Summary
Uncoupling protein 1 (UCP1) mediates non-shivering thermogenesis and, upon cold exposure, is induced in BAT and subcutaneous white adipose tissue (iWAT). Here, by high-throughput screening using the UCP1 promoter, we identify Zfp516 as a novel transcriptional activator of UCP1 as well as PGC1α thereby promoting a BAT program. Zfp516 itself is induced by cold and sympathetic stimulation through the cAMP-CREB/ATF2 pathway. Zfp516 directly binds to the proximal region of the UCP1 promoter, not to the enhancer region where other transcription factors bind, and interacts with PRDM16 to activate the UCP1 promoter. Although ablation of Zfp516 causes embryonic lethality, knockout embryos still show drastically reduced BAT mass. Overexpression of Zfp516 in adipose tissue promotes browning of iWAT even at room temperature, increasing body temperature, energy expenditure, and preventing diet-induced obesity. Zfp516 may represent a future target for obesity therapeutics.
Introduction
Whereas white adipose tissue (WAT) is the primary energy storage organ, brown adipose tissue (BAT) is specialized to perform nonshivering thermogenesis to maintain body temperature in mammals. Although the contribution of white fat to thermogenesis is not fully understood, inguinal WAT (iWAT) but not visceral WAT can undergo browning upon acute cold exposure. Nonshivering thermogenesis requires uncoupling protein-1 (UCP1), which dissipates the mitochondrial proton gradient to produce heat rather than ATP. Recently, metabolically relevant amounts of functional BAT or BAT-like tissue have been shown to be present in adult humans (Cypess et al., 2009; Farmer, 2009; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009). This finding has generated considerable interest as an increase in BAT activity in mice has been shown to have anti-obesity and anti-diabetic effects (Cederberg et al., 2001; Leonardsson et al., 2004).
Lineage-tracing has shown that brown adipocytes may arise from Myf5+/Sca-1+/Pax7+ cells of the dermomyotome, progenitors of skeletal muscle, cartilage, and dermis (Gensch et al., 2008; Lepper and Fan, 2010; Schulz et al., 2011; Seale et al., 2008). In addition to BAT, upon cold exposure, brown adipocyte-like cells termed “brite” or “beige” cells, which originate from Myf5- cells, may arise in inguinal WAT (iWAT) depots or there may be interconversion between white and brown adipocytes (Rosenwald et al., 2013; Wu et al., 2012). Contrary to classic BAT cells, brown adipocyte-like cells express very low levels of UCP1 and other BAT-enriched genes in non-stimulated conditions and are induced greatly following cold exposure (Waldén et al., 2012; Wu et al., 2012). As the thermogenic program in classic BAT and emergence of brown adipocyte-like cells in iWAT are both induced by cold, a common regulatory mechanism may operate.
For transcriptional activation of UCP1, several transcription factors and coregulators, including PPARs, PGC1α, and ATF2, have been implicated, but are found in both WAT and BAT as well as in other tissues (Collins et al., 2010; Kang et al., 2005). These factors have been shown to act through the well-characterized enhancer element located 2.5kb upstream of the UCP1 gene. In this regard, brown fat specific expression of UCP1 in vivo has been demonstrated by using a construct that contained the UCP1 proximal promoter region as well as the enhancer region (Ricquier and Bouillaud, 1997). Therefore, a yet to be identified cold inducible transcription factor(s) may bind the proximal promoter region to participate in activating UCP1 and other thermogenic genes.
Here, by performing high throughput screening of putative transcription factors for activation of the UCP1 promoter, we report that Zfp516, a novel, cold inducible transcription factor enriched in BAT compared to WAT, binds to the promoter region of UCP1. Zfp516 directly interacts with PRDM16 for transcriptional activation of thermogenic genes. Whereas ablation of Zfp516 causes embryonic lethality with drastically diminished BAT, overexpression of Zfp516 in adipose tissue causes browning of iWAT, increasing body temperature and energy expenditure to prevent diet induced obesity.
Results
To identify potential transcriptional activators of the UCP1 gene, we screened over 1100 transcription factor expression vectors representing two-thirds of known or putative transcription factors (Fulton et al., 2009). These expression vectors were individually cotransfected with a GFP reporter driven by the −5.5kb UCP1 promoter. Previous studies have shown that the −4.55kb promoter region to be sufficient for BAT-specific and regulated expression in transgenic mice (Cassard-Doulcier et al., 1993). Positive clones, identified by the detection of GFP signal, were subjected to a secondary screen employing luciferase instead of GFP as a reporter to quantitate promoter activation. During screening, known transcriptional activators of UCP1 that are widely expressed, such as CREB and ATF2, were identified as positives demonstrating the effectiveness of our screening. We identified 18 candidate genes independently capable of activating the UCP1 promoter more than 3-fold. We further selected those that were expressed at higher levels in BAT compared to WAT by RT-qPCR (See flowchart, Fig. S1). Here, we studied one of the best candidates, the previously uncharacterized Krüppel-like zinc finger transcription factor, Zfp516 (Gene ID: 329003), for its role in the transcriptional regulation of UCP1.
Zfp516 directly binds and activates UCP1 promoter
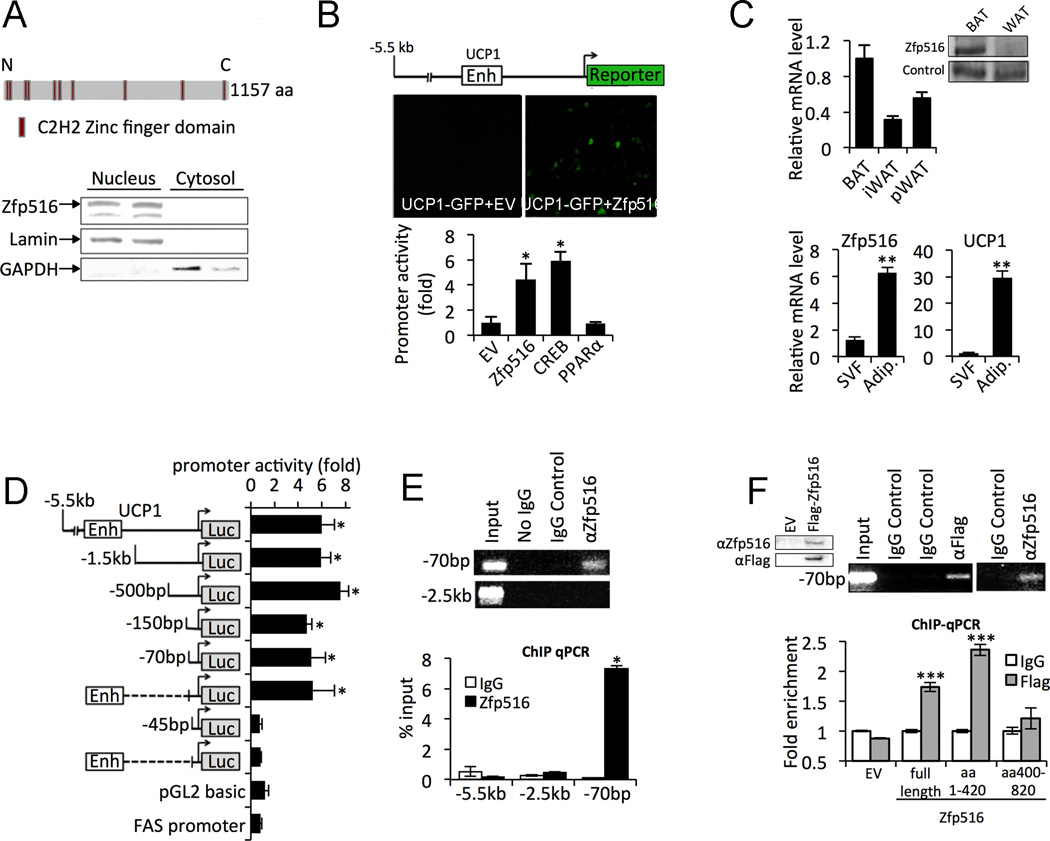
As Zfp516 activity or function has not been studied previously, we performed a motif analysis of Zfp516, which identified ten C2H2-type zinc fingers, widespread DNA binding motifs of eukaryotic transcription factors (Fig. 1A top) (Laity et al., 2001). Cell fractionation experiments to examine the localization of Zfp516 in the brown preadipocyte cell line, HIB-1B, showed that Zfp516 was present exclusively in the nucleus (Fig. 1A bottom). As detected in the initial screening, we established that Zfp516 robustly activated the UCP1 promoter-GFP reporter (Fig. 1B top). As compared to empty vector control, cotransfection of Zfp516 with the −5.5kb UCP1 promoter-luciferase resulted in a 4-fold activation of the UCP1 promoter (Fig. 1B bottom). This degree of UCP1 promoter activation was similar to that observed by cotransfection of CREB. We found that by both RT-qPCR and immunoblotting Zfp516 was enriched in BAT in comparison to WAT depots (Fig. 1C top). When BAT was fractionated into adipocytes and stromal vascular fraction (SVF) that contain brown preadipocytes, as expected, we detected UCP1 expression to be 30-fold higher in adipocytes than SVF. We found that Zfp516 expression was 6-fold higher in adipocytes compared to SVF (Fig. 1C bottom).
