SUMMARY
mTORC1 plays a key role in autophagy as a negative regulator. The currently-known targets of mTORC1 in the autophagy pathway mainly function at early stages of autophagosome formation. Here, we identify that mTORC1 inhibits later stages of autophagy by phosphorylating UVRAG. Under nutrient-enriched conditions, mTORC1 binds and phosphorylates UVRAG. The phosphorylation positively regulates the association of UVRAG with RUBICON, thereby enhancing the antagonizing effect of RUBICON on UVRAG-mediated autophagosome maturation. Upon dephosphorylation, UVRAG is released from RUBICON to interact with the HOPS complex, a component for the late endosome and lysosome fusion machinery, and enhances autophagosome and endosome maturation. Consequently, the dephosphorylation of UVRAG facilitates the lysosomal degradation of epidermal growth factor receptor (EGFR), reduces EGFR signaling, and suppresses cancer cell proliferation and tumor growth. These results demonstrate that mTORC1 engages in late stages of autophagy and endosome maturation, defining a broader range of mTORC1 functions in the membrane-associated processes.
INTRODUCTION
Autophagy is an evolutionarily-conserved process through which eukaryotic cells degrade intracellular organelles and biomolecules in the lysosome. The intracellular degradation is important to maintain the balance between synthesis, degradation and recycling of cellular constituents in response to changes in nutrient levels and other cellular environments. At the cellular level, autophagy is important to maintain the integrity of cellular structures during differentiation and under stress conditions. At the organism level, autophagy is important for the normal physiology and development of the body. Dys-regulation of autophagy has been implicated in aging, innate immunity, and many human diseases including cancers, neurodegenerative diseases, and immune disorders (Levine and Kroemer, 2008; Mizushima et al., 2008).
Autophagy is negatively regulated by mTOR (mechanistic target of rapamycin), the master controller of cell growth (Chang and Neufeld, 2009; Ganley et al., 2009; Hosokawa et al., 2009; Jung et al., 2009). mTOR forms a multiprotein complex called mTORC1 by interacting with raptor, GβL, PRAS40 and DEPTOR to regulate cell growth and autophagy in response to nutritional conditions and growth factor stimulation (Jung et al., 2010; Kim et al., 2002). A key event in the autophagy pathway regulated by mTORC1 is phosphorylation of ULK1 (Unc51-like kinase 1), a serine/threonine kinase that functions upstream in the autophagy pathway (Ganley et al., 2009; Hosokawa et al., 2009; Jung et al., 2009; Kim et al., 2011). Through phosphorylating ULK1, mTORC1 inhibits ULK1 activation by AMPK (AMP-activated protein kinase) (Egan et al., 2011; Kim et al., 2011; Shang and Wang, 2011). mTORC1 also targets proteins other than ULK1, such as Atg13 (Chang and Neufeld, 2009; Ganley et al., 2009; Hosokawa et al., 2009; Jung et al., 2009), Atg14L (Yuan et al., 2013), and Ambra1 (Autophagy/beclin-1 regulator 1) (Nazio et al., 2013). These mTORC1 targets are mainly known to function at early stages of autophagosome formation. Whether mTORC1 regulates autophagy at later stages, such as autophagosome maturation, remains unknown.
UVRAG (UV radiation resistance-associated gene product) is a protein localized in the endoplasmic reticulum (ER) and endosomes (He et al., 2013; Itakura et al., 2008; Liang et al., 2008). UVRAG is known to regulate autophagosome maturation (Liang et al., 2008) as well as early stages of autophagy (He et al., 2013; Liang et al., 2006; Takahashi et al., 2007). UVRAG regulates autophagosome maturation by binding to the HOPS (homotypic fusion and vacuole protein sorting) complex, which consists of the class C Vps complex (Vps11-Vps16-Vps18-Vps33) and two additional proteins (Vps39 and Vps41) (Liang et al., 2008). UVRAG binding to the HOPS complex stimulates lysosomal fusion with autophagosome and endosome (Liang et al., 2008; Sun et al., 2010). The function of UVRAG to regulate the HOPS complex is antagonized by RUBICON (RUN domain Beclin 1- interacting and cysteine-rich containing protein) (Sun et al., 2010). RUBICON also suppresses the UVRAG function in stimulating the kinase activity of Vps34 that contributes to autophagosome maturation (Sun et al., 2011).
In this study, we have identified that mTORC1 binds and phosphorylates UVRAG at Ser498 under nutrient-enriched conditions. We determined that the UVRAG phosphorylation has a positive effect on the interaction between UVRAG and RUBICON, whereas it has a negative effect on the kinase activity of Vps34 and the interaction between UVRAG and the HOPS complex. Preventing the UVRAG phosphorylation increased autophagosome maturation and lysosomal degradation of EGFR, reduced EGFR signaling, and suppressed cancer cell proliferation and tumor growth in vivo. These results demonstrate that mTORC1 has a broader range of functions in autophagy and endosomal pathways.
RESULTS
mTORC1 interacts with UVRAG under nutrient-enriched conditions
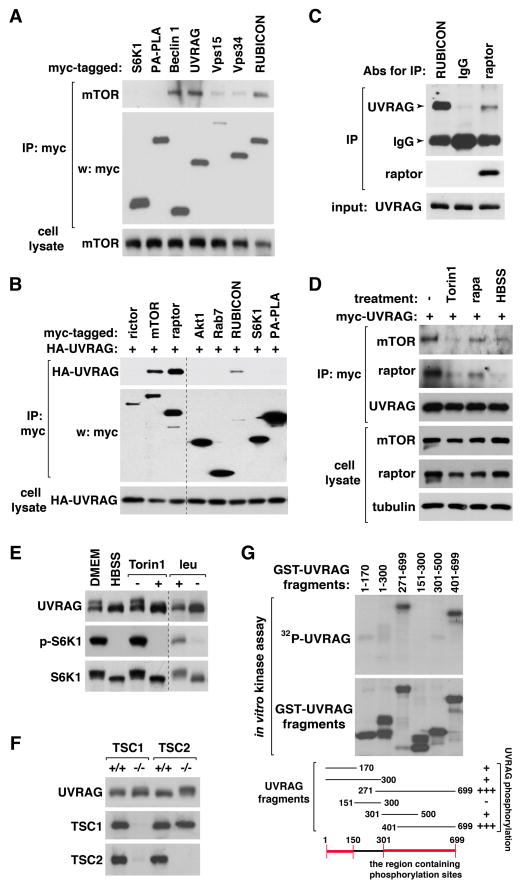
To understand the roles of mTOR in the autophagy pathway, we tested several autophagy proteins for their interaction with mTOR. We found that endogenous mTOR is co-immunoprecipitated with Beclin 1, UVRAG, and RUBICON (Figure 1A). mTOR also showed a relatively weak binding affinity toward Vps34 and Vps15. We were not able to detect any interaction of mTOR with its substrate S6K1 or a membrane-associated control protein PA-PLA, suggesting that the UVRAG-containing Vps34 complex might bind to mTOR relatively strongly compared to S6K1 binding to mTORC1 in the experimental condition. mTOR forms two distinct protein complexes, mTORC1 and mTORC2, for which raptor and rictor are the representative components, respectively (Kim et al., 2002; Sarbassov et al., 2004). UVRAG was co-immunoprecipitated with raptor (Figure 1B and Figure S1A), but barely with rictor. Consistently, rictor knockdown did not affect the interaction between mTOR and UVRAG (Figure S1B). We confirmed the interaction between raptor and UVRAG at endogenous levels (Figure 1C). We were not able to detect any interaction between UVRAG and RagB (Figure S1A), indicating that mTORC1 might bind to UVRAG independently of Rag GTPases that mediate amino acid-mTORC1 signaling on lysosomes (Kim et al., 2008a; Sancak et al., 2008).
Figure 1. mTORC1 binds to UVRAG and induces UVRAG phosphorylation.
(A) mTOR binds to the UVRAG-containing Vps34 complex. Myc-tagged proteins were transiently expressed in HEK293T cells. Endogenous mTOR recovered with myc immunoprecipitate (IP) was analyzed by western blotting (WB). (B) mTORC1 interacts with UVRAG. Myc-tagged proteins were transiently expressed with HA-UVRAG in HEK293T cells. UVRAG recovered with myc IP was analyzed by WB. An irrelevant lane in the middle was eliminated. (C) Endogenous UVRAG recovered with RUBICON, raptor, and control IgG IP was analyzed by WB. (D) The UVRAG-mTORC1 interaction is negatively regulated by Torin1 or HBSS, but not by rapamycin. HEK293T cells were transiently transduced to express myc-UVRAG, and incubated in HBSS for 2 h or treated with Torin1 or rapamycin for 1 h. Endogenous mTOR or raptor isolated with myc-UVRAG was analyzed by WB. (E) mTOR inhibition induces a mobility shift of UVRAG band on SDS-PAGE. HEK293T cells were incubated in DMEM or HBSS for 2 h, or treated with Torin1 for 1 h. Cells were also incubated in leucine-deprived medium (−) then supplemented with leucine (+). (F) Deficiency of either TSC1 or TSC2 in MEFs induces a mobility shift of UVRAG. (G) mTOR phosphorylates UVRAG in vitro. GST-tagged UVRAG fragments were prepared from bacteria and incubated with the active mTOR fragment in the presence of 32P-ATP. Incorporation of 32P into UVRAG fragments was analyzed by autoradiography. See also Figure S1.
To clarify whether the interaction is regulated by conditions that induce autophagy, we treated HEK293T cells with mTOR inhibitors, such as Torin1 or rapamycin, or incubated in nutrient-deprived Hank’s balanced saline solution (HBSS). Torin1 or HBSS disrupted the interaction between mTORC1 and UVRAG (Figure 1D). Rapamycin moderately reduced the mTOR-UVRAG interaction while drastically reducing the raptor-UVRAG interaction. The drastic effect of rapamycin on the raptor-UVRAG interaction might be because rapamycin disturbs the mTOR-raptor interaction (Kim et al., 2002). mTOR required the full length of UVRAG for their stable association, whereas all the tested fragments except the C-terminal 501–699 fragment showed a weak binding affinity toward mTOR (Figure S1C). This indicates that the whole structural conformation of UVRAG might be important for the stable interaction. Alternatively, multiple sites of UVRAG scattered apart might be involved in the interaction with mTOR.
mTORC1 induces UVRAG phosphorylations
We observed that HBSS or Torin1, but not rapamycin, induced a downshift of UVRAG band on a low percent polyacrylamide gel (Figure 1E and Figure S1D). The downshift was also induced by deprivation of leucine, a potent stimulator of mTORC1 activity, in medium. Depletion of TSC1 (tuberous sclerosis complex 1) or TSC2, which negatively regulates mTORC1, in mouse embryonic fibroblasts (MEFs) or overexpression of Rheb, a positive regulator of mTORC1, in HEK293T cells caused an upshift of UVRAG band (Figure 1F and Figure S1E), confirming that mTORC1 is responsible for the mobility change. To determine whether the mobility shift is due to phosphorylation, we prepared several truncated fragments of UVRAG using a bacterial expression system (Figure S1F) and incubated the fragments with an active form of mTOR containing residues 1362–2549 in the presence of 32P-ATP. The active form of mTOR strongly induced phosphorylation of a fragment containing residues 271–699 and a fragment containing residues 401–699 (Figure 1G). mTOR also weakly phosphorylated fragments containing residues 1–170, 1–300, and 301–500, respectively. This result suggests that mTOR might phosphorylate multiple residues on UVRAG that are spread over the whole area of the protein, especially near the C-terminus.
