Abstract
Objective
To ascertain the importance of a single regulatory element in the control of Cnn1 expression using CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) genome editing.
Approach and Results
The CRISPR/Cas9 system was used to produce 3/18 founder mice carrying point mutations in an intronic CArG box of the smooth muscle cell (SMC)-restricted Cnn1 gene. Each founder was bred for germ line transmission of the mutant CArG box and littermate interbreeding to generate homozygous mutant (Cnn1ΔCArG/ΔCArG) mice. Quantitative RT-PCR, Western blotting, and confocal immunofluorescence microscopy showed dramatic reductions in Cnn1 mRNA and CNN1 protein expression in Cnn1ΔCArG/ΔCArG mice with no change in other SMC-restricted genes and little evidence of off-target edits elsewhere in the genome. In vivo chromatin immunoprecipitation assay revealed a sharp decrease in binding of SRF to the mutant CArG box. Loss of CNN1 expression was coincident with an increase in Ki-67 positive cells in the normal vessel wall.
Conclusion
CRISPR/Cas9 genome editing of a single CArG box nearly abolishes Cnn1 expression in vivo and evokes increases in SMC DNA synthesis. This facile genome editing system paves the way for a new generation of studies designed to test the importance of individual regulatory elements in living animals, including regulatory variants in conserved sequence blocks linked to human disease.
Keywords: CRISPR, SRF, CArG box, transgenic mouse, smooth muscle
Traditional approaches to studying the in vivo functionality of regulatory elements controlling gene expression involve pronuclear injection of promoter/enhancer sequences linked to a reporter gene (e.g., lacZ). These experiments are limited by the out-of-genomic-context in which the sequences are studied and the necessity of generating multiple lines of mice to address issues surrounding copy number- and position-dependent effects on reporter gene activity as well as untoward consequences of foreign DNA sequences on the genomic landscape that could perturb endogenous gene expression1. The emergence of artificial chromosome-mediated transgenic mice represented an important step towards the study of gene expression in proper genomic context2, but these large-capacity cloning vectors are difficult to work with and still require the generation of multiple independent lines of mice, which can take months to years to complete. Thus, newer facile methods of analyzing regulatory elements in their native genomic milieu are needed.
The CRISPR/Cas9 system, defined originally as a heritable, adaptive immune system in bacteria and archaea3, has recently been harnessed as an RNA-guided nuclease method of genome editing in several animal species including zebrafish4, pig5, rat6, mouse7, rabbit8, and monkey9. In what we shall refer to as three-component CRISPR (3cCRISPR), a guide RNA (gRNA) – comprising a 20 nucleotide CRISPR RNA (crRNA) plus the invariant transactivating RNA (tracrRNA) – is combined with Cas9 endonuclease mRNA and a single-strand oligonucleotide overlapping the gRNA targeted sequence for delivery to fertilized eggs. Precise genome editing occurs following sequential gRNA-Cas9-mediated DNA double-strand break at the genomic region of interest and homology-directed repair (HDR) of the double-strand break by a single-strand oligonucleotide harboring ~60 nucleotide homology arms flanking DNA substitutions (for mutating endogenous DNA sequences) or exogenous sequences (e.g., restriction site, loxP) that are stably integrated at the site of repair10, 11. While this programmable endonuclease approach to genome editing has been deployed successfully to modify protein coding genes in vivo, there has yet to be a report on its use in altering regulatory elements controlling gene expression in a living animal12. In this context, there is an emerging need to elucidate the functionality of regulatory single nucleotide polymorphisms because most genomic variation associated with human disease occurs in noncoding sequence space where millions of regulatory elements reside13. Herein, we demonstrate CRISPR-Cas9-mediated mutation of a regulatory element that nearly abolishes expression of the endogenous Cnn1 gene in mouse tissues.
Material and Methods
Materials and Methods are available in the online-only Data Supplement.