Figure 1. Zfp516 is a Brown Fat- Enriched Transcription Factor that binds and activates UCP1 promoter.
A. Top, diagram of Zfp516 structure. Bottom, immunoblotting for Zfp516, lamin (nuclear), and GAPDH (cytosolic) in nuclear and cytosolic fractions of HIB-1B cells. B. Top, GFP fluorescence of 293FT cells transfected with −5.5kb UCP1-GFP and either empty vector (EV) or Zfp516. Bottom, relative luciferase activity of 293FT cells cotransfected with −5.5kb UCP1-Luc and the indicated expression vector. C. Top and inlay, RT-qPCR and immunoblotting for Zfp516 mRNA and protein levels in BAT and WAT tissues from 6 week-old C57BL/6 mice (n=4 mice). Bottom, RT-qPCR for Zfp516 and UCP1 mRNA levels in the adipocyte fraction and SVF from BAT. D. Schematic representation of 5’ deletion constructs of the UCP1 promoter-luciferase and relative luciferase activity in 293FT cells cotransfected with indicated promoter construct and Zfp516 (expressed as fold of vector control). E. Top, ChIP for Zfp516 association to the UCP1 promoter in BAT. Bottom, qPCR for the ChIP DNA for Zfp516 association to the UCP1 promoter in BAT. F. Top-left, immunoblotting of lysates from 293FT cells transfected with −5.5kb UCP1-GFP with Flag-Zfp516 or vector. Top-right, ChIP for Flag-Zfp516 association to the UCP1 promoter using both αFlag and αZfp516 antibodies. Bottom, qPCR quantification of ChIP DNA for the association of full length Zfp516 or truncations of Zfp516 to the UCP1 promoter in 293FT cells. See also Figure S1, S2, and S3. *p<0.05; **p<0.01; ***p<0.001
To start defining how Zfp516 activates the UCP1 promoter, 5’ deletions of the UCP1 promoter driving a luciferase reporter were generated and cotransfected along with Zfp516 into 293FT cells. Contrary to the majority of known transcriptional activators of UCP1 that act at the enhancer element at −2.5kb upstream of the transcription start site (Cassard-Doulcier et al., 1998; Kozak et al., 1994), activation of the UCP1 promoter by Zfp516 was maintained even after deletion of the −2.5kb enhancer element. Further, all UCP1 promoter constructs deleted down to −70bp showed over 4.5-fold activation upon Zfp516 cotransfection. However, UCP1 promoter activation by Zfp516 was lost when the promoter was deleted to −45bp indicating that Zfp516 worked through the sequence from −70 to −45bp. Linking the −2.5kb UCP1 enhancer element to the −70bp to −45bp sequence did not show any further effect (Fig. 1D). Gel mobility shift assay showed a complex formation between Zfp516 and −70 to −45bp sequence, which could be supershifted by Zfp516 antibodies (Fig. S2A). Chromatin immunoprecipitation (ChIP) of interscapular BAT from C57BL/6 mice showed clear evidence of Zfp516 binding to the proximal UCP1 promoter region in vivo (Fig. 1E). We conclude that Zfp516 functions through the sequence from −70 to −45bp. As indicated in Fig.1A, Zfp516 contains 10 Zinc fingers primarily at the N-terminus. Therefore in addition to full length, we generated truncations of Zfp516 containing either the first one third (AA1-420) (7 Zinc fingers) or second third (AA400-820) (2 Zinc fingers) to use in ChIP assays. As expected, use of the antibody showed the binding of full length Zfp516 at the proximal UCP1 promoter region. ChIP using Zfp516 antibody showed the same results (Fig. 1F top). ChIP qPCR showed that in addition to full length Zfp516, the first one third of the N-terminus (AA1-420) but not the second third (AA400-820) showed binding to the UCP1 promoter indicating that Zfp516 interacted with UCP1 promoter through the first third from the N-terminus (Fig. 1F bottom). Similarly, gel shift assay showed that full-length Zfp516 and Zfp516 (AA1-420) but not Zfp516 (AA400-820) form DNA-protein complexes (Fig. S2B,C). These data indicate that Zfp516, through its N-terminal domain binds to the UCP1 promoter region from −70 to −45bp to activate transcription.
In addition to UCP1, we also examined several BAT-enriched gene promoters for sequence similarity to −70 to −45bp of UCP1 promoter. Indeed several of BAT-enriched genes contained the DNA sequence, CCACT, present in the UCP1 promoter. We chose PGC1α and Cox8b, which contained sequences with the highest alignment score, for further examination. ChIP analysis of the 293FT cells transfected with Zfp516 and the PGC1α or Cox8b promoter-reporters showed that, indeed, Zfp516 was bound to the −2.4kb region of the PGC1α promoter and −2.8kb region of Cox8b promoter. Cotransfection of Zfp516 with the −2.4kb PGC1α promoter-luciferase construct in 293FT cells resulted in a 15-fold activation of the PGC1α promoter (Fig. S3). These results show that Zfp516 binds and activates not only UCP1 but other BAT gene promoters as well.
Zfp516 interacts with PRDM16
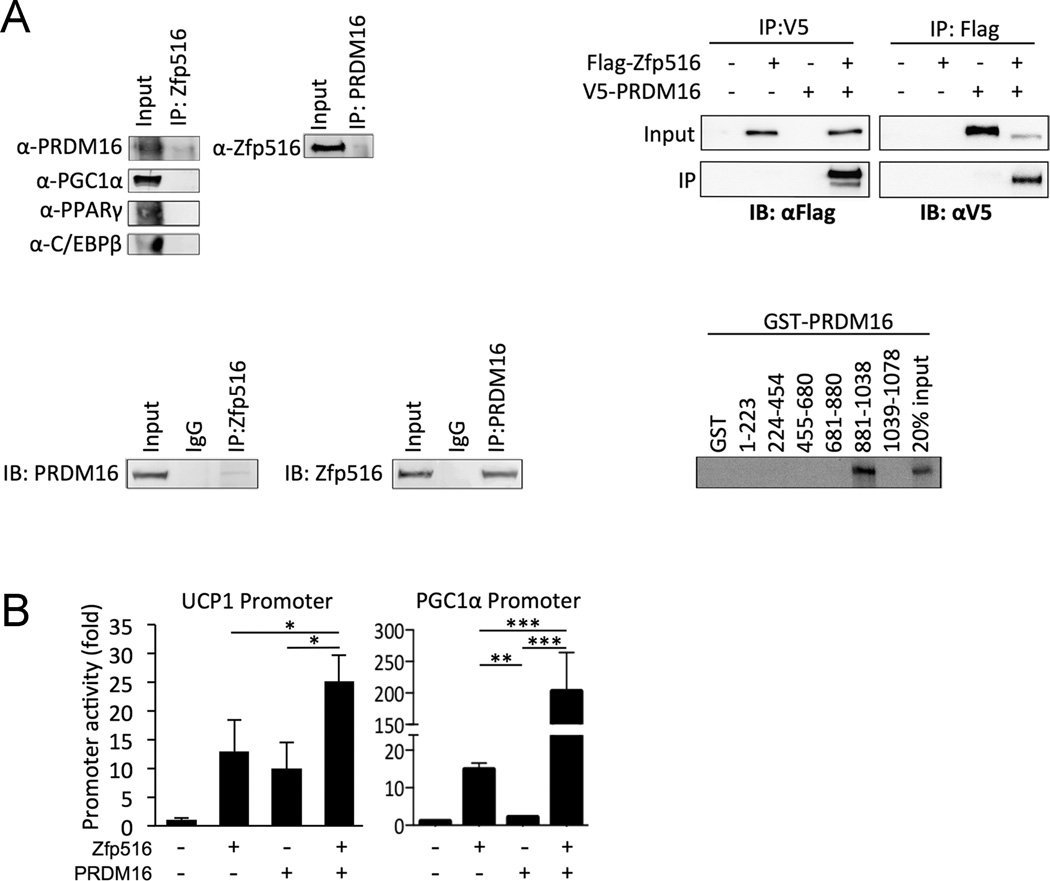
Next we asked whether Zfp516 interacts with known transcription factors or cofactors that have been reported to be involved in UCP1 activation. PGC1α, PPARγ, C/EBPβ, and PRDM16 were cotransfected with Zfp516 and immunoprecipitated with the Zfp516 antibody. Immunoblotting with antibodies against these transcription factors identified PRDM16, but not other factors, as an interacting partner of Zfp516 (Fig. 2A top-left). Interaction between Zfp516 and PRDM16 was confirmed by using Flag-tagged Zfp516 and V5-tagged PRDM16 as well as using endogenous proteins from mouse BAT (Fig. 2A top-right, bottom-left). Use of various GST-PRDM16 fusion proteins indicated that Zfp516 can directly interact with PRDM16 AA881-1038, a region shown to interact with other transcription factors (Kajimura et al., 2008) (Fig. 2A bottom-right). To examine the functional significance of Zfp516 and PRDM16 interaction in the activation of UCP1 and other BAT-enriched genes, we cotransfected Zfp516 and PRDM16 with the −5.5kb UCP1-luciferase reporter construct. Zfp516 and PRDM16 individually activated the UCP1 promoter 13- and 10- fold, respectively while, cotransfection of both resulted in a 25-fold activation (Fig. 2B left). Cotransfection of Zfp516 and PRDM16 together with the PGC1α promoter caused a robust 200-fold increase in promoter activity whereas Zfp516 and PRDM16 individually activated the PGC1α promoter 15- and 3-fold respectively (Fig. 2B right). These data show that Zfp516 and PRDM16, upon their interaction, further activate transcription of BAT genes. Since the DNA binding activity of PRDM16 is not required for BAT gene induction, Zfp516 may recruit PRDM16 to the promoter regions of BAT-enriched genes. In this regard, Re-Chip indicated that Zfp516 and PRDM16 were bound to the UCP1 promoter in a similar region (data not shown).