Identification of mTORC1-dependent phosphorylation sites of UVRAG
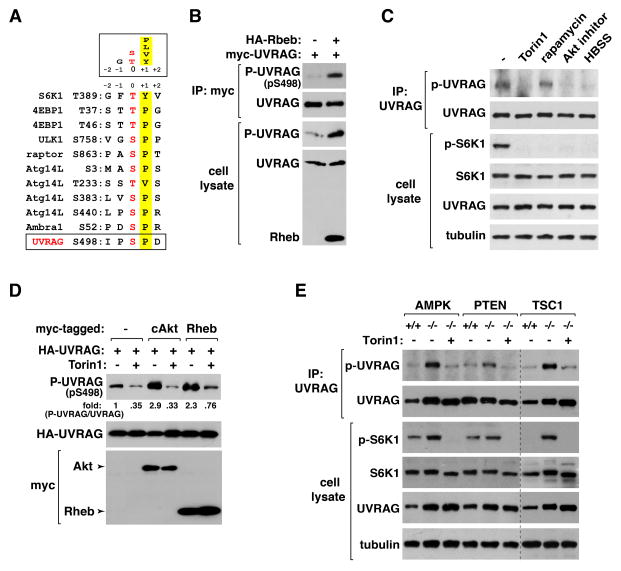
To identify mTORC1-dependent phosphorylation sites of UVRAG, we expressed myc-UVRAG with HA-Rheb in HEK293T cells and isolated UVRAG by immunoprecipitation. As a control, myc-UVRAG was isolated from HEK293T cells that were without Rheb overexpression and treated with Torin1 for 1 h. The immunoprecipitated UVRAG was resolved by SDS-PAGE, and the UVRAG band was isolated from the gel. After trypsinization, phosphorylated peptides were enriched and analyzed by mass spectrometry. Total seven phosphorylation sites (S493, S498, S508, S522, S549, S550 and S582) were identified with UVRAG from Rheb-expressing cells but not from Torin1-treated cells. The surrounding sequences of S498, S522 and S550 showed a feature of the mTORC1 target consensus motifs (Kang et al., 2013) (Figure 2A and Figure S2A). We were able to successfully generate polyclonal antibodies that recognize the phosphorylated state of UVRAG S498 (Figure S2B and S2C).
Figure 2. UVRAG Ser498 is an mTORC1-dependent phosphorylation site.
(A) Alignment of the sequence surrounding UVRAG S498 with those of the known mTORC1 target sites. (B) Rheb induces UVRAG S498 phosphorylation. HA-Rheb was expressed with myc-UVRAG in HEK293T cells. UVRAG S498 phosphorylation was analyzed by WB. (C) UVRAG phosphorylation in MEFs is inhibited by Torin1, Akt inhibitor VIII or HBSS, but not by rapamycin. Immunoprecipitated UVRAG was analyzed for the phosphorylation by WB. (D) The S498 phosphorylation is enhanced by Akt and Rheb. HA-UVRAG was transiently expressed in HEK293T cells alone (−) or together with myc-tagged constitutively active Akt mutant (cAkt) or Rheb. Two days post-transfection, cells were treated with vehicle (−) or Torin1 (+) for 1 h. (E) Deficiency of AMPK, PTEN, or TSC1 in MEFs enhances UVRAG phosphorylation. MEFs were treated with vehicle (−) or Torin1 (+) for 1 h. The UVRAG phosphorylation was analyzed by immunoprecipitation followed by WB. See also Figure S2.
mTORC1 induces UVRAG Ser498 phosphorylation
Using the phospho-specific antibody we developed, we found that UVRAG S498 phosphorylation is highly increased by Rheb overexpression in HEK293T cells (Figure 2B). Treatment of MEFs with Torin1 almost completely suppressed the phosphorylation of UVRAG at S497, the mouse equivalent of human UVRAG S498 (Figure 2C). The phosphorylation was also completely suppressed by an Akt inhibitor or HBSS, the conditions that inhibit mTORC1 (Figure 2C). By contrast, rapamycin barely suppressed the phosphorylation even at high concentrations (Figure 2C and Figure S2D). This indicates that Ser498 might be a rapamycin-insensitive site, like a few other mTORC1 target sites (Kang et al., 2013). Torin1 suppressed the induction of S498 phosphorylation by Rheb or a constitutively active mutant of Akt in HEK293T cells (Figure 2D), confirming that the Ser498 phosphorylation depends on mTORC1. Consistently, we found that deficiency of AMPK, PTEN (phosphatase and tensin homolog) or TSC1, which are negative regulators of mTORC1, in MEFs largely increased the UVRAG phosphorylation, and the increase was suppressed by Torin1 (Figure 2E and Figure S2E). Deficiency of ULK1, which functions both upstream and downstream of mTORC1 (Dunlop et al., 2011; Jung, 2011), in MEFs only marginally increased the UVRAG phosphorylation (Figure S2F). The increase was still suppressed by HBSS, indicating that mTORC1 induces the UVRAG phosphorylation independently of ULK1.
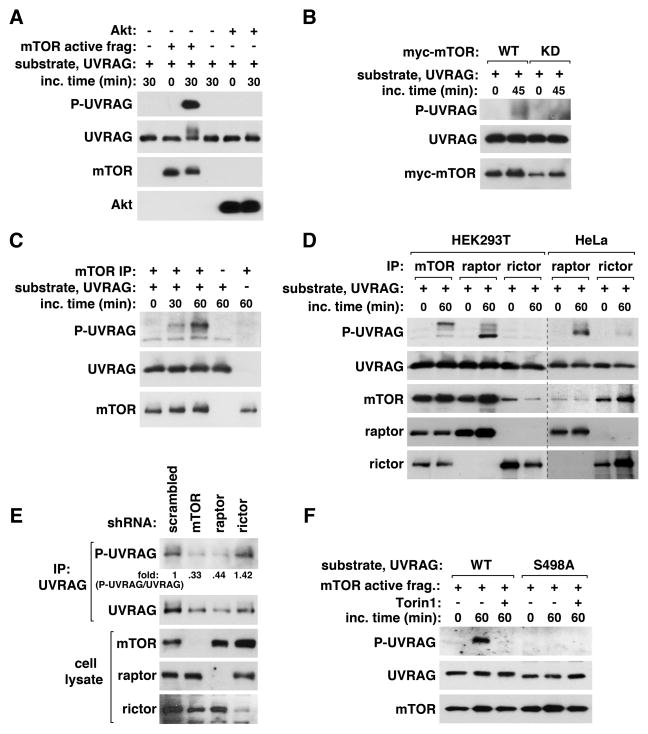
mTORC1 phosphorylates UVRAG Ser498
To determine whether mTOR phosphorylates UVRAG S498, we conducted in vitro kinase assays. The active mTOR 1362–2549 fragment induced a large increase of S498 phosphorylation of UVRAG purified from bacteria (Figure 3A). We also found that mTOR wild type (WT), but not its kinase-dead mutant (KD), showed the kinase activity toward the UVRAG phosphorylation in vitro (Figure 3B). Endogenous mTOR IP also largely increased the phosphorylation of UVRAG 271–699 fragment in vitro (Figure 3C). mTOR or raptor IP induced multiple forms of UVRAG harboring S498 phosphorylation, which showed different mobilities on SDS-PAGE (Figure 3D), implying there might be other mTOR-mediated phosphorylations. mTOR IP, relative to raptor IP, induced more of slowly-migrating UVRAG forms, implying that mTOR IP might contain a molecular factor important for UVRAG phosphorylations at sites other than S498. When rictor IP was isolated from HeLa cells that express rictor highly, we were able to detect a marginal level of S498 phosphorylation, but which was much less than that induced by raptor IP (Figure 3D). Knockdown of mTOR or raptor, but not rictor, in HEK293T cells drastically reduced S498 phosphorylation (Figure 3E). The in vitro phosphorylation of UVRAG by mTOR was not detected when S498 was replaced with alanine (S498A) or when Torin1 was added during the reaction (Figure 3F and Figure S3). Combined, these results demonstrate that mTORC1 phosphorylates UVRAG at S498.
Figure 3. mTORC1 phosphorylates UVRAG Ser498.
(A) mTOR phosphorylates UVRAG S498 in vitro. The active form of mTOR was incubated with puririfed UVRAG in the presence of ATP. Akt1 was used as a negative control. UVRAG S498 phosphorylation was analyzed by WB. (B) Myc-mTOR WT or its kinase dead mutant (KD), transiently expressed in HEK293T cells, was isolated by immunoprecipitation using anti-myc antibody and incubated with UVRAG and ATP. (C) Endogenous mTOR IP was isolated from HEK293T cells, and its kinase activity was analyzed using UVRAG 271–699 fragment as substrate. (D) mTOR, raptor, or rictor IPs were obtained from HEK293T or HeLa cells, and the kinase reaction was analyzed as in (C). (E) The phosphorylation state of S498 was analyzed for endogenous UVRAG isolated from shRNA-transduced HEK293T cells. (F) Torin1 inhibits UVRAG S498 phosphorylation in vitro. The active fragment of mTOR was incubated with UVRAG WT or S498A purified from bacteria. The kinase reaction was performed in the presence or absence of Torin1. See also Figure S3.
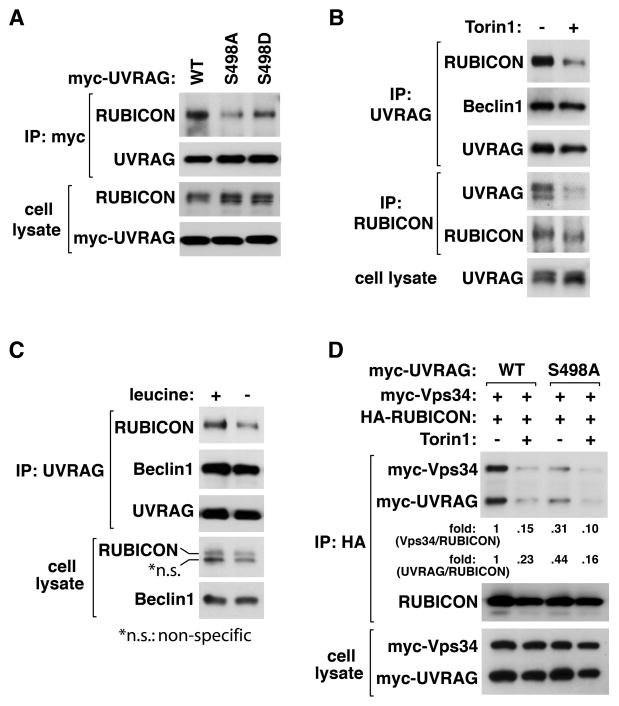
UVRAG Ser498 phosphorylation positively regulates the UVRAG-RUBICON interaction
To understand the functional consequence of the UVRAG phosphorylation, we investigated whether the phosphorylation status could regulate the mTORC1-UVRAG interaction. The S498A mutation only marginally reduced the mTOR-UVRAG interaction, and the interaction was still disrupted by Torin1 (Figure S4A). This indicates that S498 phosphorylation might not be critical for the interaction but rather an event following their association under nutrient-rich conditions. Next, we examined whether S498A mutation has an effect on the UVRAG-RUBICON interaction. We expressed UVRAG WT, S498A, or S498D (a mutant with replacement of S498 with aspartate) in HEK293T cells where endogenous UVRAG was silenced by shRNA targeting UVRAG mRNA 3′-UTR (Figure S4B). S498A mutant interacted with RUBICON with a weaker affinity compared to WT (Figure 4A and Figure S4C). S498D mutant interacted with RUBICON more strongly than S498A mutant. However, S498D mutant could not fully recover the affinity of WT, indicating that aspartate is not a perfect mimic of phosphoserine at the residue. Combined, the results suggest that the dephosphorylation state of S498 might disturb the UVRAG-RUBICON interaction.