Results
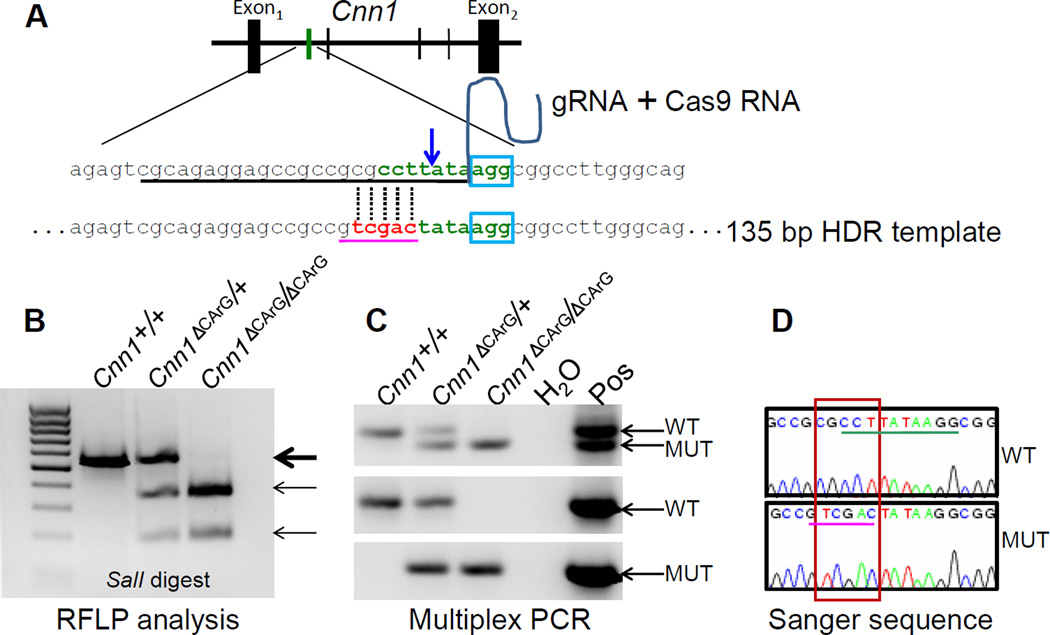
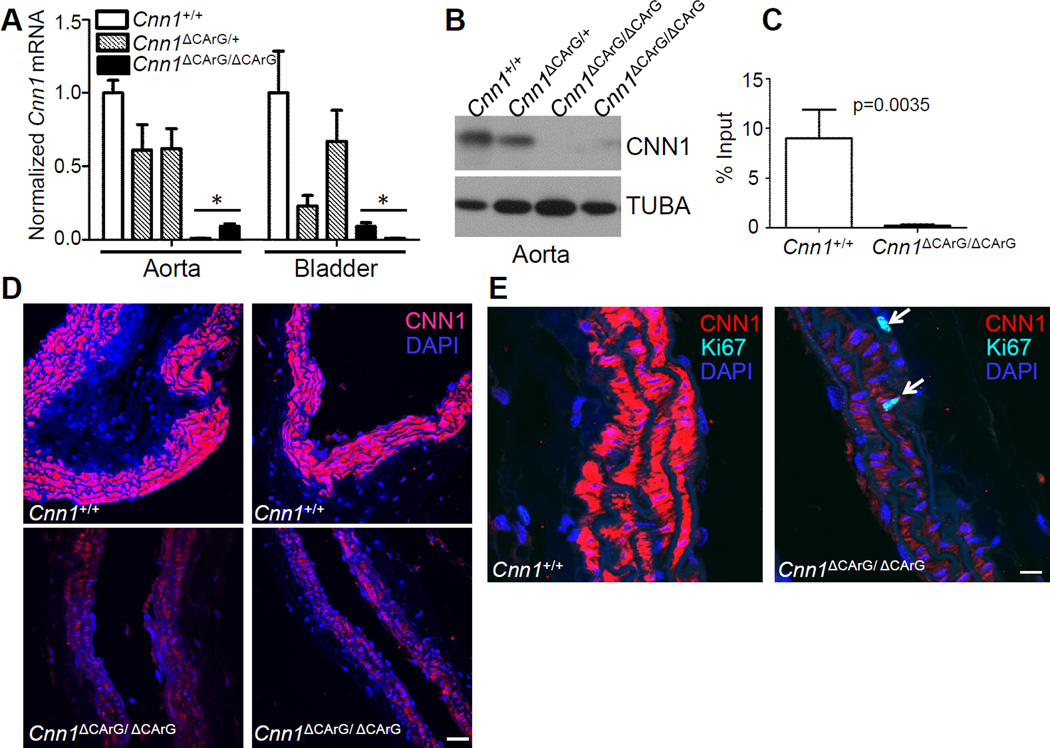
CArG boxes bind serum response factor (SRF) around a growing number of target genes, including most SMC-restricted genes14. We used 3cCRISPR to introduce nucleotide substitutions within a consensus intronic CArG box of the endogenous mouse Cnn1 gene (Fig. 1A). This particular CArG box is suspected to play an important role in the endogenous expression of Cnn115, 16. Initial pro-nuclear injections of plasmid DNA carrying Cas9 and the gRNA were unsuccessful in generating mutant mice. However, cytoplasmic injection of Cas9 mRNA, purified gRNA, and a single-strand oligonucleotide HDR template resulted in 3/18 pups with either one (n=2) or both (n=1) alleles correctly edited as indicated by a restriction fragment length polymorphism (RFLP) assay (Figure 1B) and a novel multiplex PCR assay we have developed for the unambiguous genotyping of subtle mutations introduced into the genome (Figure 1C and Figure I in the online-only Data Supplement). Off-target mutations of related sequences in the genome were limited and, when present (2/10) in founder mice, affected only non-conserved intergenic sequences (Table I in the on-line Data Supplement). Importantly, repeat sequencing of these sites in F1 mice revealed the absence of mutations indicating the off-target edits were successfully bred out of the genome. Quantitative RT-PCR analysis of the bi-allelic targeted founder mouse showed significant decreases in Cnn1 mRNA expression in aorta and bladder (Figure II in the online-only Data Supplement). Each of the founders was successfully bred for germ line transmission of the CArG box mutation and F1 littermate interbreeding. Levels of Cnn1 mRNA in aorta and bladder were reduced ~50% in Cnn1ΔCArG/+ mice and virtually extinguished in Cnn1ΔCArG/ΔCArG mice (Figure 2A and Figure III in the online-only Data Supplement). Sanger sequencing verified precise genome editing (Figure 1D) with no mutations in sequences surrounding the CArG box, including the entire cDNA and 5’ promoter region of Cnn1 (data not shown), indicating reduced Cnn1 mRNA expression was not the result of spurious mutations outside of the targeted CArG box. In agreement with mRNA expression analysis, Western blotting revealed low levels of CNN1 protein in the aorta of Cnn1ΔCArG/ΔCArG mice (Figure 2B and Figure III in the online-only Data Supplement). As expected from previous CArG mutagenesis experiments15, in vivo chromatin immunoprecipitation