Figure 2. Zfp516 directly interacts with PRDM16.
A. Top-left (left column), immunoblot for various brown fat specific transcription factors after IP with αZfp516 antibody in 293FT cells cotransfected with Zfp516 and the indicated TF. Top-left (right side), immunoblot for Zfp516 protein after IP with αPRDM16 of lysates coexpressing Zfp516 and PRDM16. Top-right, immunoblot with αV5 or αFlag after IP with either Flag or V5 respectively of 293FT lysates transfected the indicated vectors. Bottom-left, immunoblot for either Zfp516 or PRDM16 after IP with indicated antibody of 50µg of BAT nuclear extracts. Bottom-right, Autoradiograph of GST pulldown using GST fusion proteins containing the indicated domains of PRDM16 and 35S-labelled in vitro translated Zfp516. B. Left, luciferase activity of 293FT cells cotransfected with the −5.5kb UCP1 promoter with Zfp516 and PRDM16 either together or separately. Right, luciferase activity of 293 FT cells cotransfected with the −2.4kb PGC1α promoter with Zfp516 and PRDM16 either together or separately. *p<0.05; **p<0.01; ***p<0.001
Zfp516 is cold induced via cAMP-CREB/ATF pathway
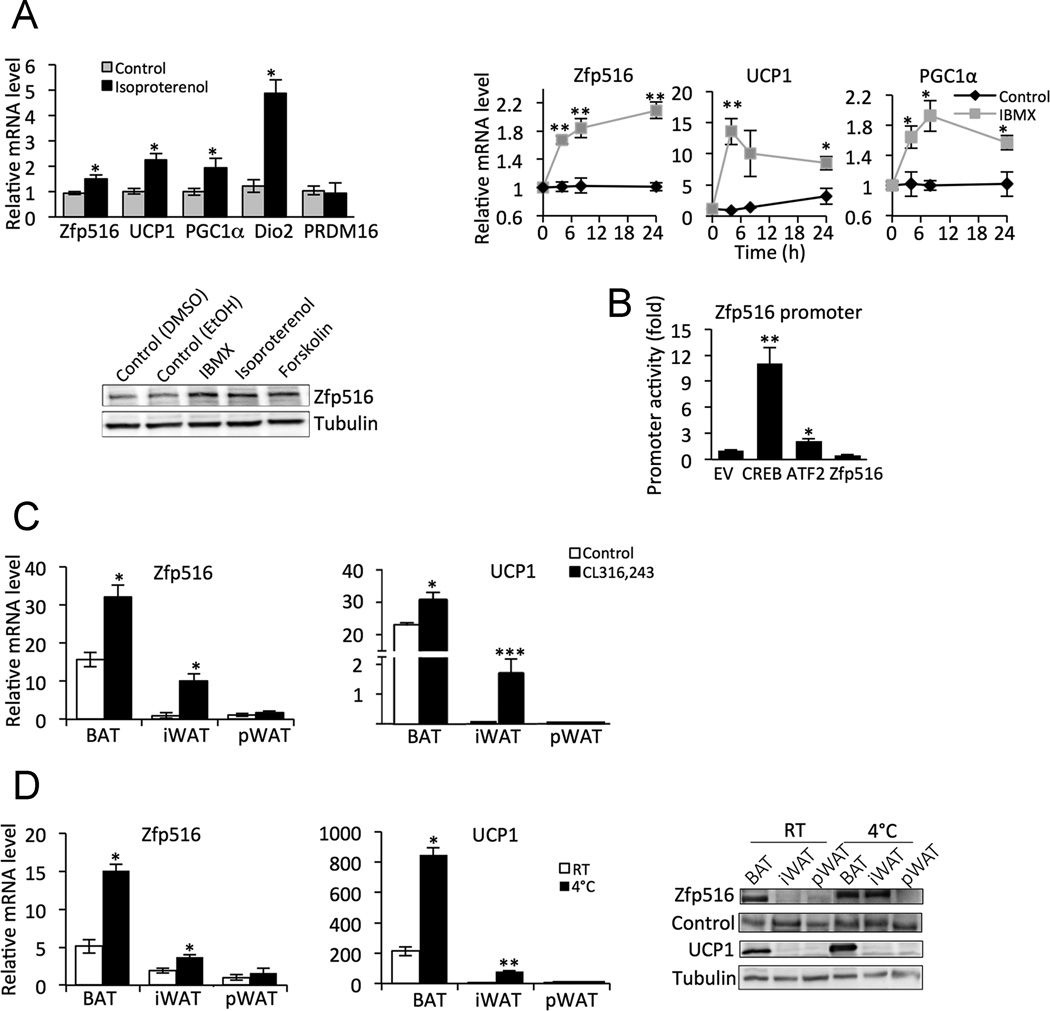
Expression of many BAT-enriched genes as well as BAT development are known to be induced by cold (Bachman et al., 2002; Cannon and Nedergaard, 2004; Farmer, 2008; Nedergaard and Cannon, 2010). Therefore, we next assessed possible regulation by the β-adrenergic receptor-cAMP pathway of Zfp516 itself. We treated HIB-1B cells with IBMX, Isoproterenol, or Forskolin, agents that can increase intracellular cAMP levels. Treatment of cells with the β-adrenergic agonist, isoproterenol resulted in approximately 2-fold increase in both Zfp516 and UCP1 mRNA levels. Other known cold responsive genes such as PGC1α and Dio2 were also induced while PRDM16 remained unchanged (Fig. 3A top-left). Treatment of HIB-1B cells with IBMX, a phosphodiesterase inhibitor, resulted in a rapid increase in Zfp516 mRNA level reaching maximal increase of 2-fold after 6h treatment which was maintained up to 24h. Expression of other known targets such as UCP1 and PGC1α showed similar pattern during IBMX treatment but increased by 13- and 2-fold, respectively (Fig. 3A top-right). Treatment with these agents resulted in increased Zfp516 protein levels in all treatment conditions (Fig. 3A bottom). We next generated a reporter construct linking −2.0kb of the Zfp516 promoter to a luciferase reporter and cotransfected it with CREB, ATF2, and other known BAT transcription factors. Motif search of the proximal −2.0kb of the Zfp516 promoter region revealed two half CREs at −1.1kb and −1.8kb upstream of the Zfp516 transcription start site. Indeed, cotransfection of CREB or ATF2 with the Zfp516 promoter-reporter construct increased the luciferase activity by 11- and 2-fold, respectively, whereas other transcription factors, such as PRDM16, C/EBPβ, PGC1α, PPARα, and PPARγ, did not show any effect (Fig. 3B; Fig. S4). These data show that Zfp516 is induced via cAMP-CREB/ATF2 pathway.
Figure 3. Zfp516 is regulated by Cold through CREB/ATF.
