. [Google Scholar]
- 25.Cockerill PN. Structure and function of active chromatin and DNase I hypersensitive sites. FEBS J. 278(13):2182–2210. doi: 10.1111/j.1742-4658.2011.08128.x. [DOI] [PubMed] [Google Scholar]
- 26.Gaulton KJ, Nammo T, Pasquali L, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42(3):255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLean CY, Bristor D, Hiller M, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39(2):232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 29.Shimosato D, Shiki M, Niwa H. Extra-embryonic endoderm cells derived from ES cells induced by GATA factors acquire the character of XEN cells. BMC Dev Biol. 2007;7:80. doi: 10.1186/1471-213X-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Familari M. Characteristics of the endoderm: Embryonic and extra embryonic in mouse. The scientific world journal. 2006;6:1815–1827. doi: 10.1100/tsw.2006.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown K, Legros S, Artus J, et al. A comparative analysis of extra-embryonic endoderm cell lines. PLoS One. 2010;5(8):e12016. doi: 10.1371/journal.pone.0012016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherwood RI, Jitianu C, Cleaver O, et al. Prospective isolation and global gene expression analysis of definitive and visceral endoderm. Dev Biol. 2007;304(2):541–555. doi: 10.1016/j.ydbio.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet. 2012;13(7):469–483. doi: 10.1038/nrg3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 35.Rada-Iglesias A, Bajpai R, Swigut T, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visel A, Blow MJ, Li Z, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Curatola AM, Basilico C. Expression of the K-fgf proto-oncogene is controlled by 3' regulatory elements which are specific for embryonal carcinoma cells. Mol Cell Biol. 1990;10(6):2475–2484. doi: 10.1128/mcb.10.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dailey L, Yuan H, Basilico C. Interaction between a novel F9-specific factor and octamer-binding proteins is required for cell-type-restricted activity of the fibroblast growth factor 4 enhancer. Mol Cell Biol. 1994;14(12):7758–7769. doi: 10.1128/mcb.14.12.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan H, Corbi N, Basilico C, et al. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9(21):2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 40.Stergachis AB, Neph S, Reynolds A, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154(4):888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xi H, Shulha HP, Lin JM, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3(8):e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J, Shao Z, Glass K, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell. 2012;23(4):796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J, Adli M, Zou JY, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152(3):642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossant J. Stem cells and early lineage development. Cell. 2008;132(4):527–531. doi: 10.1016/j.cell.2008.01.039. [DOI] [PubMed] [Google Scholar]
- 46.Senner CE, Krueger F, Oxley D, et al. DNA methylation profiles define stem cell identity and reveal a tight embryonic-extraembryonic lineage boundary. Stem Cells. 2012;30(12):2732–2745. doi: 10.1002/stem.1249. [DOI] [PubMed] [Google Scholar]
- 47.Veillard AC, Marks H, Bernardo AS, et al. Stable methylation at promoters distinguishes Epiblast Stem Cells from Embryonic Stem Cells and the in vivo epiblast. Stem Cells Dev. 2014 doi: 10.1089/scd.2013.0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostuni R, Piccolo V, Barozzi I, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152(1–2):157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 49.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21(8):1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han DW, Tapia N, Joo JY, et al. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 2010;143(4):617–627. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 51.Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 52.Hutchins AP, Choo SH, Mistri TK, et al. Co-motif discovery identifies an Esrrb-Sox2-DNA ternary complex as a mediator of transcriptional differences between mouse embryonic and epiblast stem cells. Stem Cells. 2013;31(2):269–281. doi: 10.1002/stem.1279. [DOI] [PubMed] [Google Scholar]
- 53.Ma Z, Swigut T, Valouev A, et al. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat Struct Mol Biol. 2011;18(2):120–127. doi: 10.1038/nsmb.2000. [DOI] [PubMed] [Google Scholar]
- 54.Jiang J, Chan YS, Loh YH, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10(3):353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 55.Gillich A, Bao S, Grabole N, et al. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell. 2012;10(4):425–439. doi: 10.1016/j.stem.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo G, Yang J, Nichols J, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136(7):1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buecker C, Srinivasan R, Wu Z, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14(6):838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayashi K, Ohta H, Kurimoto K, et al. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146(4):519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 59.Acampora D, Di Giovannantonio LG, Simeone A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development. 2013;140(1):43–55. doi: 10.1242/dev.085290. [DOI] [PubMed] [Google Scholar]
- 60.Iwafuchi-Doi M, Matsuda K, Murakami K, et al. Transcriptional regulatory networks in epiblast cells and during anterior neural plate development as modeled in epiblast stem cells. Development. 2012;139(21):3926–3937. doi: 10.1242/dev.085936. [DOI] [PubMed] [Google Scholar]
- 61.Festuccia N, Osorno R, Wilson V, et al. The role of pluripotency gene regulatory network components in mediating transitions between pluripotent cell states. Curr Opin Genet Dev. 2013;23(5):504–511. doi: 10.1016/j.gde.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeo JC, Ng HH. The transcriptional regulation of pluripotency. Cell Res. 2013;23(1):20–32. doi: 10.1038/cr.2012.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mouse EC, Stamatoyannopoulos JA, Snyder M, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13(8):418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murtha M, Tokcaer-Keskin Z, Tang Z, et al. FIREWACh: high-throughput functional detection of transcriptional regulatory modules in mammalian cells. Nat Methods. 2014;11(5):559–565. doi: 10.1038/nmeth.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.