Figure 4. UVRAG Ser498 phosphorylation positively regulates the UVRAG-RUBICON interaction.
(A) S498 phosphorylation positively regulates the UVRAG-RUBICON interaction. Myc-UVRAG WT or mutant was transiently expressed in UVRAG-silenced HEK293T cells. Endogenous RUBICON recovered with myc-UVRAG IP was analyzed by WB. (B) Torin1 negatively regulates the UVRAG-RUBICON interaction. HEK293T cells were treated with Torin1 for 4 h. The interaction between endogenous UVRAG and RUBICON was analyzed by immunoprecipitation and WB. (C) Leucine deprivation reduces the UVRAG-RUBICON interaction. HEK293T cells were incubated in medium with (+) or without leucine (−) for 2 h. Endogenous RUBICON recovered with UVRAG IP was analyzed by WB. (D) S498A mutation has a negative effect on the interaction between RUBICON and the UVRAG-containing Vps34 complex. The indicated proteins were expressed in UVRAG-silenced HEK293T cells. Two days post-transfection, the cells were treated with Torin1 as in (B). See also Figure S4.
If S498 phosphorylation positively regulates the UVRAG-RUBICON interaction, we predicted that Torin1 might suppress the interaction. As predicted, Torin1 suppressed the UVRAG-RUBICON interaction (Figure 4B). Similarly, leucine deprivation in medium suppressed the UVRAG-RUBICON interaction (Figure 4C). Neither Torin1 nor leucine altered the interaction of UVRAG or Atg14L with Beclin 1 (Figure 4B and C and Figure S4D). The S498A mutation also drastically suppressed the RUBICON-Vps34 interaction (Figure 4D). These results suggest that UVRAG S498 phosphorylation might facilitate binding of RUBICON to the UVRAG-containing Vps34 complex. The suppressive effects of Torin1 on the interactions of RUBICON with UVRAG and Vps34 were still observed with S498A mutant (Figure 4D). This suggests that there might be other mTOR-dependent phosphorylations that regulate the interaction between RUBICON and the Vps34 complex.
UVRAG Ser498 phosphorylation negatively regulates Vps34
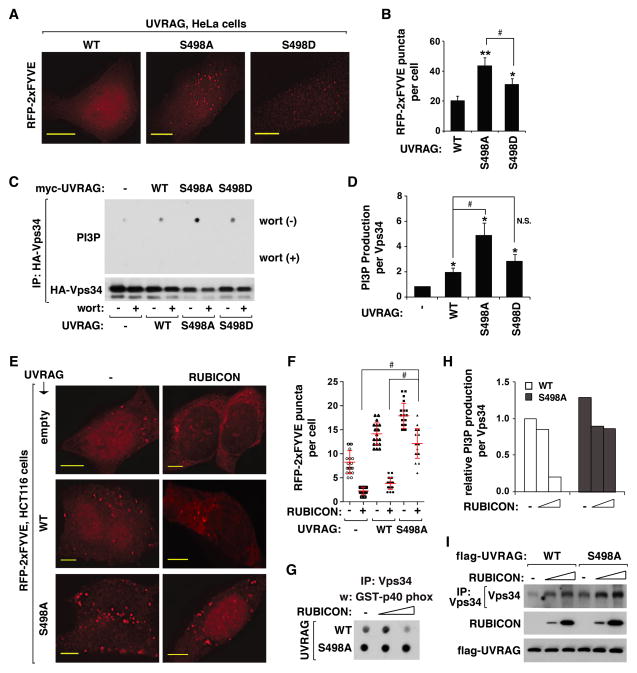
Knowing that S498 phosphorylation positively regulates the RUBICON-Vps34 interaction, we wondered whether the phosphorylation might regulate the kinase activity of Vps34. We expressed RFP-tagged 2xFYVE in WT or mutant UVRAG-reconstituted cells, and monitored the formation of RFP-FYVE puncta, which reflects the production of phosphatidylinositide-3-phosphate (PI3P) (Sun et al., 2011; Zhong et al., 2009). We observed a higher induction of RFP-FYVE puncta in S498A cells compared to WT cells (Figure 5A and B and Figure S5A–S5C). The FYVE puncta formation was also increased in S498D cells but to a significantly less extent compared to S498A cells. Although we could not observe any drastic change in the cellular distribution pattern of FYVE puncta between WT and mutant cells, FYVE positive areas co-stained with the endosomal marker proteins, such as Rab5 and Rab7, were highly increased in S498A cells compared to WT cells (Figure S5D). This might be due to the higher accumulation of FYVE puncta in the mutant cells compared to WT cells.
Figure 5. UVRAG Ser498 phosphorylation is important for the inhibitory effect of RUBICON on Vps34.
(A) Prevention of S498 phosphorylation increased the accumulation of PI3P in HeLa cells. RFP-tagged 2xFYVE, transiently expressed in WT or mutant UVRAG-reconstituted HeLa cells, was monitored by fluorescence microscope. Scale bar, 10 μm. (B) Quantitative analysis of FYVE puncta formation. Results are represented as means ± standard deviation (SD) (*, p<0.05 vs WT; **, p<0.01 vs WT; #, p<0.01; n≥17). (C) Prevention of S498 phosphorylation increased the kinase activity of Vps34. HA-Vps34 was expressed alone or together with myc-UVRAG in UVRAG-depleted HEK293T cells. HA-Vps34 IP was obtained and incubated with phosphatidylinositol (PI) and ATP in the presence or absence of wortmannin (200 nM). The amount of PI3P was analyzed as described in Experimental Procedures. (D) Quantitative analysis of Vps34 kinase activity. Results are represented as means ± SD (*, p<0.01 vs empty vector; #, p<0.01; NS, Non-Significant; n=3). (E) RUBICON depends on S498 phosphorylation to suppress UVRAG-induced production of PI3P. RFP-2xFYVE expressed alone (−) or together with RUBICON in empty vector- or UVRAG-transduced HCT116 cells was analyzed as described in (A). Scale bar, 5 μm. (F) Quantitative analysis of PI3P puncta formation. The error bars represent means ± SD (#, p<0.01; n=20). (G and H) RUBICON depends on S498 phosphorylation to suppress UVRAG-mediated stimulation of Vps34 kinase activity. UVRAG-reconstituted HEK293T cells were transiently transfected with empty vector or RUBICON construct. The in vitro kinase assay was conducted as in (C). The graph is representative of two independent experiments. (I) Vps34 recovered with Vps34 IP was analyzed by WB. See also Figure S5.
To clarify whether the increase of PI3P in cells is due to an increase in the activity of Vps34, we analyzed the kinase activity of Vps34 in vitro. We isolated Vps34 from WT- and mutant-reconstituted HEK293T cells. The catalytic activity of Vps34 was analyzed by measuring the amount of a p40 phox domain-containing polypeptide that binds to PI3P following the procedure established previously (Farkas et al., 2011). Vps34 isolated from WT cells showed a higher production of PI3P compared to that from empty vector-transduced cells (Figure 5C and D and Figure S5E). This result is consistent with the positive effect of UVRAG on Vps34 as shown by other studies (Liang et al., 2006; Sun et al., 2011). PI3P production was dramatically increased with Vps34 isolated from S498A cells compared with that from WT cells (Figure 5C and D and Figure S5E). By contrast, Vps34 isolated from S498D cells showed an activity similar to that from WT cells. This suggests that S498 phosphorylation might suppress the UVRAG-mediated stimulation of the Vps34 kinase activity.
UVRAG Ser498 phosphorylation is important for the suppressive effect of RUBICON on Vps34
Knowing that S498 phosphorylation has a negative effect on the Vps34 activity, we hypothesized that the phosphorylational effect might be through enhancing RUBICON binding to Vps34. Expression of RUBICON in WT-reconstituted HCT116 cells significantly reduced the number of FYVE puncta (Figure 5E and F and Figure S5F), a result consistent with the negative role of RUBICON on Vps34 (Sun et al., 2011). Such a suppressive effect of RUBICON was significantly attenuated in S498A cells without any drastic change in the puncta sizes (Figure 5E and F and Figure S5F and S5G). This result suggests that RUBICON might depend on S498 phosphorylation to suppress the Vps34 activity. We confirmed the result in vitro by observing that the kinase activity of Vps34 isolated from WT cells was drastically reduced by RUBICON, while such a drastic reduction was not observed with Vps34 isolated from S498A cells (Figure 5G–5I). Consistently, RUBICON knockdown largely increased the kinase activity in WT cells but the knockdown effect was reduced in S498A cells (Figure S5H–S5M). These results suggest that S498 phosphorylation might play an important role in RUBICON-mediated suppression of the Vps34 kinase activity.