A. Top-left, RT-qPCR for selected genes in HIB-1B cells with or without 4h treatment with 10µM isoproterenol. Top-right, RT-qPCR for Zfp516, UCP1, and PGC1α mRNA in HIB-1B cells during 500µM IBMX treatment. Bottom, immunoblotting for Zfp516 or tubulin in HIB-1B cells treated with vehicle or indicated compound for 12h. Values are normalized to nontreated cells. B. Relative luciferase activity of 293FT cells transfected with the −2.0kb Zfp516-Luc and indicated transcription factor expression vector. C. RT-qPCR for Zfp516 and UCP1 mRNA in various tissues of wild-type mice following 10d of intraperitoneal injection with either CL316,243 or saline. D. RT-qPCR for Zfp516 and UCP1 mRNA (left and center) and immunoblotting for indicated proteins (right) in various adipose depots of wild-type mice exposed to cold (4°C) for 6h. See also Figure S4. *p<0.05; **p<0.01; ***p<0.001
To test cAMP pathway in vivo we treated mice with the β-adrenergic agonist, CL316,243 for 10d. Expression of UCP1 and Zfp516 was increased 1.5- to 2-fold, respectively in BAT (Fig. 3C). There was no change in UCP1 or Zfp516 expression levels in perigonadal WAT (pWAT), a visceral WAT depot. In contrast, in iWAT, UCP1 and Zfp516 were drastically increased by 30- and 10-fold, respectively (Fig. 3C). Overall, these results indicate that Zfp516 expression in iWAT, although its absolute levels are still lower than in BAT, is induced to a greater extent by β-adrenergic agonist treatment.
Next we subjected mice to a 6h cold challenge. Similarly to β-adrenergic agonist treatment, we detected an increase of UCP1 and Zfp516 mRNA and protein levels in BAT upon cold exposure. Zfp516 expression in pWAT was lower and remained low, even after cold exposure. In iWAT, UCP1 and Zfp516 expression increased 40- and 2.5-fold, respectively, with Zfp516 expression approaching the expression levels detected in BAT at room temperature (Fig. 3D). Overall, we conclude that although Zfp516 level is higher in BAT than WAT, the degree of induction by β-agonist or cold exposure was much higher in iWAT compared to BAT while there was no change in pWAT. These data suggest that Zfp516 may be involved in the browning of iWAT.
Zfp516 promotes browning of white adipose tissue
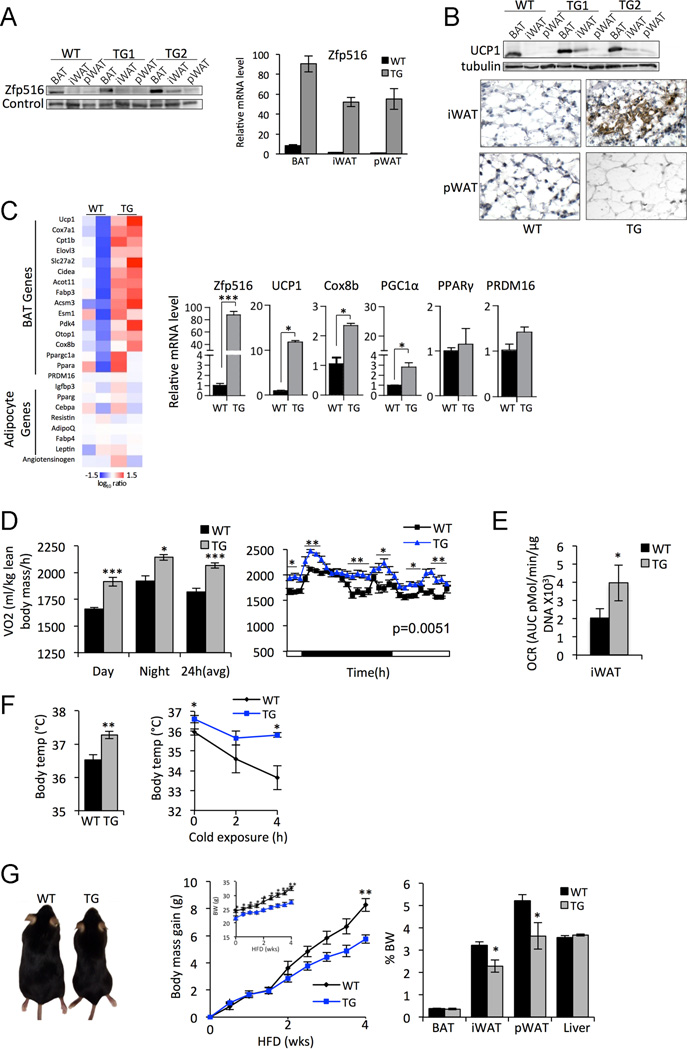
Next, to test the effect of Zfp516 overexpression in adipose tissue in vivo, we generated transgenic mouse lines expressing Zfp516 driven by the −5.4kb aP2 promoter. Although endogenous Zfp516 expression is significantly higher in BAT than WAT (Fig. 1C), the fold increase of Zfp516 in iWAT and pWAT of transgenic mice was greater than in BAT (50-fold versus 10-fold) (Fig. 4A). Zfp516 expression in other tissues was unaffected (Fig. S5H). Similarly, Zfp516 protein levels were elevated in two independent transgenic lines in all adipose depots compared to wild-type mice. We then examined UCP1 levels by immunoblotting. UCP1 levels were increased approximately 3–4 fold in BAT of aP2-Zfp516 mice compared to wild-type littermates. Strikingly, although its level was lower than that in BAT, the fold increase of UCP1 was more drastic in iWAT, but not pWAT, of aP2-Zfp516 mice even in the absence of cold or adrenergic stimuli (Fig. 4B). Noticeably, iWAT of aP2-Zfp516 mice that were maintained at room temperature showed large clustered populations of cells with smaller multilocular lipid droplets, a characteristic of the browning of WAT depots whereas pWAT of aP2-Zfp516 mice showed no difference in morphology compared to wild-type littermates (Fig. S5A, 5B ). UCP1 immunostaining of UCP1 in iWAT sections from aP2-Zfp516 mice maintained at room temperature also showed robust UCP1 staining (Fig. 4B), while no significant staining was observed in wild type littermates.
Figure 4. Zfp516 Promotes Browning of White Adipose Tissue.
A. Left, immunoblotting for Zfp516 in adipose depots of WT and aP2-Zfp516 mice (TG) with different transgene copy number. Right, RT-qPCR for Zfp516. B. Top, immunoblotting for UCP1 in adipose depots of WT and TG mice. Bottom, immunostaining for UCP1 in sections of iWAT and pWAT from WT and TG mice. C. Left, microarray analysis from iWAT of WT or TG mice. Right, RT-qPCR of genes from microarray in WT or TG mice (n= 3 per group). D. VO2 assayed by indirect calorimetry in WT and TG mice on chow diet (CD) (n=6 mice per group). E. Oxygen consumption rate of iWAT from WT or TG mice. F. Rectal temperature measured in 15wk old WT and TG mice fed CD at room temperature (left) and after 4 h cold exposure (right) (n=7–8 mice per group). G. Left, representative photograph of 26wk old WT or TG mice fed HFD for 16wk. Center, body mass gain for the mice from left. Inlay, body weight in WT and TG mice fed HFD starting at 6wk old (n=6–8 mice per group). Right, mass of each adipose tissue depot and liver represented in percentage of body weight in the mice mentioned above. See also Figure S5. *p<0.05; **p<0.01; ***p<0.001
To examine overall gene expression changes in iWAT, RNA from iWAT of wild-type and aP2-Zfp516 mice were subjected to Affymetrix microarray analysis. We found that transgenic expression of Zfp516 led to upregulation of a broad program of BAT-enriched genes while common adipocyte markers were not affected significantly (Fig. 4C left). RT-qPCR analysis of these samples showed that expression levels of BAT-enriched genes, such as UCP1, PGC1α, and Cox8b, were all significantly elevated in iWAT of aP2-Zfp516 mice, while PPARγ and PRDM16 remain unaffected (Fig. 4C top-right). Considering the striking increase in UCP1 levels as well as extensive browning of iWAT of aP2-Zfp516 mice, we next assessed the metabolic effect in the transgenic animals. We observed 10% higher oxygen consumption rate (VO2) in transgenic mice during both day and night cycles (Fig. 4D). We next tested whether changes in respiratory activity in iWAT could contribute to the altered respiratory activity in these transgenic mice. Indeed, upon using a Seahorse XF-24 Extracellular flux analyser, we found a 70% increase in oxygen consumption rate (OCR) in iWAT of transgenic mice (Fig. 4E). We did not detect significant alterations of OCR in BAT of transgenic mice (data not shown). Strikingly, aP2-Zfp516 mice fed a standard chow diet and maintained at room temperature had 0.7°C higher core body temperature than their wild-type littermates, although its implication in thermogenesis is not clear (Nedergaard and Cannon, 2014) (Fig. 4F). However, when subjected to an acute cold exposure, these transgenic mice showed rectal temperature to be dropped by 1°C after 4h at 4°C whereas the wild-type mice showed drop of 3°C, showing improved thermogenic capacity in Zfp516 transgenic mice. Taken together, these data show that Zfp516 overexpression causes browning of iWAT that results in increase in oxygen consumption and increased cold resistance.