assay showed essentially no binding of SRF to the mutant intronic CArG box (Figure 2C). Confocal immunofluorescence microscopy confirmed a clear reduction in expression of CNN1 protein in SMC of the carotid artery (Figure 2C) and other SMC-containing tissues (data not shown). There was no change in mRNA or protein expression of several SMC-restricted markers under similar CArG-SRF regulatory control (Figure III in the online-only Data Supplement). CNN1 has been reported to function as a tumor suppressor gene17. Consistent with this idea, we noted a significant increase in the number of Ki67 positive cells per longitudinal section of carotid artery in Cnn1ΔCArG/ΔCArG mice (21.7 ± 2.1) as compared to Cnn1+/+ mice (7.7 ± 4.0) (mean Ki67+ cells ± std, n=3, p=0.0334, Figure 2E). Since Cnn1ΔCArG/ΔCArG mice are viable, they represent a valuable resource to further characterize the phenotypic changes associated with marked reductions in CNN1 under normal and pathological conditions.
Figure 1. 3cCRISPR genome editing of a CArG box.
(A) Strategy for targeting one of the 4 conserved intron 1 CArG boxes (vertical green and black lines) in mouse Cnn1 gene. A synthetic crRNA (underlined sequence) was designed to include the first seven nucleotides of the CArG box (green) immediately 5’ of a protospacer adjacent motif (PAM, blue box) comprising the remaining three nucleotides of the CArG box. The blue arrow indicates the predicted double strand break three nucleotides upstream of the PAM sequence. The crRNA was cloned upstream of the invariant tracrRNA (curved line) and this gRNA was then combined with Cas9 mRNA and the HDR template harboring three nucleotide substitutions of the CArG box following two nucleotide changes (red) that together create a novel SalI restriction site (pink underlined sequence), for cytoplasmic injection of fertilized mouse eggs. Genotyping of F1 intercrossed mice was done using a SalI RFLP digest (B), a novel multiplex PCR assay (C) and Sanger sequencing (D). Thick arrow in panel B represents the wildtype PCR product and thin arrows represent the expected size products of one or both alleles with correct SalI site. Arrows in panel C represent the PCR products with multiplex PCR (top gel) and PCR using wildtype (middle) or mutant (bottom) forward primers (see Figure I in the online-only Data Supplement). The red box in panel D shows the confirmed sequence edit creating novel SalI site (pink underlined sequence) that changes the normal CArG box (green underlined sequence).