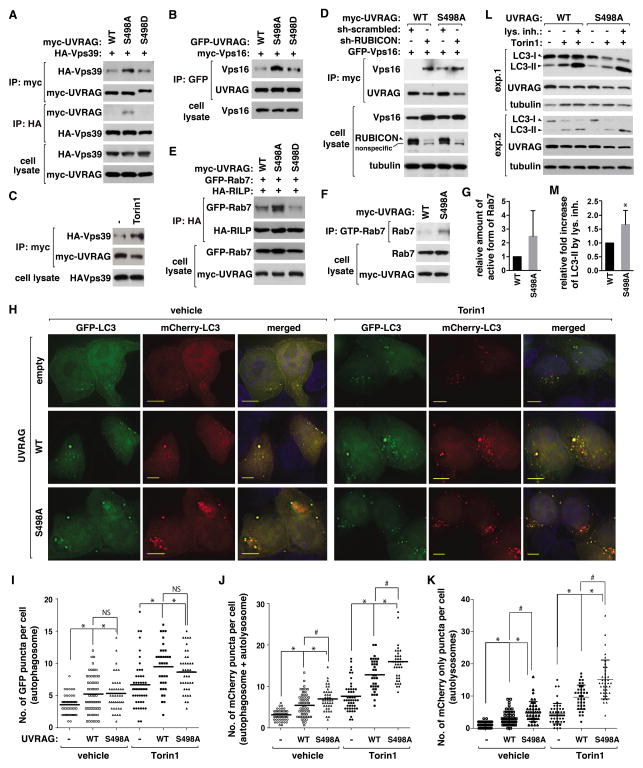
UVRAG Ser498 phosphorylation negatively regulates the UVRAG-HOPS interaction and Rab7
RUBICON is known to prevent UVRAG from binding to the HOPS complex (Sun et al., 2010). Since S498A mutation suppressed the UVRAG-RUBICON interaction, we predicted that the mutation might increase the UVRAG-HOPS interaction. As predicted, higher levels of Vps39 and Vps16 of the HOPS complex were co-immunoprecipitated with S498A mutant compared to WT (Figure 6A and 6B). Relative to S498A mutant, S498D mutant interacted with Vps39 and Vps16 with a weaker affinity. The positive effect of S498A mutation on the UVRAG-HOPS interaction was mimicked by Torin1 (Figure 6C). Consistently, the colocalization of UVRAG with HOPS was increased in S498A cells compared to WT cells (Figure S6A). The colocalization was further increased by Torin1 in both WT and S498A cells (Figure S6B), suggesting that other mTOR-dependent phosphorylations might also be involved in regulating the colocalization. RUBICON knockdown largely increased the binding of Vps16 with WT but only marginally with S498A mutant (Figure 6D and Figure S6C). Consistently, RUBICON overexpression drastically suppressed the interaction of Vps39 with WT but not with S498A mutant (Figure S6D). Combined, these results demonstrate that S498 phosphorylation is important for RUBICON-mediated suppression of the UVRAG-HOPS interaction.
Figure 6. Prevention of UVRAG Ser498 phosphorylation facilitates autophagosome maturation.
(A) S498 phosphorylation negatively regulates the UVRAG-Vps39 interaction. HA-Vps39 was transiently expressed in UVRAG-reconstituted HEK293T cells, and their interaction was analyzed by immunoprecipitation and WB. (B) S498 phosphorylation negatively regulates the UVRAG-Vps16 interaction. Myc-Vps16 recovered with GFP-UVRAG IP from HEK293T cells was analyzed by WB. (C) Torin1 enhances the UVRAG-Vps39 interaction. HEK293T cells transiently expressing HA-Vps39 and myc-UVRAG were treated with Torin1 or vehicle (−). HA-Vps39 recovered with myc-UVRAG IP was analyzed by WB. (D) RUBICON knockdown enhances the UVRAG-Vps16 interaction in WT cells but not in S498A cells. GFP-Vps16 recovered with myc-UVRAG IP from shRNA-transduced HEK293T cells was analyzed by WB. (E) S498 phosphorylation negatively regulates the Rab7-RILP interaction. Rab7 recovered with HA-RILP from WT or mutant UVRAG-reconstituted HEK293T cells was analyzed by WB. (F) S498 phosphorylation negatively regulates Rab7. The GTP-bound Rab7 was detected as described in Experimental Procedures. (G) Quantitative analysis of (F). Data are means ± SD (n=2). (H) S498 phosphorylation negatively regulates autophagosome maturation. mCherry-GFP-LC3 was expressed in empty vector- or UVRAG-transduced HCT116 cells. Cells were treated with Torin1 or vehicle. LC3 was monitored by fluorescence microscope. Scale bar, 5 μm. (I and J) Quantitative analysis of GFP puncta (I) and mCherry puncta (J) (*, p<0.01 vs empty vector; #, p<0.01; NS; n≥35). Mean value is shown as a horizontal bar. (K) Quantitative analysis of mCherry only puncta (*, p<0.01 vs empty vector; #, p<0.01; n≥35). Mean and SD are shown as horizontal bars. (L) S498A mutation enhances autophagy flux. HEK293T cells reconstituted with UVRAG constructs were treated with Torin1 in the presence or absence of Bafilomycin A1 (exp1) or E-64/Pepstatin A (exp2). (M) Quantitative analysis of the fold increase of LC3-II level induced by the lysosomal inhibitors in Torin1-treated cells. Data are means ± SD (*, p<0.05; n=5). See also Figure S6.
UVRAG binding is known to enhance the HOPS activity as a guanine nucleotide exchange factor (GEF) toward Rab7, a small GTPase whose activation is important for autophagosome and endosome maturation (Liang et al., 2008; Sun et al., 2010). We assessed the active state of Rab7 by monitoring the recruitment of RILP (Rab7-interacting lysosomal protein) to Rab7. We found that the RILP-Rab7 interaction was largely increased in S498A cells compared to WT or S498D cells (Figure 6E). Consistently, a higher level of GTP-bound Rab7 was detected in S498A cells (Figure 6F and G and Figure S6E). We also asked whether S498 phosphorylation might regulate Rab7 via the Mon1-Ccz1 complex that was recently shown to act as a GEF toward Rab7 in yeast and plant cells (Cui et al., 2014; Nordmann et al., 2010). We found that UVRAG interacts with Ccz1 and Mon1B (Figure S6F and Figure S6G), but the interaction, unlike the UVRAG-HOPS interaction, was not altered by S498 mutations. Although the interaction implies a function link between UVRAG and the Ccz1 complex, our result suggests that S498 phosphorylation likely has specific effects on HOPS rather than on the Ccz1 complex.
Prevention of UVRAG Ser498 phosphorylation facilitates autophagosome maturation
Knowing that S498 phosphorylation negatively regulates Vps34 and HOPS, we predicted that the phosphorylation might regulate autophagosome maturation. To address this possibility, we used the dual-tagged LC3 construct (mCherry-GFP-LC3) (Zhong et al., 2009). The formation of mCherry-LC3 puncta, which may reflect LC3 destined in autophagosomes or lysosomes, was significantly increased in S498A cells compared to WT cells (Figure 6H–J and Figure S6H–S6J). Torin1 induced the formation of green and yellow LC3 puncta, which may reflect autophagosomes, to similar extents between WT and S498A cells, whereas Torin1 induced the formation of mCherry-LC3-only puncta to a greater extent in S498A cells compared to WT cells (Figure 6H–K and Figure S6H–S6J). The higher induction of mCherry-LC3-only puncta in S498A cells compared to WT cells in response to Torin1 might be due to a residual amount of WT UVRAG that was not dephosphorylated at S498. Supporting this, Torin1 could not completely suppress S498 phosphorylation of recombinant WT UVRAG (Figure 2D), and Torin1 induced dephosphorylation of recombinant UVRAG S498 at a reduced rate relative to endogenous UVRAG (Figure S6K and S6L). Relevant to this, Torin1 more drastically impaired the interactions mediated by UVRAG for binding to RUBICON and mTOR when S498 was mutated to alanine (Figure 4D and Figure S4A). Thus, S498A mutation might somehow facilitate Torin1-induced changes in the protein interactions involved in UVRAG-mediated autophagosome maturation. It is possible that those additional phosphorylations of UVRAG we identified (Figure S2A) might be involved in the Torin1 effects.
To further confirm that S498 phosphorylation negatively regulates UVRAG-mediated autophagosome maturation, we conducted an autophagy flux assay. Lysosomal inhibitors increased the LC3-II level to a greater extent in S498A cells compared to WT cells either in nutrient-rich or Torin1-treated conditions (Figure 6L and M and Figure S6M and S6N). This result, in accordance with those from the analysis of LC3 puncta, suggests that autophagosomal maturation might be facilitated in mutant cells. The higher increase of LC3-II levels in mutant cells compared to WT cells in response to Torin1 could be interpreted as above where we interpreted the results of the higher accumulation of LC3 in autolysosomes in S498A mutant cells compared to WT cells. Consistent with no significant difference in autophagosome formation between WT and S498A cells (Figure 6H and I), S498A mutation did not have any significant effect on the formation of DFCP1 and WIPI2 puncta (Figure S6O–S6Q). Thus, the effects of S498A mutation are unlikely due to up-regulation of early steps of autophagy.
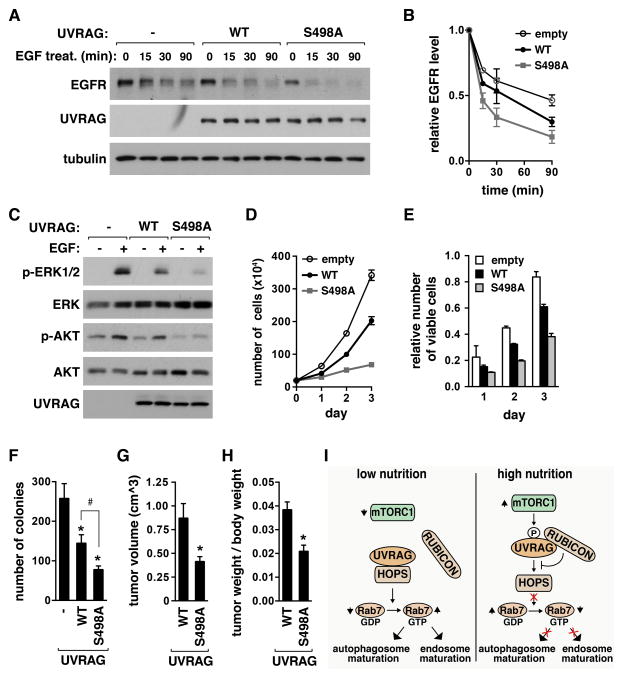
Prevention of UVRAG Ser498 phosphorylation facilitates UVRAG-mediated endosome-lysosomal degradation of EGFR and reduces EGFR signaling
UVRAG positively regulates the endosome-lysosomal fusion through enhancing the HOPS activity (Liang et al., 2008; Sun et al., 2010). A consequence of the enhanced endosomal maturation is a facilitated degradation of EGFR in lysosomes (Liang et al., 2008). Since S498A mutation enhanced the UVRAG-HOPS interaction and the active state of Rab7, we predicted that S498A mutation might facilitate the endosome maturation and the lysosomal degradation of EGFR. Reconstitution of WT UVRAG in HEK293T cells enhanced EGFR degradation in response to EGF (Figure 7A and 7B). The degradation of EGFR was significantly increased in S498A cells compared to WT cells (Figure 7A and 7B, and Figure S7A). The level of EGFR was lower in S498A cells compared to WT cells before EGF treatment, suggesting that the phosphorylation might also affect the basal endosome-lysosomal degradation of EGFR. The facilitated degradation of EGFR was accompanied by reduction in EGF-stimulated phosphorylations of ERK1/2 and AKT (Figure 7C and Figure S7B). Similarly as S498A mutation, Torin1 increased EGF-stimulated degradation of EGFR and reduced the level of EGFR even before cells were treated with EGF (Figure S7C–S7F), suggesting that mTORC1 inhibition might enhance the basal endosome maturation. The reduction of EGFR by Torin1 was marginal in S498A cells compared to WT cells (Figure S7G and S7H), supporting our interpretation that S498 dephosphorylation might contribute to Torin1-stimulated EGFR degradation. The further reduction of EGFR by Torin1 in mutant cells implies other mTORC1-dependent mechanisms involved in EGFR downregulation. Despite other potential mechanisms, the decreased basal level of EGFR in S498A cells might be due to facilitated endosome-lysosomal fusion.