We next subjected aP2-Zfp516 transgenic mice to high-fat diet (HFD) to test whether the increase in thermogenesis results in protection from diet-induced obesity. Interestingly, aP2-Zfp516 transgenic mice have significantly smaller body weight compared to wild type littermates on chow diet at room temperature, despite no change in food intake or activity level in these mice (Fig.S5C-F). When subjected to HFD feeding, aP2-Zfp516 transgenic mice gained 30% less weight than their wild-type littermates (Fig. 4G). Noticeably, the iWAT and pWAT depots of transgenic mice had approximately 30% lower weights than wild-type littermates, even when normalized to body weight while BAT as well as other organs such as liver was unaffected (Fig. 4G right). These data suggest that Zfp516 overexpressing mice are protected from diet induced obesity. After 4wks on HFD, mice were subjected to glucose and insulin tolerance tests. We found that Zfp516 transgenic mice had improved glucose tolerance and insulin sensitivity as compared to wild-type littermates (Fig. S5G). Taken together, these data show that overexpression of Zfp516 causes an increase in UCP1 and browning of iWAT resulting in increased energy expenditure, improved cold tolerance, as well as protection against diet-induced obesity.
Ablation of Zfp516 impairs BAT development in vivo
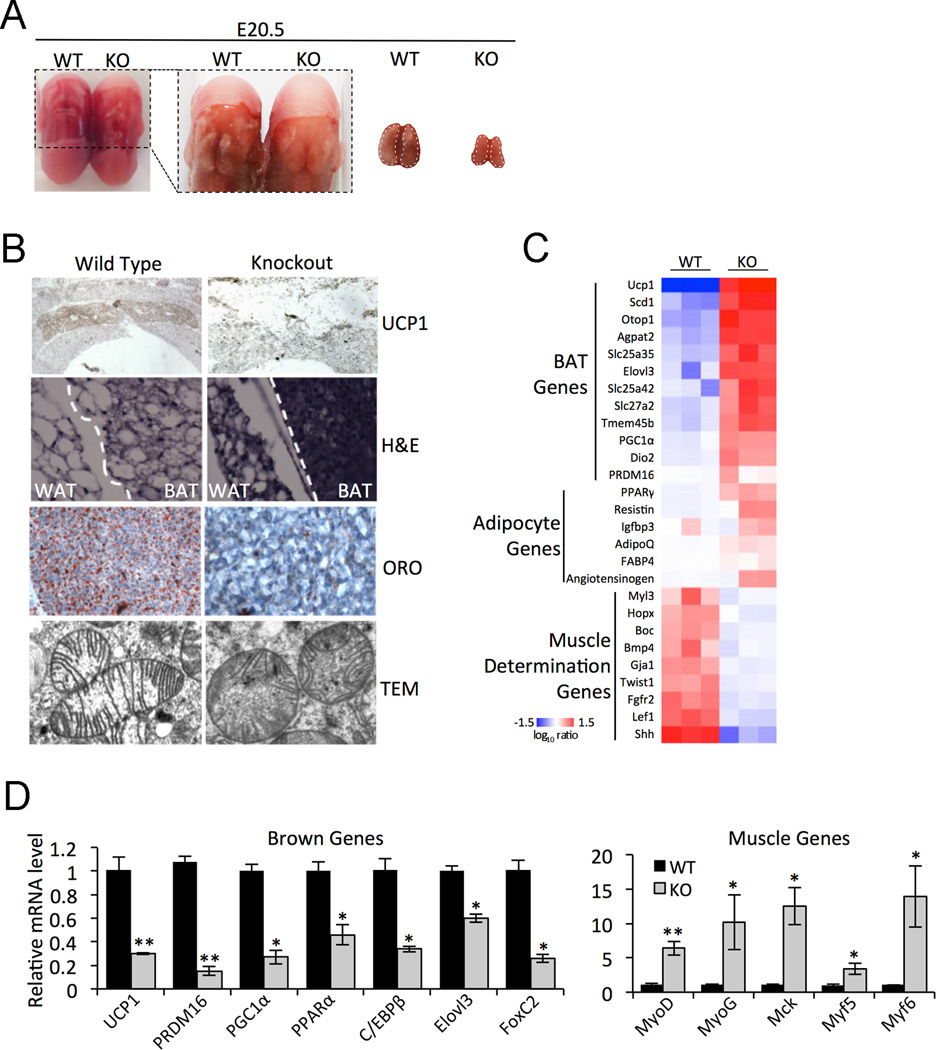
To further examine Zfp516 function in BAT program in vivo, we ablated Zfp516 in mice by using embryonic stem cells containing a gene trap inserted in intron 1 of the Zfp516 gene (Fig. S6B). Mice homozygous for the Zfp516 gene-trap were found to die immediately after birth due to a yet to be defined role of this gene during development. Therefore, we examined Zfp516−/− embryos at E20.5 when the presence of BAT, but not WAT, can be easily detected in wild-type mice. RT-qPCR showed that Zfp516 expression was undetectable in Zfp516−/− embryos compared to wild-type littermates confirming Zfp516 ablation (Fig. S6C). Although Zfp516−/− embryos were somewhat smaller at E17.5, they were visibly indistinguishable from their wild-type littermates in terms of size at E20.5, yet the defective BAT formation was easily detected (Fig. 5A). UCP1 staining on transverse sections of embryos showed a complete absence of UCP1 in the Zfp516−/− presumptive BAT (Fig. 5B top). Haematoxylin and eosin (H&E) staining of the WAT/BAT boundary indicated abnormal cell morphology in the presumptive BAT of Zfp516 deficient embryos, while nearby WAT showed decreased cell size (Fig. 5B upper-middle). Oil Red O staining of presumptive BAT from Zfp516−/− embryos revealed a near complete loss of lipid staining (Fig. 5B lower-middle). Furthermore, transmission electron microscopy revealed that mitochondria from presumptive BAT from Zfp516−/− embryos have randomly-oriented cristae as compared to the classic laminar cristae found in wild-type littermates (Fig. 5B bottom).
Figure 5. Zfp516 ablation blocks brown fat development in mice.
A. Left, E20.5 WT and Zfp516 KO embryos from back view. White dots delineate area of BAT or presumptive BAT. Right, cropped image from left. B. Top, immunostaining for UCP1 in BAT or presumptive BAT in WT and KO embryos. Top middle, H&E staining of the BAT/WAT border in WT and KO mice. Bottom middle, ORO staining of WT or KO BAT. Bottom, transmission electron microscopy of WT and KO BAT mitochondria. C. Microarray analysis from BAT or presumptive BAT of WT or KO E20.5 embryos. D. RT-qPCR for BAT enriched (left) and muscle (right) genes in BAT from E17.5 WT or KO embryos (n=3). See also Figure S6. *p<0.05; **p<0.01; ***p<0.001.
We next examined gene expression in presumptive BAT of our Zfp516 ablated embryos at the global level by performing Affymetrix microarray analysis. Clustering of significantly affected genes into functionally related gene groups revealed down regulation of a broad program of genes required for BAT function, whereas expression of muscle genes were elevated (Fig. 5C). Common adipocyte markers were not significantly altered (data not shown). The gene expression analysis of BAT from Zfp516−/− embryos by RT-qPCR revealed that expression of BAT-enriched genes, such as UCP1, PRDM16, PGC1α, PPARα, C/EBPβ, Elovl3, and FoxC2, was all significantly lower by 40–85% (Fig. 5D left). In contrast, myogenic genes, such as MyoD, MyoG, Mck, Myf5, and Myf6, were significantly upregulated by 3- to 14-fold (Fig. 5D right). We conclude that Zfp516 ablation abrogates the induction of UCP1 and other BAT genes during normal BAT development.