Figure 2. Expression of Cnn1 mRNA and CNN1 protein in F1 mice.
(A) Mouse aorta or bladder analyzed for expression of Cnn1 mRNA by quantitative RT-PCR with normalized Cnn1+/+ set to 1.0; other genotypes expressed relative to Cnn1+/+. (asterisks indicate p<0.01 compared to wildtype). See also Supplemental Figures II and III. (B) Expression of CNN1 protein in aorta by Western blotting. (C) In vivo chromatin immunoprecipitation assay of SRF binding to wildtype versus mutant intronic CArG box in bladder tissue. (D) Confocal immunofluorescence microscopy of CNN1 in wildtype versus mutant carotid arteries of two different founder lines. (E) SMC DNA synthesis (arrows) revealed with an antibody to Ki-67. Similar findings were seen in an independent founder line. Scale bars are 30 µm and 10 µm for panels D and E, respectively.
Discussion
In this report, we used 3cCRISPR to mutate a regulatory element controlling gene expression in a living animal. We found that a three nucleotide substitution within an intronic SRF-binding CArG box, suggested previously to be of potential importance in the expression of the endogenous Cnn1 gene15, 16, nearly abrogates expression of Cnn1 mRNA and CNN1 protein in SMC of the vessel wall, without altering expression of a number of other CArG-SRF-dependent genes. Detectable CNN1 protein in the vessel wall of mutant mice suggests there may be additional cis elements (such as downstream CArG boxes) directing low level expression of the gene. We also note an increase in the number of SMC undergoing DNA synthesis in the vessel wall of mice carrying the CArG box mutation. The fact that no other local mutations were present, and congruent results were found in independent founder mice, indicate the phenotypes observed are a consequence of a loss in SRF binding to the mutant intronic CArG box. To our knowledge, this study provides the first genetic proof for a decisive role of a single regulatory element inducing a target gene’s expression in vivo without the introduction of foreign DNA sequences or deletion of native sequences.
Genetically inactivating genes, whether protein-coding or otherwise, is common practice in the laboratory. For example, a CArG box in the proximal promoter of the Telokin gene was deleted with some flanking sequences using Cre-mediated excision, and this deletion resulted in the loss of Telokin expression with consequent changes in the tone of intestinal smooth muscle18. Such experimentation is crucial to study the functional consequences associated with loss of gene products and to gain insight into the potential role of regulatory elements such as CArG boxes in the setting of a living animal. Traditional methods of deleting a gene in this manner require complex design and validation of a targeting vector followed by the time-consuming steps of electroporating the targeting vector into embryonic stem cells, enrichment and screening of embryonic stem cells for correct targeting, transference of the embryonic stem cell clone into the blastocyst, and breeding of chimeric mice for germ line transmission of the targeted allele19, 20. Due to the protracted time in generating genetically altered mice with this traditional approach, coupled with the complexity and redundancy of gene regulation, there have been far fewer regulatory knockouts as compared to genic knockouts. Further, genic control regions, such as enhancers, have almost always been deleted via Cre recombinase-mediated excision of 100s-1000s of nucleotides, effectively leaving a genomic cavity21–23. While these studies certainly provide some insight into the role of enhancers in target gene expression and animal pathophysiology, they lack resolution in narrowing down specific elements within the enhancer that mediate gene expression. Moreover, foreign DNA sequences such as loxP sites or the neomycin selectable marker can alter expression of local and/or distal genes, thus confounding accurate interpretation of knockout phenotypes24. In contrast, application of 3cCRISPR, as implemented here, provides exquisite genomic editing such that subtle nucleotide substitutions are introduced without deleting sequences or introducing any foreign DNA sequences. This revolutionary technology, therefore, offers an unprecedented experimental approach to precisely model regulatory variants, including previously identified human CArG-SNPs25, that may contribute to human disease. We predict 3cCRISPR will find widespread utility in the analysis of regulatory elements and will likely supplant traditional approaches to their functional study in transgenic animals.
Supplementary Material
Significance.