Figure 7. Prevention of UVRAG Ser498 phosphorylation enhances EGFR degradation, reduces EGFR signaling, and suppresses cancer cell proliferation and tumor growth.
(A) S498A mutation enhances EGFR degradation. HEK293T cells stably transduced by empty vector (−) or UVRAG construct were starved of serum overnight, and treated EGF. EGFR level in cell lysate was analyzed by WB. (B) EGFR level was presented relative to that at time zero. Data are means ± SD (n=4). (C) S498A mutation suppresses EGFR signaling. HEK293T cells, transduced as in (A), were serum-starved overnight and treated with EGF. pERK1/2 (Thr202/Tyr204) or pAKT (Thr308) were analyzed by WB. (D) S498A mutation suppresses cancer cell proliferation. HCT116 cells stably transduced by empty vector (−) or UVRAG construct were cultured in DMEM. The number of viable cells was counted by hemacytometer. Data are means ± SD (n=3). (E) Viable cells were assayed using MTT. Data are means ± SD (n=3). (F) S498A mutation significantly inhibits anchorage-independent growth of cancer cells. HCT116 cells prepared as in (D) were used. Data are shown as mean ± SD (*, p<0.01 vs empty vector; #, p<0.01; n=3). (G) S498A mutation significantly inhibits tumor growth in mice. Values represent means ± SD (*, p=0.012; n=5). (H) Tumor weight relative to the whole body weight was analyzed. Values represent mean ± SD (*, p<0.01; n=5). (I) Regulatory function of UVRAG S498 phosphorylation in autophagosome and endosome maturation. See also Figure S7.
Prevention of UVRAG Ser498 phosphorylation suppresses cancer cell proliferation and tumor growth
Knowing that S498A mutation facilitates EGFR degradation and reduces EGFR signaling, we asked whether the mutation would suppress cancer cell proliferation. WT UVRAG expression in HCT116 colon cancer cells reduced cell proliferation (Figure 7D and E). This result is consistent with the known tumor suppressor function of UVRAG (Liang et al., 2006). The suppressive effect of UVRAG became greater when S498A mutant was expressed instead of WT. The proliferation rate of S498A cells was about 5 and 2.9 times lower than empty-vector transduced cells and WT cells, respectively (Figure 7D). Expression of S498A mutant in HCT116 cells dramatically suppressed colony formation (about 70% and 54% reduction in the number of colonies compared to empty vector-transduced cells and WT cells, respectively) in a clonogenic assay (Figure 7F and Figure S7I). Furthermore, the colonies of S498A cells were much smaller in size compared with those of empty-vector transduced cells and WT cells (Figure S7I).
To address whether the suppressive effect of S498A mutation on cell proliferation could be recapitulated in vivo, we conducted a xenograft experiment. We subcutaneously injected WT or S498A HCT116 cells into the flank of immunity-deficient Balb/c mice, and analyzed the sizes of tumors. The sizes of tumors formed by S498A cells were significantly smaller than those formed by WT cells (Figure 7G and Figure S7J). The average volume of S498A cell-derived tumor was about 47.3% of that of WT cell-derived tumor (Figure 7G). Tumor weight, relative to the whole body weight, was significantly reduced by S498A mutation (Figure 7H and Figure S7K and S7L). This result suggests that Ser498 phosphorylation might be important for the tumor suppressive function of UVRAG.
DISCUSSION
In this study, we have determined that mTORC1 phosphorylates UVRAG at Ser498 and thereby regulates late stages of autophagy and endosomal maturation. The mTORC1-mediated UVRAG phosphorylation may depend on the ability of mTORC1 to interact with UVRAG. With this regard, it is noteworthy that mTOR is localized to late endosomes and lysosomes in nutrient-enriched conditions (Sancak et al., 2008). The nutrient-regulated localization of mTOR fits our model that mTOR inhibits autophagosomal and endosomal maturation by binding and phosphorylating UVRAG in association with the HOPS complex, which also localizes to late endosomes and lysosomes (Itakura et al., 2008; Pols et al., 2013). According to our results, the phosphorylation positively regulates the UVRAG-RUBICON interaction and suppresses the UVRAG functions in autophagosome and endosome maturation (Figure 7I). Although our results support the role of S498 phosphorylation in regulating the lysosomal fusion machinery, we do not rule out a possibility that the effect of S498A mutation on autophagy could be secondary to a change in the lysosome function due to disturbance of the endocytic pathways.
Hyperactivation of mTORC1 is frequently found in many cancers (Laplante and Sabatini, 2012). The contribution of mTORC1 to cancer is mainly attributed to increases in protein synthesis. Our study suggests that mTORC1 might contribute to cancer by phosphorylating UVRAG and thereby protecting EGFR from the lysosomal degradation. Many genetic mutations that suppress the lysosomal degradation of EGFR have been implicated in oncogenesis and tumor progression (Grandal and Madshus, 2008; Kuan et al., 2001). Thus, mTORC1 inhibition might contribute to suppression of tumor growth at least in part through facilitating the lysosomal degradation of EGFR, although the extent of the contribution might vary depending on cancer types.
Several recent studies have raised a question about the function of UVRAG in the autophagosome-lysosome fusion (Farre et al., 2010; Jiang et al., 2014; Takats et al., 2014). UVRAG participates in early autophagy events, such as recruitment of Bif-1 (N-BAR domain-containing protein) to the Beclin 1-Vps34 complex, which promotes the membrane curvature necessary for autophagosome formation (Takahashi et al., 2007). UVRAG also plays a role in coordinating the Golgi-ER retrograde and autophagic membrane trafficking in early stages of the autophagy processes (He et al., 2013). However, since UVRAG S498A mutation did not alter the formation of omegasome and phagophore, S498 phosphorylation is unlikely involved in the early processes of autophagy. UVRAG also has a non-autophagic function, such as the maintenance of the chromosomal stability (Zhao et al., 2012). It remains to be explored whether mTORC1 regulates the non-autophagic function of UVRAG via the phosphorylation.
Our finding defines the mTORC1-UVRAG pathway as an important regulatory axis through which cells coordinate autophagy and the endosome-lysosomal degradation pathway. The coordinate regulation might be important to maintain normal cellular physiology under changing environments of growth factors and nutrients. Some cancer cells might disturb the coordination by reducing the level of UVRAG and suppressing the endosomal degradation pathway as implicated by frequent alterations of the UVRAG gene locus in breast, colorectal and gastric cancers (Kim et al., 2008b; Knaevelsrud et al., 2010). Alternatively, cancer cells might maintain a favorable cell growth condition through the mTORC1-mediated UVRAG phosphorylation. Elucidating further mTORC1-regulated interactions and phosphorylations in the membrane processes may shed light on the molecular network that coordinate autophagy and the endosomal pathway and provide insight into how the network is disturbed in cancers and other diseases.
EXPERIMENTAL PROCEDURES
Identification of UVRAG Phosphorylation Sites
Myc-UVRAG was expressed with HA-Rheb in HEK293T cells and immunoprecipitated using anti-myc antibody. As a control, myc-UVRAG was expressed alone and treated with Torin1 for 1 h. Immunoprecipitated UVRAG was resolved by SDS-PAGE. UVRAG band was subjected to trypsinization followed by enrichment of phosphorylated peptides. Eluted fractions of tryptic digests of peptides were analyzed by mass spectrometry. A detailed procedure for mass spectrometry is described in the Supplemental Experimental Procedures.
Plasmids
Human UVRAG cDNA (IMAGE: 6186851) and its DNA fragments were cloned into pRK5 vector by PCR amplification. pLKO.1 vector was used to knock down UVRAG or RUBICON. CSII-EF-MCS was used to stably express UVRAG. The cDNAs for UVRAG, Vps16, Rab5a, Rab7, DFCP1 and WIPI2 were cloned into pEGFP-N3 and/or pmCherry-C1 (Clontech Lab. Inc., Palo Alto, CA) for cell imaging experiments. The mCherry-GFP-LC3 and the RFP-2xFYVE constructs were kindly provided by T. Johansen and N. Mizushima, respectively. All other constructs have been previously described (Jung et al., 2009; Vander Haar et al., 2007).
Cell Image Analysis
Autophagosome maturation, omegasome and phagophore formation, and the protein colocalization were analyzed by monitoring the fluorescence protein-tagged constructs using a Deltavision fluorescence microscope. A detailed procedure is described in the Supplemental Experimental Procedures.
In Vitro Assay of UVRAG phosphorylation by mTOR
The mTOR kinase activity toward UVRAG phosphorylation was analyzed in vitro using immunoprecipitated mTOR or a purified form of mTOR containing residues 1362–2549 (Millipore, Bedford, MA). As substrates, UVRAG fragments and its full length WT or S498A were purified using the ArcticExpress system (Stratagene, LaJolla, CA). The kinase reaction was performed as described previously (Jung et al., 2009).
In Vitro Assay of Vps34 kinase activity
The Vps34 kinase activity was assayed as described previously (Farkas et al., 2011). Briefly, Vps34 complex obtained by immunoprecipitation was incubated with PI and ATP. The amount of PI3P produced from the reaction was analyzed by use of GST-tagged p40 phox domain-containing polypeptide that binds to PI3P. A detailed procedure is described in the Supplemental Experimental Procedures.
Autophagy Flux Assay
HEK293T cells stably expressing UVRAG WT or S498A were treated with Torin1 (250 nM) for 4 h in the presence or absence of Bafilomycin A1 (100 nM) or E-64/Pepstatin (10 μg/mL). The amounts of LC3-I and LC3-II in cell lysate were analyzed by WB.
Analysis of EGFR Degradation and EGFR signaling
Endocytic degradation of EGFR was assayed as described previously (Liang et al., 2008; Sun et al., 2010). Briefly, cells starved of serum overnight were treated with EGF (100 ng/ml) for the indicated periods of time and lysed in 0.3% Chaps-containing lysis buffer. To analyze the activity of EGFR signaling, serum-starved HCT116 or HEK293T cells were stimulated with EGF for 10 min at 20 ng/ml or 10 ng/ml, respectively. The phosphorylation states of ERK1/2 and AKT were analyzed by WB.
Xenograft Analysis of Tumor Formation
WT and S498A HCT116 stable cells were injected subcutaneously into the flank of athymic nude male mice. Three weeks after the injection, tumors were removed and weighed. Tumor volume was evaluated by measurement of the length and width of the tumor mass in millimeters using electronic vernier calipers. A detailed procedure is described in the Supplemental Experimental Procedures.
Other Experimental Procedures
Other experimental procedures, including materials, cell culture, transfection, mutagenesis, immunoprecipitation, western blotting, lentiviral preparation, stable cell generation, preparation of recombinant proteins, cell proliferation assay, analysis of Rab7 active state, colony formation assay, and statistical analysis, are described in the Supplemental Experimental Procedures.