Zfp516 promotes brown adipogenesis and suppresses myogenesis in vitro
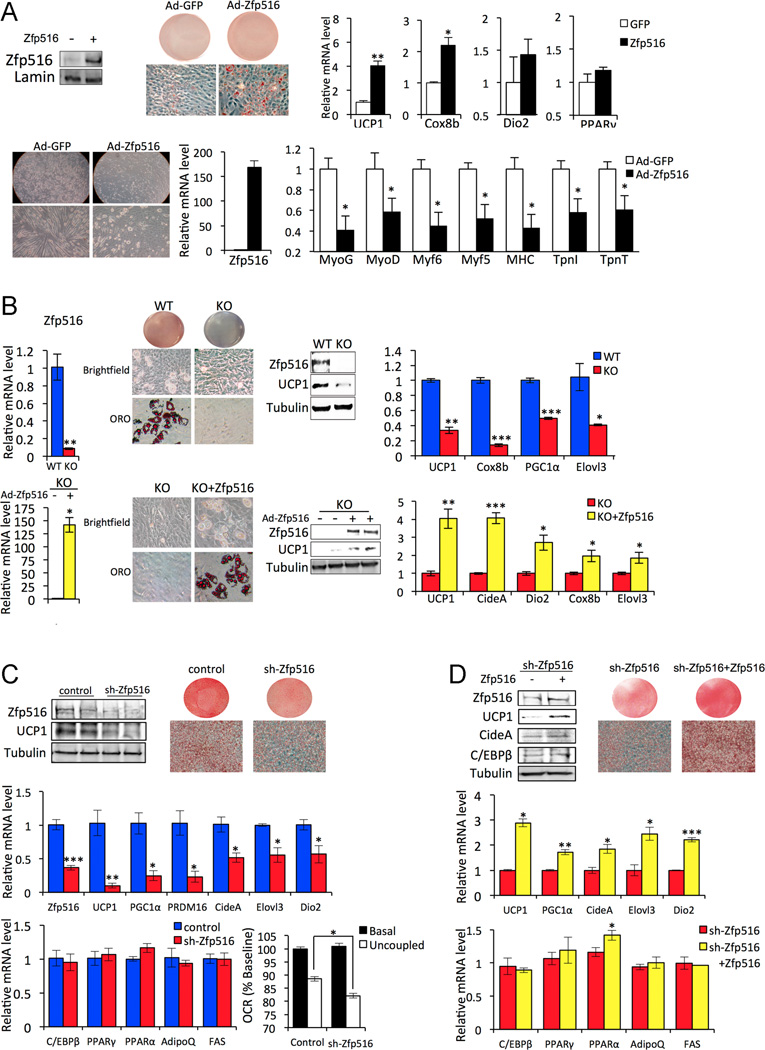
As stated above, BAT develops from Myf5+ precursors that can differentiate to both muscle and BAT. Since we detected an increase in myogenic genes accompanied by decrease in BAT enriched genes in presumptive BAT of Zfp516−/− embryos, we employed C2C12 cells to test the ability of Zfp516 to induce UCP1 and other thermogenic genes to promote brown adipocyte differentiation. C2C12 cells were transduced with adenovirus expressing Zfp516 and treated with brown adipogenic cocktail. Endogenous expression of Zfp516 in C2C12 was very low but was substantially increased by adenoviral infection as verified at the protein level by immunoblotting (Fig. 6A top-left) and at the mRNA level by RT-qPCR (data not shown). Upon infection with Zfp516, we observed an activation of the brown adipogenic program in C2C12 cells as evidenced by enhanced lipid accumulation (Fig. 6A top-center) and increased expression of UCP1 and Cox8b (respectively by 4- and 2- fold; Fig. 6A top-right). In contrast, in myogenic conditions, Zfp516-overexpressing cells failed to undergo efficient myogenic differentiation, exhibiting a rounded cell shape, whereas control C2C12 cells underwent drastic morphological modifications toward multinucleated and elongated characteristic of myotubes (Fig. 6A bottom-left). Consistent with the cell morphology, in comparison to control C2C12 cells, ectopic Zfp516 expressing cells had a significant reduction in the expression of myogenic genes, including myogenic transcription factors, MyoG, MyoD, Myf6 and Myf5, as well as late markers MHC, TpnI and TpnT (Fig. 6A bottom right). These data show that ectopic expression of Zfp516 represses the myogenic program and drives transcription of a BAT gene program to promote brown adipocyte differentiation in C2C12 cells.
Figure 6. Zfp516 promotes brown adipogenesis and suppresses myogenesis.
A. Top, immunoblotting for indicated proteins (left), ORO staining (center) and RT-qPCR for BAT enriched genes and PPARγ (right) in C2C12 cells transduced with either GFP or Zfp516 adenovirus after 6 days of brown adipogenic differentiation. Bottom, Brightfield view at 20X (top left) and 40X (bottom left) magnification, RT-qPCR for Zfp516 mRNA (center), and RT-qPCR for muscle specific genes (right) in C2C12 cells transduced with GFP or Zfp516 adenovirus after 6d of myogenic differentiation. B. Top, RT-qPCR for Zfp516 mRNA (left), ORO staining and brightfield view pictures (center left), immunoblotting for indicated proteins (center right) and RT-qPCR for BAT enriched genes (right) in WT and KO MEFs after 5d of adipogenic differentiation. Bottom, RT-qPCR for Zfp516 mRNA (left), ORO staining and brightfield view pictures (center left), immunoblotting for indicated proteins (center right) and RT-qPCR for BAT enriched genes (right) in KO MEFs infected with GFP or Zfp516 adenovirus after 5d of adipogenic differentiation. C. Top-left, immunoblotting for indicated proteins in HIB-1B cells infected with control or sh-Zfp516 lentivirus after 5d of adipogenic differentiation. Top-right, ORO staining at day 5 of differentiation. Middle, RT-qPCR for mRNA level of Zfp516 and BAT enriched genes these cells. Bottom-left, RT-qPCR for mRNA level of common adipogenic genes. Bottom-right, Relative OCR rates in cells before and after oligomycin treatment. OCR measurements before drug injection in the control cells were set as 100%. (N=9–10). D. Top-left, Immunoblotting for indicated proteins in sh-Zfp516 HIB-1B cells or sh-Zfp516 infected with Zfp516 adenovirus after 5 days of adipogenic differentiation. Top-right, ORO staining at day 5 of differentiation. Middle, RT-qPCR for BAT enriched genes. Bottom, RT-qPCR for common adipogenic genes *p<0.05; **p<0.01; ***p<0.001.
Next, we tested whether ablation of Zfp516 causes impairment of brown adipocyte differentiation in vitro by using mouse embryo fibroblasts (MEFs) isolated from Zfp516 −/−embryos (Tseng et al., 2008). In contrast to wild-type that accumulated lipid droplets, knockout MEFs failed to differentiate as evidenced by lack of Oil red O (ORO) staining (Fig. 6B center-left). While the wild-type MEFs underwent a morphological change to the rounded polygonal shape characteristic of brown adipocytes, the knockout MEF retained fibroblast-like morphology (Fig. 6B center-left). UCP1 at both the protein and mRNA levels was greatly lower in the knockout MEFs (Fig. 6B center-left). Expression levels of other BAT-enriched genes, such as Cox8b, PGC1α, and Elovl3, were 50–80% lower in the knockout compared to wild-type MEFs (Fig. 6B top right). We next performed a rescue experiment using Zfp516 adenovirus in knockout MEFs. Compared to Zfp516 −/− MEFs, transduction with Zfp516 adenovirus completely rescued the phenotype in regard to morphological changes, lipid accumulation, and expression of BAT-enriched genes (Fig. 6B bottom). These data show that Zfp516 expression is necessary for differentiation of multipotent MEFs to brown adipocytes.
We next investigated whether knockdown of Zfp516 can inhibit BAT gene expression in brown preadipocyte cell line. HIB-1B cells were transduced with sh-Zfp516 or control lentivirus and the cells selected for stable integration were subjected to brown adipocyte differentiation. Zfp516 knockdown cells, which had approximately 50% lower Zfp516 mRNA (Fig. 6C center) and protein levels (Fig. 6C top-left) at confluence, showed drastically lower ORO staining after 5d of differentiation (Fig. 6C top-right). Similarly, Zfp516 knockdown resulted in a marked decrease in the expression of various BAT genes, including UCP1, PRDM16, PGC1α decreased by 70–90% and CideA, Elovl3 and Dio2 by 40% (Fig. 6C middle) further indicating inhibition of brown adipocyte differentiation by Zfp516 knockdown. Gene expression levels of PPARγ, PPARα, AdipoQ and FAS, common markers for both WAT and BAT, were not affected (Fig. 6C bottom-left). We next measured the respiratory rates of the Zfp516 knockdown cells, to test if there was altered uncoupling in Zfp516 knockdown cells. While the basal respiratory rates were similar, the Zfp516 knockdown cells had significantly reduced uncoupling following oligomycin treatment compared to control cells (Fig. 6C bottom-right). These data show that Zfp516 is required for proper brown adipocyte differentiation in vitro, which can be reflected in mitochondrial uncoupling. Furthermore, in order to eliminate off-target effects of our shRNA knockdown, we performed a rescue experiment by infecting Zfp516 knockdown cells with adenoviral Zfp516. We observed a rescue of differentiation as evidenced by enhanced lipid accumulation (Fig. 6D top-right), increased expression of BAT enriched genes such as UCP1, CideA, PGC1α, Elovl3 and Dio2 at the mRNA level (Fig. 6D center) as well as at the protein level, while common adipocyte markers were unaffected (Fig. 6D bottom). Together these studies demonstrate that Zfp516 transcriptionally activates UCP1 and other BAT genes resulting in cell autonomous brown adipocyte differentiation.