The CRISPR/Cas9 system is a revolutionary approach to rapidly generate precision-guided mutations in any genome. There have been a number of reports detailing CRISPR/Cas9-mediated edits in protein coding and noncoding genes in vivo. However, no study has yet to report on the use of CRISPR/Cas9 to effectuate a mutation in a regulatory element controlling gene expression in an animal model system. This type of genome editing is highly significant given the increasing recognition of disease-associated variants linked to regulatory elements that determine the extent and duration of a target gene’s expression and the need to model such mutations in a living organism. Here, we provide the first case study of CRISPR/Cas9 directing a subtle mutation within a key regulatory element controlling gene expression in vivo. This study provides a foundation for future experiments that will swiftly interrogate regulatory elements and their variants in animal models.
Acknowledgements
We thank Dr. Lin Gan and Dr. Min Deng of the University of Rochester Transgenic Facility for their expert RNA micro-injections and embryo transfers.
Source of Funding
This work was supported by NIH grant HL-117907 to JMM.
Abbreviations
- 3cCRISPR
Three-component CRISPR
- CRISPR
Clustered regularly interspaced short palindromic repeats
- Cas9
CRISPR-associated protein 9
- crRNA
CRISPR RNA
- gRNA
Guide RNA
- HDR
Homology directed repair
- PAM
Protospacer adjacent motif
- RFLP
Restriction fragment length polymorphism
- SMC
Smooth muscle cell(s)
- SRF
Serum response factor
- tracrRNA
Transactivating RNA
Footnotes
Disclosures
None.
References
- 1.Palmiter RD, Brinster RL. Germ-line transformation of mice. Annu Rev Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giraldo P, Montoliu L. Size matters: Use of YACs, BACs, and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372–375. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol. 2013;31:684–686. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang D, Xu J, Zhu T, Fan J, Lai L, Zhang J, Chen YE. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol. 2014;6:97–99. doi: 10.1093/jmcb/mjt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu Y, Shen B, Cui Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 10.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miano JM. Serum response factor: Toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 15.Miano JM, Carlson MJ, Spencer JA, Misra RP. Serum response factor-dependent regulation of the smooth muscle calponin gene. J Biol Chem. 2000;275:9814–9822. doi: 10.1074/jbc.275.13.9814. [DOI] [PubMed] [Google Scholar]
- 16.Long X, Slivano OJ, Cowan SL, Georger MA, Lee TH, Miano JM. Smooth muscle calponin: An unconventional CArG-dependent gene that antagonizes neointimal formation. Arterioscler Thromb Vasc Biol. 2011;31:2172–2180. doi: 10.1161/ATVBAHA.111.232785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniguchi S. Suppression of cancer phenotypes through a multifunctional actin-binding protein, calponin, that attacks cancer cells and simultaneously protects the host from invasion. Cancer Sci. 2005;96:738–746. doi: 10.1111/j.1349-7006.2005.00118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khromov AS, Wang H, Choudhury N, McDuffie M, Herring BP, Nakamoto R, Owens GK, Somlyo AP, Somlyo AV. Smooth muscle of telokin-deficient mice exhibits increased sensitivity to Ca2+ and decreased cGMP-induced relaxation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2440–2445. doi: 10.1073/pnas.0508566103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capecchi MR. Gene targeting in mice: Functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 20.Davis J, Maillet M, Miano JM, Molkentin JD. Lost in transgenesis: A user's guide for genetically manipulating the mouse in cardiac research. Circ Res. 2012;111:761–777. doi: 10.1161/CIRCRESAHA.111.262717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sur IK, Hallikas O, Vaharautio A, Yan J, Turunen M, Enge M, Taipale M, Karhu A, Aaltonen LA, Taipale J. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science. 2012;338:1360–1363. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- 22.Attanasio C, Nord AS, Zhu Y, et al. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 2013;342:1241006. doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M, Zhang W, Lu X, Hoggatt AM, Gunst SJ, Kassab GS, Tune JD, Herring BP. Regulation of 130-kda smooth muscle myosin light chain kinase expression by an intronic CArG element. J Biol Chem. 2013;288:34647–34657. doi: 10.1074/jbc.M113.510362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier ID, Bernreuther C, Tilling T, Neidhardt J, Wong YW, Schulze C, Streichert T, Schachner M. Short DNA sequences inserted for gene targeting can accidentally interfere with off-target gene expression. FASEB J. 2010;24:1714–1724. doi: 10.1096/fj.09-140749. [DOI] [PubMed] [Google Scholar]
- 25.Benson CC, Zhou Q, Long X, Miano JM. Identifying functional single nucleotide polymorphisms in the human CArGome. Physiol Genomics. 2011;43:1038–1048. doi: 10.1152/physiolgenomics.00098.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.