Supplementary Material
Figure S1. mTOR interacts with UVRAG and induces a mobility shift of UVRAG on SDS-PAGE. (A) mTOR and raptor bind to UVRAG. HEK293T cells were transiently transduced with plasmids encoding the indicated genes. Two-days post-transfection, myc immunoprecipitates were analyzed for the interaction. Irrelevant lanes in the middle were removed. (B) Rictor knockdown does not affect the interaction between UVRAG and mTORC1. (C) The full-length of UVRAG is required for stable binding to mTOR. Myc-tagged full-length and fragments of UVRAG were expressed with HA-mTOR in HEK293T cells where endogenous UVRAG was silenced. The amount of HA-mTOR co-immunoprecipitated with the UVRAG constructs was analyzed by WB. (D) Rapamycin does not alter the mobility of UVRAG band on SDS-PAGE. HEK293T cells were treated with either rapamycin (rapa; 100 nM) or Torin1 (250 nM) for 1 h. As vehicle (−), DMSO was used in the same volume. The mobility of UVRAG band on SDS-PAGE as well as the phosphorylation states of S6K1 (Thr389) and 4EBP1 (Thr37/46) were analyzed by WB. (E) Rheb overexpression induces a mobility shift of UVRAG. Myc-Rheb was transiently overexpressed in HEK293T cells. Cell lysate was analyzed by WB. (F) UVRAG fragments purified by use of a bacterial expression system. Proteins were stained with Coomassie blue G-250.
Figure S2. UVRAG Ser498 is an mTORC1-dependent phosphorylation site. (A) Sequence alignments of the residues surrounding the phosphorylation sites of UVRAG identified by mass spectrometry. The serine residues in red show a common feature of the known mTORC1 substrates (Kang et al., 2013). (B) Validation of the polyclonal antibody specific to Ser498 phosphorylation. Flag-tagged UVRAG was isolated by immunoprecipitation using anti-flag antibody-conjugated agarose. The phosphorylation state of UVRAG Ser498 was analyzed by WB using the phospho-specific antibody. The polyclonal antibody detected UVRAG WT but not S498A mutant. Treatment of HEK293T cells with Torin1 (250 nM) for 1 h drastically suppressed the detection of the UVRAG band. (C) mTORC1 activation specifically induces Ser498 phosphorylation. HA-tagged Rheb was expressed together with myc-tagged UVRAG WT or S498A in HEK293T cells, where endogenous UVRAG was silenced. Myc immunoprecipitates were analyzed by WB with use of the polyclonal antibody. (D) Rapamycin does not inhibit the Ser498 phosphorylation. HEK293T cells were treated with rapamycin (0, 0.1, 0.25, 0.5, or 1 μM) or Torin1 (250 nM) for 1 h. The phosphorylation of UVRAG Ser498 was analyzed by WB. (E) The polyclonal antibody detected the phosphorylation of endogenous UVRAG in cell lysate. TSC1 KO MEFs were treated with Torin1 as described in (B). Cell lysate was obtained, and the phosphorylation state of UVRAG and S6K1 (pT389) were analyzed by WB. (F) ULK1 is not required for the UVRAG phosphorylation. ULK1 MEFs were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) or Hank’s Buffered Salt Solution (HBSS) for 2 h. The phosphorylation of Ser498 was analyzed as described in (B).
Figure S3. mTOR phosphorylates UVRAG at Ser498. The active fragment of mTOR containing residues 1362–2549 was incubated in the presence or absence of ATP with the UVRAG 271–699 fragment purified from a bacterial expression system. Torin1 (250 nM) was added during the reaction, and the phosphorylation state of UVRAG Ser498 was analyzed by WB.
Figure S4. UVRAG S498A mutation reduces the interaction between UVRAG and RUBICON. (A) Ser498 phosphorylation state is not critical for the regulation of the UVRAG-mTORC1 interaction. Myc-UVRAG WT or S498A was transiently expressed with HA-mTOR in endogenous UVRAG-depleted HEK293T cells. The transfected cells were treated with Torin1 for 1 h. The amount of HA-mTOR bound to myc-UVRAG was analyzed by WB. (B) Knockdown of UVRAG in HEK293T and HeLa cells. The knockdown was achieved by lentivirally transducing the cells with shRNA targeting a 3′-UTR sequence of UVRAG mRNA (see details in Experimental Procedures). (C) Ser498 phosphorylation state regulates the interaction between UVRAG and RUBICON. HA-tagged RUBICON was co-expressed with myc-tagged UVRAG WT or mutant constructs in HEK293T cells, where endogenous UVRAG was silenced. The amount of HA-RUBICON recovered with myc-UVRAG immunoprecipitates was analyzed by WB. (D) Torin1 does not alter the binding affinity of the interaction between Beclin 1 and Atg14L. HEK293T cells were treated with vehicle (DMSO) or Torin1 (250 nM) for the indicated period of time. The amounts of Atg14L and Beclin 1 recovered with Atg14L immunoprecipitates were analyzed by WB.
Figure S5. Abolishing the UVRAG Ser498 phosphorylation increases the kinase activity of Vps34. (A) The RFP-2xFYVE was transiently expressed together with myc-tagged UVRAG (WT, S498A, or S498D) in HeLa cells where endogenous UVRAG was silenced. 2xFYVE puncta were monitored by fluorescence microscope. Scale bar, 10 μm. (B) The RFP-2xFYVE was transiently expressed in HCT116 cells stably expressing UVRAG (empty vector, WT, or S498A). The accumulation of FYVE was analyzed as described in Figure 5A. Scale bar, 5 μm. (C) Quantitative analysis of RFP-2xFYVE puncta formation of (B). The error bars represent mean ± SD (n≥55). (*, p < 0.01 vs empty vector; #, p < 0.01 WT vs S498A. (D) 2xFYVE positive area colocalizes with Rab5 and Rab7. HCT116 WT and S498A mutant cells were transiently transduced by RFP-2xFYVE construct together with either GFP-Rab5 or GFP-Rab7. The RFP and GFP positive areas were monitored by fluorescence microscope. Scale bar, 5 μm. (E) Preventing UVRAG phosphorylation increases the lipid kinase activity of Vps34. The lipid kinase assay was conducted as described in Figure 5C in the absence or presence of LY294002 (100 μM) for 30 min. (F) UVRAG Ser498 phosphorylation is important for the inhibitory effect of RUBICON on UVRAG-induced production of PI3P in cells. HCT116 cells, which were stably transduced by UVRAG constructs, were transiently transduced to express RFP-2xFYVE alone (−) or both RFP-2xFYVE and RUBICON (+). 2xFYVE positive area was monitored by fluorescence microscope. Scale bar, 5 μm. (G) RUBICON does not significantly alter the sizes of 2xFYVE positive areas. The error bars represent means ± SD [WT alone (n=121); WT with RUBICON (n=77); S498A alone (n=220); S498A with RUBICON (n=116)]. (H–K) The increase of Vps34 kinase activity by RUBICON knockdown is significantly reduced with UVRAG S498A mutation. HEK293T cells that were stably transduced by shRNA were transiently transduced to express the indicated proteins. Two days post-transfection, Vps34 complex was isolated by immunoprecipitation using HA antibody. The in vitro kinase assay for Vps34 was conducted as in Figure 5C. (L, M) Quantitative analysis of Vps34 kinase activity from H–K. The error bars represent mean ± SD (*, p<0.05 between WT and S498A; n=2).
Figure S6. Abolishing the UVRAG Ser498 phosphorylation increases autophagosome maturation. (A, B) UVRAG colocalizes with Vps16 more highly in S498A mutant cells compared to WT cells, which is further enhanced by Torin1 treatment. HeLa cells, where endogenous UVRAG was silenced, were transiently transduced to express GFP-Vps16 and mCherry-UVRAG constructs. The transduced cells were analyzed under fluorescence microscope. Intensity line scanes on the right-hand side indicate the degree of colocalization between Vps16 and UVRAG. The line scans were obtained along the yellow lines drawn in the inset boxes of the merged images. Arrows indicate colocalization of UVRAG and Vps16. Scale bar, 5 μm. (C) RUBICON knockdown enhances the interaction between UVRAG WT and Vps16 but barely the interaction between UVRAG S498A and Vps16. HEK293T cells stably transduced by shRNA were transiently transduced to express myc-UVRAG and GFP-Vps16 constructs together. The amount of GFP-Vps16 co-immunoprecipitated with myc-UVRAG was analyzed by WB. (D) RUBICON overexpression reduces the interaction between UVRAG WT and Vps39 but barely the interaction between UVRAG S498A and Vps39. HEK293T cells, where endogenous UVRAG was silenced, were transiently transduced to express myc-UVRAG and HA-Vps39 constructs together with or without HA-RUBICON. The amounts of HA-RUBICON and HA-Vps39 co-immunoprecipitated with myc-UVRAG were analyzed by WB. (E) Validation of the monoclonal antibody that specifically recognizes Rab7-GTP but not Rab7-GDP (Abcam, ab173250). The GFP-Rab7 mutants were transiently expressed in HEK293T cells, and GTP bound Rab7 was immunoprecipitated with the antibody specific to the active state of Rab7 and analyzed by WB using anti-GFP antibody. The active-state mutant of Rab7 (Q67L), but not the inactive-state mutant of Rab7 (T22N), was immunoprecipitated by the antibody. (F) Ccz1 interacts with UVRAG with an affinity similar to that of the UVRAG-Vps16/39 interaction. HEK293T cells were transiently transduced to express the indicated myc-tagged constructs together with GFP-UVRAG. The amount of GFP-UVRAG bound to the myc-tagged constructs in myc-immunoprecipitates was analyzed by WB. (G) UVRAG Ser498 phosphorylation does not have effects on the interaction between UVRAG and the Mon1/Ccz1 complex. HEK293T cells were transiently transduced to express myc-tagged UVRAG construct together with HA-tagged Mon1B and Ccz1. Two days post-transfection, myc immunoprecipitates were obtained. The amount of HA-Mon1B and HA-Ccz1 in myc immunoprecipitates were analyzed by WB. (H–J) UVRAG S498A mutation increases autophagosome maturation. The mCherry-GFP-LC3 construct was transiently expressed in HCT116 cells that were stably transduced by empty vector (H), wild type (I), or S498A (J). The transduced cells were treated with Torin1 (250 nM) for 4 h, and mCherry and GFP puncta were monitored by fluorescence microscope. Scale bar, 5 μm. (K) The dephosphorylation of Ser498 of recombinant UVRAG in response to Torin1 is moderately delayed relative to that of endogenous UVRAG. HEK293T cells stably expressing flag-UVRAG were treated with Torin1 (250 nM) for the indicated periods of time. The phosphorylation state of Ser498 of flag-UVRAG in flag immunoprecipitates was analyzed by WB. For comparison, endogenous UVRAG was isolated by immunoprecipitation using anti-UVRAG antibody from HEK293T cells treated with Torin1. (L) Quantitative analysis of the relative levels of UVRAG phosphorylation between endogenous and recombinant UVRAG. (M) Lysosomal fusion inhibitor BAFA1 increases the level of LC3-II in S498A cells compared to WT cells. HEK293T cells stably transduced by UVRAG WT or S498A were incubated in DMEM in the presence or absence of Bafilomycin A1 (100 nM) for 4 h. (N) Quantitative analysis of the fold increase of LC3-II level induced by the treatment with lysosomal inhibitors in Torin1-treated WT and S498A mutant cells. (O) Ser498 phosphorylation does not have effects on omegasome and phagophore formation. GFP-DFCP1 was transiently expressed in HCT116 WT and S498A mutant cells, whereas GFP-WIPI2 was transiently expressed with myc-tagged UVRAG (WT or S498A) in HeLa cells, where endogenous UVRAG is silenced. Cells were treated with Torin1 (250 nM) for 4 h, and GFP-DFCP1 and GFP-WIPI2 were monitored by fluorescence microscope. (P, Q) Quantitative analysis of DFCP1 puncta (P) and WIPI2 puncta from (Q). Mean value and SD are shown as a horizontal bar. Scale bars are 5 μm and 10 μm for DFCP1- and WIPI2-stained images, respectively.