Discussion
Due to its potential as a target of anti-obesity therapeutics, arising from evidenced presence of BAT like cells in humans, BAT development and activity have recently become an area of intense interest. While widely expressed transcription factors, such as PRDM16, PGC1α, C/EBPβ, and Ebf2, have been identified to promote the BAT program, specific brown fat enriched transcription factors may operate to activate BAT genes and BAT development (Rajakumari et al., 2013). By unbiased genome-wide transcription factor library screening, we identified the zinc-finger protein, Zfp516, as a novel brown fat enriched, cold-inducible DNA-binding transcriptional activator of UCP1 and other BAT genes, which promotes browning of iWAT and is required for BAT development. Importantly, we show that Zfp516 is induced by sympathetic signaling, which to our knowledge, represents the first sympathetically regulated BAT-enriched transcription factor promoting a BAT program in iWAT and BAT.
The above-mentioned transcriptional activators act through the well-characterized enhancer element at −2.5kb upstream of UCP1 promoter. Indeed, the enhancer region found at −2.5kb along with the proximal −400bp of the UCP1 promoter has been found to be sufficient to drive reporter expression in a tissue specific and sympathetically regulated manner (Ricquier and Bouillaud, 1997). Here, we identified Zfp516 as a novel activator of the UCP1 promoter, through binding at the proximal promoter region. Interestingly, CREB has previously been identified to interact with the proximal promoter region at −140bp to activate the UCP1 promoter. However, even the proximal −120bp of the UCP1 promoter that does not contain a CRE has still shown to respond to sympathetic stimuli (Rim and Kozak, 2002) clearly suggesting presence of a yet to be identified cold responsive transcription factors to function through binding to the proximal promoter region. Our finding that expression of Zfp516 is induced by cold and sympathetic stimulation and binds −70 to −45bp proximal region may represent such a transcription factor. In addition, Zfp516 can also bind PGC1α and Cox8b promoter regions for transcriptional activation. Zfp516 clearly acts as a transcriptional activator by binding to the promoter regions of BAT-enriched genes.
Recent research on the transcriptional factors critical for BAT development has centered on PRDM16. However, PRDM16 is broadly expressed and promotion of a BAT program by PRDM16 is independent of its DNA-binding activity (Ohno et al., 2013; Seale et al., 2007). Thus PRDM16 must act through other transcription factors that bind promoter regions of BAT-enriched genes. In this regard, PPARγ and C/EBPβ, which bind to the −2.5kb enhancer region, have been reported to function with PRDM16 to activate a BAT program (Kajimura et al., 2009; Seale et al., 2008) but these transcription factors also function in WAT development. Since the BAT program is induced upon cold exposure, DNA binding transcription factor(s) that are enriched in BAT and also cold inducible may act by interacting with PRDM16. We found that Zfp516, which binds to proximal promoter region of UCP1, directly interacts with PRDM16. In addition Zfp516 interacts with the PRDM16 ZF-2 domain that has been shown to interact with C/EBPβ (Kajimura et al., 2009). The relationship between these transcription factors interacting with PRDM16 needs further studies. Regardless, unlike other PRDM16 interacting proteins, Zfp516 is cold inducible by the classical β-AR-PKA-CREB pathway. Zfp516 expression is induced by β-adrenergic stimulation in both BAT and iWAT depots by cold exposure as well as by β-agonist treatment. These results suggest that the transcriptional regulation of UCP1 in response to cold stimuli may require Zfp516 activity. Our finding that Zfp516 drives promoter activity of UCP1 and PGC1α, two genes both known to be essential for proper BAT response to cold (Leone et al., 2005; Lin et al., 2004), establishes the key role Zfp516 plays in transcriptional activation of BAT-genes during cold exposure.
We show here that overexpression of Zfp516 in vivo causes browning of iWAT, even at room temperature. While we cannot disregard a potential effect of Zfp516 in the central nervous system due to use of the aP2-promoter driving the transgene (Martens et al., 2010), the fact that Zfp516 is inducable by cold exposure for thermogenic genes activation indicates that Zfp516 may play a critical role in browning of iWAT. Sympathetic stimulation in mice causes Zfp516 levels to be an order of magnitude higher in iWAT but not in pWAT. However, we did not detect significant alterations in BAT phenotype in our transgenic mice. In this regard, although Zfp516 is also induced in BAT, the fold induction was significantly lower than that detected in iWAT. This may explain the inability of detecting changes in BAT in our transgenic mice. However, it is also possible that the endogenous Zfp516 level in BAT is sufficient for a BAT phenotype. Similar to our aP2-Zfp516 transgenic mice, overexpression of PRDM16 in adipose tissue caused browning of iWAT without alteration in BAT phenotype (Seale et al., 2011).
As Zfp516 directly interacts with PRDM16, this suggests that Zfp516 cooperatively works along with PRDM16 for browning of iWAT. Previously, PRDM16 presumptive BAT from null embryos was reported to have lower BAT enriched and higher muscle specific gene expression, with higher lipid stores. A recent report of a Myf5-cre PRDM16 conditional KO, however, indicated no defect in early BAT development but impaired BAT function in adults (Harms et al., 2014). Although Zfp516 null embryonic presumptive BAT showed similar gene expression pattern to PRDM16 null, they also showed significantly decreased lipid staining, with altered genome-wide gene expression pattern as well as abnormal mitochondrial morphology. It is also possible that as a DNA binding protein, which often shows more severe phenotype of null mutation than coregulators, Zfp516 may interact with coregulator(s) other than PRDM16 during early BAT development.
Regardless, PRDM16 is expressed constitutively and, thus, the cold-inducible nature of Zfp516 could be a key driving BAT-gene program in browning of iWAT. Since iWAT is similar to human subcutaneous adipocytes, Zfp516, which induces browning of iWAT even at room temperature preventing diet induced obesity, may represent a compelling target for future anti-obesity therapeutics.
Experimental Procedures
Animals, and cell culture
All protocols for mice studies were approved from the University of California at Berkeley Animal Care and Use Committee. Mice were fed a chow diet ad libitum. Oxygen consumption (VO2) was measured using the Comprehensive Laboratory Animal Monitoring System (CLAMS). Data were normalized to lean body mass as determined by MRI. Body temperatures were assessed using a RET-3 rectal probe for mice (Physitemp). CL316,243 (Sigma) was intraperitoneally injected into mice at 1mg/kg. GTT and ITT performed upon intraperitoneal injection of D-glucose 1mg/g or of insulin 0.75mU/g of body weight as previously described (Ahmadian et al., 2011). Tail vein blood was collected for measurements. Derivation of mice is described in the supplementary methods.
Brown adipocyte differentiation was performed as described in (Kajimura et al., 2009). To induce thermogenic genes, cells were treated for 6h with 10µM forskolin or 10µM isoproterenol. For myogenic differentiation, confluent cells were treated with DMEM containing 2% Horse serum and 25mM Hepes, pH 7.4. Primary MEFs were isolated as previously described (Wang and Sul, 2009). MEF differentiation was performed as described in (Tseng et al., 2008). Cellular and tissue explant respiration was measured using a XF24 Analyzer (Seahorse Bioscience).
Functional Screen
cDNA clones from the Mammalian Genome Collection were cotransfected with the −5.5kb UCP1-eGFP-N1 into 293FT cells. Cells were assayed at 24 and 48hr post transfection for GFP signal. Positive clones were cotransfected with the −5.5kB UCP1- Luciferase construct into 293FT cells and assayed 24hr post transfection using the Dual-Luciferase Reporter assay (Promega). A more detailed protocol is included in the supplementary methods.