Figure S7. Prevention of UVRAG Ser498 phosphorylation enhances endosome-lysosomal degradation of EGFR, reduces EGFR signaling, and suppresses cancer cell and tumor growth. (A) Preventing UVRAG Ser498 phosphorylation increases the lysosomal degradation of EGFR. HCT116 cells that stably express flag-UVRAG (WT or S498A) were treated with EGF (100 ng/ml) for the indicated periods of time. The level of EGFR was monitored by WB. (B) Preventing UVRAG Ser498 phosphorylation suppresses EGF-stimulated ERK1/2 phosphorylation. HCT116 cells that stably expressing flag-UVRAG WT or S498A were starved overnight and treated with EGF (20 ng/ml) for 10 min. (C) Torin1 facilitates the degradation of EGFR. HEK293T cells were starved in serum-free media overnight and treated with vehicle (DMSO) or Torin 1 (250 nM) for 2 h, then stimulated with EGF (100 ng/ml). (D and E) Quantitative analysis of EGFR degradation in Figure S7C. The level of EGFR was presented relative to that at time zero for DMSO (D) or relative to those at time zero for DMSO and Torin1, respectively (E). (F) Torin1 reduces the basal level of EGFR. HEK293T cells were serum starved overnight, incubated DMSO or Torin (250 nM) for 4 h, and followed by treatment of EGF (10 ng/ml). (G) Torin1 marginally reduces the basal level of EGFR in S498A mutant cells relative to WT cells. The cells were treated with Torin1 and EGF as described in (F). (H) Quantitative analysis of the levels of EGFR of (G). (I) UVRAG S498A mutation significantly inhibited cancer cell colony formation. HCT116 cells (1×104/6 well plate), prepared as described in Figure 7D, were seeded and incubated in soft agar for 2 weeks. The colony was stained with 0.005% crystal violet as described in Methods. (J-L) Abolishing UVRAG phosphorylation suppressed tumor growth in vivo. HCT116 cells stably expressing flag-UVRAG WT or S498A were subcutaneously injected into flank of Balb/c nu/nu mice (n=5). Tumor-bearing mice were pictured (J) and weighed (K) before they were sacrificed. Tumors were removed from the mice and weighed (L). The error bars represent mean ± SD (n = 5). (*, p < 0.01, WT vs S498A).
Acknowledgments
We thank M. Kundu for ULK1 MEFs; N. Mizushima for 2xFYVE construct; T. Johansen for dual-tagged LC3 construct; D. Kwiatkowski for TSC1 and TSC2 MEFs; V. Stambolic for PTEN MEFs; B. Viollet for AMPK MEFs; T. Neufeld and Kim lab members for helpful discussion; L. Higgins for mass spec analysis; R. Foncea for lentivirus prep. This study was supported by E0143033654 (to CHJ); A120879 (to SSB); P30-DK050456, GM097057, AG039758, and ADA 7–12-BS-093 (to DHK).
Footnotes
Supplemental Information includes seven figures, Supplemental Experimental Procedures, and Supplemental References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Zhao Q, Gao C, Ding Y, Zeng Y, Ueda T, Nakano A, Jiang L. Activation of the Rab7 GTPase by the MON1-CCZ1 Complex Is Essential for PVC-to-Vacuole Trafficking and Plant Growth in Arabidopsis. Plant Cell. 2014;26:2080–2097. doi: 10.1105/tpc.114.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop EA, Hunt DK, Acosta-Jaquez HA, Fingar DC, Tee AR. ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy. 2011;7:737–747. doi: 10.4161/auto.7.7.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T, Daugaard M, Jaattela M. Identification of small molecule inhibitors of phosphatidylinositol 3-kinase and autophagy. J Biol Chem. 2011;286:38904–38912. doi: 10.1074/jbc.M111.269134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Mathewson RD, Manjithaya R, Subramani S. Roles of Pichia pastoris Uvrag in vacuolar protein sorting and the phosphatidylinositol 3-kinase complex in phagophore elongation in autophagy pathways. Autophagy. 2010;6:86–99. doi: 10.4161/auto.6.1.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandal MV, Madshus IH. Epidermal growth factor receptor and cancer: control of oncogenic signalling by endocytosis. J Cell Mol Med. 2008;12:1527–1534. doi: 10.1111/j.1582-4934.2008.00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Ni D, Ma B, Lee JH, Zhang T, Ghozalli I, Pirooz SD, Zhao Z, Bharatham N, Li B, et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol. 2013;15:1206–1219. doi: 10.1038/ncb2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Seo M, Otto NM, Kim DH. ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy. 2011;7:1212–1221. doi: 10.4161/auto.7.10.16660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341:1236566. doi: 10.1126/science.1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008a;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Human pathology. 2008b;39:1059–1063. doi: 10.1016/j.humpath.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Knaevelsrud H, Ahlquist T, Merok MA, Nesbakken A, Stenmark H, Lothe RA, Simonsen A. UVRAG mutations associated with microsatellite unstable colon cancer do not affect autophagy. Autophagy. 2010;6:863–870. doi: 10.4161/auto.6.7.13033. [DOI] [PubMed] [Google Scholar]
- Kuan CT, Wikstrand CJ, Bigner DD. EGF mutant receptor vIII as a molecular target in cancer therapy. Endocr Relat Cancer. 2001;8:83–96. doi: 10.1677/erc.0.0080083. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- Nordmann M, Cabrera M, Perz A, Brocker C, Ostrowicz C, Engelbrecht-Vandre S, Ungermann C. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol. 2010;20:1654–1659. doi: 10.1016/j.cub.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Pols MS, ten Brink C, Gosavi P, Oorschot V, Klumperman J. The HOPS proteins hVps41 and hVps39 are required for homotypic and heterotypic late endosome fusion. Traffic. 2013;14:219–232. doi: 10.1111/tra.12027. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Shang L, Wang X. AMPK and mTOR coordinate the regulation of Ulk1 and mammalian autophagy initiation. Autophagy. 2011;7:924–926. doi: 10.4161/auto.7.8.15860. [DOI] [PubMed] [Google Scholar]
- Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci U S A. 2010;107:19338–19343. doi: 10.1073/pnas.1010554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S, Zhong Q. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem. 2011;286:185–191. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats S, Pircs K, Nagy P, Varga A, Karpati M, Hegedus K, Kramer H, Kovacs AL, Sass M, Juhasz G. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014;25:1338–1354. doi: 10.1091/mbc.E13-08-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Yuan HX, Russell RC, Guan KL. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy. 2013;9:1983–1995. doi: 10.4161/auto.26058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Oh S, Li D, Ni D, Pirooz SD, Lee JH, Yang S, Lee JY, Ghozalli I, Costanzo V, et al. A dual role for UVRAG in maintaining chromosomal stability independent of autophagy. Dev Cell. 2012;22:1001–1016. doi: 10.1016/j.devcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. mTOR interacts with UVRAG and induces a mobility shift of UVRAG on SDS-PAGE. (A) mTOR and raptor bind to UVRAG. HEK293T cells were transiently transduced with plasmids encoding the indicated genes. Two-days post-transfection, myc immunoprecipitates were analyzed for the interaction. Irrelevant lanes in the middle were removed. (B) Rictor knockdown does not affect the interaction between UVRAG and mTORC1. (C) The full-length of UVRAG is required for stable binding to mTOR. Myc-tagged full-length and fragments of UVRAG were expressed with HA-mTOR in HEK293T cells where endogenous UVRAG was silenced. The amount of HA-mTOR co-immunoprecipitated with the UVRAG constructs was analyzed by WB. (D) Rapamycin does not alter the mobility of UVRAG band on SDS-PAGE. HEK293T cells were treated with either rapamycin (rapa; 100 nM) or Torin1 (250 nM) for 1 h. As vehicle (−), DMSO was used in the same volume. The mobility of UVRAG band on SDS-PAGE as well as the phosphorylation states of S6K1 (Thr389) and 4EBP1 (Thr37/46) were analyzed by WB. (E) Rheb overexpression induces a mobility shift of UVRAG. Myc-Rheb was transiently overexpressed in HEK293T cells. Cell lysate was analyzed by WB. (F) UVRAG fragments purified by use of a bacterial expression system. Proteins were stained with Coomassie blue G-250.
Figure S2. UVRAG Ser498 is an mTORC1-dependent phosphorylation site. (A) Sequence alignments of the residues surrounding the phosphorylation sites of UVRAG identified by mass spectrometry. The serine residues in red show a common feature of the known mTORC1 substrates (Kang et al., 2013). (B) Validation of the polyclonal antibody specific to Ser498 phosphorylation. Flag-tagged UVRAG was isolated by immunoprecipitation using anti-flag antibody-conjugated agarose. The phosphorylation state of UVRAG Ser498 was analyzed by WB using the phospho-specific antibody. The polyclonal antibody detected UVRAG WT but not S498A mutant. Treatment of HEK293T cells with Torin1 (250 nM) for 1 h drastically suppressed the detection of the UVRAG band. (C) mTORC1 activation specifically induces Ser498 phosphorylation. HA-tagged Rheb was expressed together with myc-tagged UVRAG WT or S498A in HEK293T cells, where endogenous UVRAG was silenced. Myc immunoprecipitates were analyzed by WB with use of the polyclonal antibody. (D) Rapamycin does not inhibit the Ser498 phosphorylation. HEK293T cells were treated with rapamycin (0, 0.1, 0.25, 0.5, or 1 μM) or Torin1 (250 nM) for 1 h. The phosphorylation of UVRAG Ser498 was analyzed by WB. (E) The polyclonal antibody detected the phosphorylation of endogenous UVRAG in cell lysate. TSC1 KO MEFs were treated with Torin1 as described in (B). Cell lysate was obtained, and the phosphorylation state of UVRAG and S6K1 (pT389) were analyzed by WB. (F) ULK1 is not required for the UVRAG phosphorylation. ULK1 MEFs were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) or Hank’s Buffered Salt Solution (HBSS) for 2 h. The phosphorylation of Ser498 was analyzed as described in (B).
Figure S3. mTOR phosphorylates UVRAG at Ser498. The active fragment of mTOR containing residues 1362–2549 was incubated in the presence or absence of ATP with the UVRAG 271–699 fragment purified from a bacterial expression system. Torin1 (250 nM) was added during the reaction, and the phosphorylation state of UVRAG Ser498 was analyzed by WB.
Figure S4. UVRAG S498A mutation reduces the interaction between UVRAG and RUBICON. (A) Ser498 phosphorylation state is not critical for the regulation of the UVRAG-mTORC1 interaction. Myc-UVRAG WT or S498A was transiently expressed with HA-mTOR in endogenous UVRAG-depleted HEK293T cells. The transfected cells were treated with Torin1 for 1 h. The amount of HA-mTOR bound to myc-UVRAG was analyzed by WB. (B) Knockdown of UVRAG in HEK293T and HeLa cells. The knockdown was achieved by lentivirally transducing the cells with shRNA targeting a 3′-UTR sequence of UVRAG mRNA (see details in Experimental Procedures). (C) Ser498 phosphorylation state regulates the interaction between UVRAG and RUBICON. HA-tagged RUBICON was co-expressed with myc-tagged UVRAG WT or mutant constructs in HEK293T cells, where endogenous UVRAG was silenced. The amount of HA-RUBICON recovered with myc-UVRAG immunoprecipitates was analyzed by WB. (D) Torin1 does not alter the binding affinity of the interaction between Beclin 1 and Atg14L. HEK293T cells were treated with vehicle (DMSO) or Torin1 (250 nM) for the indicated period of time. The amounts of Atg14L and Beclin 1 recovered with Atg14L immunoprecipitates were analyzed by WB.
Figure S5. Abolishing the UVRAG Ser498 phosphorylation increases the kinase activity of Vps34. (A) The RFP-2xFYVE was transiently expressed together with myc-tagged UVRAG (WT, S498A, or S498D) in HeLa cells where endogenous UVRAG was silenced. 2xFYVE puncta were monitored by fluorescence microscope. Scale bar, 10 μm. (B) The RFP-2xFYVE was transiently expressed in HCT116 cells stably expressing UVRAG (empty vector, WT, or S498A). The accumulation of FYVE was analyzed as described in Figure 5A. Scale bar, 5 μm. (C) Quantitative analysis of RFP-2xFYVE puncta formation of (B). The error bars represent mean ± SD (n≥55). (*, p < 0.01 vs empty vector; #, p < 0.01 WT vs S498A. (D) 2xFYVE positive area colocalizes with Rab5 and Rab7. HCT116 WT and S498A mutant cells were transiently transduced by RFP-2xFYVE construct together with either GFP-Rab5 or GFP-Rab7. The RFP and GFP positive areas were monitored by fluorescence microscope. Scale bar, 5 μm. (E) Preventing UVRAG phosphorylation increases the lipid kinase activity of Vps34. The lipid kinase assay was conducted as described in Figure 5C in the absence or presence of LY294002 (100 μM) for 30 min. (F) UVRAG Ser498 phosphorylation is important for the inhibitory effect of RUBICON on UVRAG-induced production of PI3P in cells. HCT116 cells, which were stably transduced by UVRAG constructs, were transiently transduced to express RFP-2xFYVE alone (−) or both RFP-2xFYVE and RUBICON (+). 2xFYVE positive area was monitored by fluorescence microscope. Scale bar, 5 μm. (G) RUBICON does not significantly alter the sizes of 2xFYVE positive areas. The error bars represent means ± SD [WT alone (n=121); WT with RUBICON (n=77); S498A alone (n=220); S498A with RUBICON (n=116)]. (H–K) The increase of Vps34 kinase activity by RUBICON knockdown is significantly reduced with UVRAG S498A mutation. HEK293T cells that were stably transduced by shRNA were transiently transduced to express the indicated proteins. Two days post-transfection, Vps34 complex was isolated by immunoprecipitation using HA antibody. The in vitro kinase assay for Vps34 was conducted as in Figure 5C. (L, M) Quantitative analysis of Vps34 kinase activity from H–K. The error bars represent mean ± SD (*, p<0.05 between WT and S498A; n=2).
Figure S6. Abolishing the UVRAG Ser498 phosphorylation increases autophagosome maturation. (A, B) UVRAG colocalizes with Vps16 more highly in S498A mutant cells compared to WT cells, which is further enhanced by Torin1 treatment. HeLa cells, where endogenous UVRAG was silenced, were transiently transduced to express GFP-Vps16 and mCherry-UVRAG constructs. The transduced cells were analyzed under fluorescence microscope. Intensity line scanes on the right-hand side indicate the degree of colocalization between Vps16 and UVRAG. The line scans were obtained along the yellow lines drawn in the inset boxes of the merged images. Arrows indicate colocalization of UVRAG and Vps16. Scale bar, 5 μm. (C) RUBICON knockdown enhances the interaction between UVRAG WT and Vps16 but barely the interaction between UVRAG S498A and Vps16. HEK293T cells stably transduced by shRNA were transiently transduced to express myc-UVRAG and GFP-Vps16 constructs together. The amount of GFP-Vps16 co-immunoprecipitated with myc-UVRAG was analyzed by WB. (D) RUBICON overexpression reduces the interaction between UVRAG WT and Vps39 but barely the interaction between UVRAG S498A and Vps39. HEK293T cells, where endogenous UVRAG was silenced, were transiently transduced to express myc-UVRAG and HA-Vps39 constructs together with or without HA-RUBICON. The amounts of HA-RUBICON and HA-Vps39 co-immunoprecipitated with myc-UVRAG were analyzed by WB. (E) Validation of the monoclonal antibody that specifically recognizes Rab7-GTP but not Rab7-GDP (Abcam, ab173250). The GFP-Rab7 mutants were transiently expressed in HEK293T cells, and GTP bound Rab7 was immunoprecipitated with the antibody specific to the active state of Rab7 and analyzed by WB using anti-GFP antibody. The active-state mutant of Rab7 (Q67L), but not the inactive-state mutant of Rab7 (T22N), was immunoprecipitated by the antibody. (F) Ccz1 interacts with UVRAG with an affinity similar to that of the UVRAG-Vps16/39 interaction. HEK293T cells were transiently transduced to express the indicated myc-tagged constructs together with GFP-UVRAG. The amount of GFP-UVRAG bound to the myc-tagged constructs in myc-immunoprecipitates was analyzed by WB. (G) UVRAG Ser498 phosphorylation does not have effects on the interaction between UVRAG and the Mon1/Ccz1 complex. HEK293T cells were transiently transduced to express myc-tagged UVRAG construct together with HA-tagged Mon1B and Ccz1. Two days post-transfection, myc immunoprecipitates were obtained. The amount of HA-Mon1B and HA-Ccz1 in myc immunoprecipitates were analyzed by WB. (H–J) UVRAG S498A mutation increases autophagosome maturation. The mCherry-GFP-LC3 construct was transiently expressed in HCT116 cells that were stably transduced by empty vector (H), wild type (I), or S498A (J). The transduced cells were treated with Torin1 (250 nM) for 4 h, and mCherry and GFP puncta were monitored by fluorescence microscope. Scale bar, 5 μm. (K) The dephosphorylation of Ser498 of recombinant UVRAG in response to Torin1 is moderately delayed relative to that of endogenous UVRAG. HEK293T cells stably expressing flag-UVRAG were treated with Torin1 (250 nM) for the indicated periods of time. The phosphorylation state of Ser498 of flag-UVRAG in flag immunoprecipitates was analyzed by WB. For comparison, endogenous UVRAG was isolated by immunoprecipitation using anti-UVRAG antibody from HEK293T cells treated with Torin1. (L) Quantitative analysis of the relative levels of UVRAG phosphorylation between endogenous and recombinant UVRAG. (M) Lysosomal fusion inhibitor BAFA1 increases the level of LC3-II in S498A cells compared to WT cells. HEK293T cells stably transduced by UVRAG WT or S498A were incubated in DMEM in the presence or absence of Bafilomycin A1 (100 nM) for 4 h. (N) Quantitative analysis of the fold increase of LC3-II level induced by the treatment with lysosomal inhibitors in Torin1-treated WT and S498A mutant cells. (O) Ser498 phosphorylation does not have effects on omegasome and phagophore formation. GFP-DFCP1 was transiently expressed in HCT116 WT and S498A mutant cells, whereas GFP-WIPI2 was transiently expressed with myc-tagged UVRAG (WT or S498A) in HeLa cells, where endogenous UVRAG is silenced. Cells were treated with Torin1 (250 nM) for 4 h, and GFP-DFCP1 and GFP-WIPI2 were monitored by fluorescence microscope. (P, Q) Quantitative analysis of DFCP1 puncta (P) and WIPI2 puncta from (Q). Mean value and SD are shown as a horizontal bar. Scale bars are 5 μm and 10 μm for DFCP1- and WIPI2-stained images, respectively.
Figure S7. Prevention of UVRAG Ser498 phosphorylation enhances endosome-lysosomal degradation of EGFR, reduces EGFR signaling, and suppresses cancer cell and tumor growth. (A) Preventing UVRAG Ser498 phosphorylation increases the lysosomal degradation of EGFR. HCT116 cells that stably express flag-UVRAG (WT or S498A) were treated with EGF (100 ng/ml) for the indicated periods of time. The level of EGFR was monitored by WB. (B) Preventing UVRAG Ser498 phosphorylation suppresses EGF-stimulated ERK1/2 phosphorylation. HCT116 cells that stably expressing flag-UVRAG WT or S498A were starved overnight and treated with EGF (20 ng/ml) for 10 min. (C) Torin1 facilitates the degradation of EGFR. HEK293T cells were starved in serum-free media overnight and treated with vehicle (DMSO) or Torin 1 (250 nM) for 2 h, then stimulated with EGF (100 ng/ml). (D and E) Quantitative analysis of EGFR degradation in Figure S7C. The level of EGFR was presented relative to that at time zero for DMSO (D) or relative to those at time zero for DMSO and Torin1, respectively (E). (F) Torin1 reduces the basal level of EGFR. HEK293T cells were serum starved overnight, incubated DMSO or Torin (250 nM) for 4 h, and followed by treatment of EGF (10 ng/ml). (G) Torin1 marginally reduces the basal level of EGFR in S498A mutant cells relative to WT cells. The cells were treated with Torin1 and EGF as described in (F). (H) Quantitative analysis of the levels of EGFR of (G). (I) UVRAG S498A mutation significantly inhibited cancer cell colony formation. HCT116 cells (1×104/6 well plate), prepared as described in Figure 7D, were seeded and incubated in soft agar for 2 weeks. The colony was stained with 0.005% crystal violet as described in Methods. (J-L) Abolishing UVRAG phosphorylation suppressed tumor growth in vivo. HCT116 cells stably expressing flag-UVRAG WT or S498A were subcutaneously injected into flank of Balb/c nu/nu mice (n=5). Tumor-bearing mice were pictured (J) and weighed (K) before they were sacrificed. Tumors were removed from the mice and weighed (L). The error bars represent mean ± SD (n = 5). (*, p < 0.01, WT vs S498A).