Oil red O staining
Wells were washed once with phosphate-buffered saline and subsequently fixed for 30min in 10% formalin. Cells were then stained with Oil Red O working solution for 1h. Cells were washed twice with distilled water prior to visualization.
RT-PCR Analysis and Western blotting
Total RNA was extracted using TRIzol reagent (Invitrogen). Reverse transcription was performed with 1µg of total RNA using SuperScript II (Invitrogen). RT-qPCR was performed in triplicate with an ABI PRISM 7500 sequence detection system (Applied Biosystems) to quantify the relative mRNA levels for various genes. Statistical analysis of the qPCR was obtained using the ΔΔCt method with U36B4 as the control. Primer sets used are listed in Table S1. Microarray hybridization and scanning were performed by the Functional Genomics Laboratory core facility using Affymetrix Mouse Genome 430A 2.0 Gene Chip arrays. Microarray data for both KO and TG mice are available at http://dx.doi.org/10.6084/m9.figshare.1009210 and http://dx.doi.org/10.6084/m9.figshare.1009211. For western blot analysis, total cell lysates were prepared using RIPA buffer and nuclear extracts were isolated using the NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo). Proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane and probed with the indicated antibodies.
Luciferase Assays
293FT cells were transfected with 300ng Zfp516 or empty vector and/or 300ng PRDM16 or empty vector in cotransfection assays together with 100 ng of indicated luciferase reporter construct and 0.5ng pRL-CMV in 24-well plates. Cells were lysed 24h post-transfection and assayed for luciferase activity as above.
Transmission electron microscopy
WT and KO BAT were fixed in 2% glutaraldehyde in 0.1M phosphate buffer, pH 7.3, at 4°C overnight, then postfixed in 1% OsO4 and embedded in an Epon-Araldite mixture. Ultrathin sections (0.2µm) mounted on 150-mesh copper grids were stained with lead citrate and observed under a FEI Tecnai 12 transmission electron microscope.
Statistical analysis
Data are expressed as means ± standard errors of the means (SEM). The statistical differences in mean values were assessed by Student's t test. All experiments were performed at least twice and representative data are shown.
Supplementary Material
Acknowledgements
We thank Dr. S. Kajimura for GST-PRDM16 constructs and Dr. P. Tontonoz for the V5-PRDM16 construct. We thank D. Hunerdosse, J. Niliwat, W. Orenella, N. Chien, J. Zheng, X. Liang, E. Esquivel and J. Chong for technical assistance. The work was supported in part by DK095338 to H.S.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.D. performed initial screening, characterization of Zfp516 and UCP1 promoters, Zfp516 interaction with PRDM16, characterization of Zfp516 knockout and transgenic mice. A.S. characterized the role of Zfp516 in preadipocyte and myoblast differentiation and PRDM16 interaction and edited the manuscript. O.G. characterized Zfp516 expression, its regulation and role in MEF and HIB-1B differentiation and edited the manuscript. S.M.P. characterized knockout mice and Zfp516 binding. C.S.S.H. and H.F.R. assisted in animal studies. C.K. generated knockout and transgenic mice. H.Y.K performed initial characterization of Zfp516. R.H.F.W. performed bioinformatic analysis and assisted in initial characterization of Zfp516. H.S.S. designed the project and guided experiments and analysis of data. J.D. and H.S.S. wrote the manuscript.
References
- Ahmadian M, Abbott MJ, Tang T, Hudak CSS, Kim Y, Bruss M, Hellerstein MK, Lee H-Y, Samuel VT, Shulman GI, et al. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab. 2011;13:739–748. doi: 10.1016/j.cmet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman ES, Dhillon H, Zhang C-Y, Cinti S, Bianco AC, Kobilka BK, Lowell BB. βAR Signaling Required for Diet-Induced Thermogenesis and Obesity Resistance. Science. 2002;297:843–845. doi: 10.1126/science.1073160. [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Cassard-Doulcier AM, Gelly C, Fox N, Schrementi J, Raimbault S, Klaus S, Forest C, Bouillaud F, Ricquier D. Tissue-specific and beta-adrenergic regulation of the mitochondrial uncoupling protein gene: control by cis-acting elements in the 5’-flanking region. Mol. Endocrinol. Baltim. Md. 1993;7:497–506. doi: 10.1210/mend.7.4.8388995. [DOI] [PubMed] [Google Scholar]
- Cassard-Doulcier AM, Gelly C, Bouillaud F, Ricquier D. A 211-bp enhancer of the rat uncoupling protein-1 (UCP-1) gene controls specific and regulated expression in brown adipose tissue. Biochem. J. 1998;333(Pt 2):243–246. doi: 10.1042/bj3330243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederberg A, Grønning LM, Ahrén B, Taskén K, Carlsson P, Enerbäck S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- Collins S, Yehuda-Shnaidman E, Wang H. Positive and negative control of Ucp1 gene transcription and the role of β-adrenergic signaling networks. Int. J. Obes. 2010;2005(34 Suppl 1):S28–S33. doi: 10.1038/ijo.2010.180. [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng Y-H, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR. Molecular determinants of brown adipocyte formation and function. Genes Dev. 2008;22:1269–1275. doi: 10.1101/gad.1681308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR. Obesity: Be cool, lose weight. Nature. 2009;458:839–840. doi: 10.1038/458839a. [DOI] [PubMed] [Google Scholar]
- Fulton DL, Sundararajan S, Badis G, Hughes TR, Wasserman WW, Roach JC, Sladek R. TFCat: the curated catalog of mouse and human transcription factors. Genome Biol. 2009;10:R29. doi: 10.1186/gb-2009-10-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Dev. Camb. Engl. 2008;135:1597–1604. doi: 10.1242/dev.019331. [DOI] [PubMed] [Google Scholar]
- Harms MJ, Ishibashi J, Wang W, Lim H-W, Goyama S, Sato T, Kurokawa M, Won K-J, Seale P. Prdm16 Is Required for the Maintenance of Brown Adipocyte Identity and Function in Adult Mice. Cell Metab. 2014;19:593–604. doi: 10.1016/j.cmet.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, Spiegelman BM. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Bajnok L, Longo KA, Petersen RK, Hansen JB, Kristiansen K, MacDougald OA. Effects of Wnt signaling on brown adipocyte differentiation and metabolism mediated by PGC-1alpha. Mol. Cell. Biol. 2005;25:1272–1282. doi: 10.1128/MCB.25.4.1272-1282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak UC, Kopecky J, Teisinger J, Enerbäck S, Boyer B, Kozak LP. An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol. Cell. Biol. 1994;14:59–67. doi: 10.1128/mcb.14.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Leonardsson G, Steel JH, Christian M, Pocock V, Milligan S, Bell J, So P-W, Medina-Gomez G, Vidal-Puig A, White R, et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Fan C-M. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genes. N.Y.N 2000. 2010;48:424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu P-H, Tarr PT, Lindenberg KS, St-Pierre J, Zhang C-Y, Mootha VK, Jäger S, Vianna CR, Reznick RM, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JMAFL, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJJ. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Martens K, Bottelbergs A, Baes M. Ectopic recombination in the central and peripheral nervous system by aP2/FABP4-Cre mice: Implications for metabolism research. FEBS Lett. 2010;584:1054–1058. doi: 10.1016/j.febslet.2010.01.061. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Cannon B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell Metab. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Cannon B. The Browning of White Adipose Tissue: Some Burning Issues. Cell Metab. 2014;20:396–407. doi: 10.1016/j.cmet.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;504:163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Wu J, Ishibashi J, Lim H-W, Giang A-H, Won K-J, Reed RR, Seale P. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricquier D, Bouillaud F. The mitochondrial uncoupling protein: structural and genetic studies. Prog. Nucleic Acid Res. Mol. Biol. 1997;56:83–108. doi: 10.1016/s0079-6603(08)61003-x. [DOI] [PubMed] [Google Scholar]
- Rim JS, Kozak LP. Regulatory motifs for CREB-binding protein and Nfe2l2 transcription factors in the upstream enhancer of the mitochondrial uncoupling protein 1 geneJBiol. Chem. 2002;277:34589–34600. doi: 10.1074/jbc.M108866200. [DOI] [PubMed] [Google Scholar]
- Rosenwald M, Perdikari A, Rülicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat. Cell Biol. 2013 doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc. Natl. Acad. Sci. U.S.A. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scimè A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y-H, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto N-J, Enerbäck S, et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Waldén TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, ‘brite,” and white adipose tissues. Am. J. Physiol. Endocrinol. Metab. 2012;302:E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sul HS. Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell Metab. 2009;9:287–302. doi: 10.1016/j.cmet.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Boström P, Sparks LM, Ye L, Choi JH, Giang A-H, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.