SUMMARY
Alternative polyadenylation (APA) has been implicated in a variety of developmental and disease processes. A particularly dramatic form of APA occurs in the developing nervous system of flies and mammals, whereby various developmental genes undergo coordinate 3′ UTR extension. In Drosophila, the RNA-binding protein ELAV inhibits RNA processing at proximal polyadenylation sites, thereby fostering the formation of exceptionally long 3′ UTRs. Here, we present evidence that paused Pol II promotes recruitment of ELAV to extended genes. Replacing promoters of extended genes with heterologous promoters blocks normal 3′ extension in the nervous system, while extension-associated promoters can induce 3′ extension in ectopic tissues expressing ELAV. Computational analyses suggest that promoter regions of extended genes tend to contain paused Pol II and associated cis-regulatory elements such as GAGA. ChIP-Seq assays identify ELAV in the promoter regions of extended genes. Our study provides evidence for a regulatory link between promoter-proximal pausing and APA.
Keywords: ELAV, alternative polyadenylation, 3′ UTR extension, nervous system, promoter, paused Pol II
INTRODUCTION
Nascent transcripts undergo 3′ cleavage and polyadenylation (CPA) prior to transcription termination to produce mature mRNAs. The C-terminal domain (CTD) of the large subunit of RNA Polymerase II (Pol II) serves as an interaction platform for multiple factors that control transcription initiation, elongation, and termination (Hsin and Manley, 2012). CPA factors have been detected in promoter regions (Glover-Cutter et al., 2008), where they interact with general transcription factors (Dantonel et al., 1997), transcriptional activators (Calvo and Manley, 2001; Nagaike et al., 2011) and the Pol II CTD (McCracken et al., 1997). Functional interactions between transcriptional initiation and termination have been documented (Andersen et al., 2013), for example, impaired 3′ processing can diminish initiation rates at yeast and human promoters (Mapendano et al., 2010; Zhang et al., 2012).
Transcriptome-wide studies have revealed that most genes contain multiple polyadenylation (poly(A)) signals and are subject to alternative polyadenylation (APA) (Brown et al., 2014; Elkon et al., 2013; Pelechano et al., 2013; Shi, 2012; Tian et al., 2005; Wang et al., 2008). The most common form of APA, ‘tandem 3′ UTR APA’, generates different mRNA isoforms possessing distinct 3′ untranslated regions (3′ UTRs) with identical protein-coding sequences. APA-mediated alterations of 3′ UTRs have been implicated in a variety of processes, including animal development and human disease. For example, global 3′ UTR shortening accompanies cell proliferation (Elkon et al., 2012; Ji and Tian, 2009) and can cause oncogenic transformation in cultured mammalian cells (Mayr and Bartel, 2009). Abnormal APA has been linked to oculopharyngeal muscular dystrophy (OPMD) (Jenal et al., 2012).
A particularly dramatic example of tissue-specific APA is seen in the developing nervous system of flies and vertebrates, whereby hundreds of genes exhibit 3′UTR extension. Neural-specific 3′ UTR extensions have been documented in Drosophila (Hilgers et al., 2011; Smibert et al., 2012), zebrafish (Ulitsky et al., 2012), mouse and human (Miura et al., 2013; Zhang et al., 2005), and seem to be a conserved feature of animal neurogenesis. The extended 3′ UTR sequences, which can reach tens of kilobases (kb) in length, are thought to confer post-transcriptional regulation underlying specific neuronal functions, such as axonal transport.
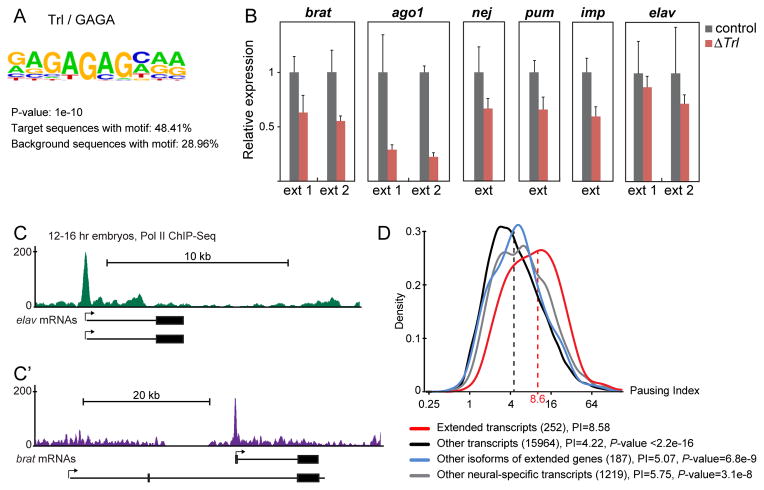
Figure 3. Extended genes contain the GAGA motif and paused Pol II.
A. A motif search among 252 neural-specific transcripts exhibiting 3′ UTR extensions yielded the GAGA motif as the most significantly enriched motif compared to background sequences (all other annotated gene promoters).
B. Quantification of indicated transcripts by qPCR using primer combinations specific to the partially extended (ext 1) or fully extended (ext 2) 3′ UTR forms of each gene. RNA was extracted from brains of yw (control) or Trl mutant (ΔTrl) third instar larvae. Extension levels were normalized to coding regions of each gene to reflect levels relative to the short isoforms. For each primer pair, expression in control larvae was set to the value 1. In Trl mutants, 3′ UTR extension of each of the six analyzed genes is significantly reduced (P-values<0.01, unpaired Student’s t-test) compared to control larvae. Error bars represent mean ± SD of three samples for each genotype.
C, C′. Normalized Pol II ChIP-Seq reads at the elav (C) and brat (C′) loci in 12–16 hr embryos (Negre et al., 2011). Short and extended isoforms are represented below the tracks. Arrows denote the start site and directionality of transcription. Pol II peaks indicate promoter-proximal pausing at the elav locus (C) and at the promoter expressing the extended form of brat, but not the short form (C′).
D. Pausing index (PI) distribution and median PI values of the promoters of the indicated groups of transcripts in whole embryos. The numbers in parentheses denote the number of transcripts in each group. Promoters of extended transcripts are significantly more paused than promoters of any control group. Wilcoxon rank sum test P-values were calculated by comparing the pausing index of extended transcripts with each group of controls. See also Fig. S4.
In Drosophila, the nuclear RNA-binding protein Embryonic Lethal Abnormal Visual system (ELAV) was shown to be a key regulator of 3′ UTR extension. ELAV is expressed in the nuclei of neurons. It inhibits CPA by binding in the vicinity of proximal poly(A) sites of nascent transcripts, thereby promoting Pol II read-through and 3′ extension. Ectopic expression of ELAV was shown to be sufficient to induce ectopic extension of endogenous genes in non-neural tissues (Hilgers et al., 2012). Studies using cultured cells suggest that ELAV homologues perform similar functions in mammals (Mansfield and Keene, 2012). We hereafter refer to genes with extended 3′ UTRs in the nervous system as “extended genes”.
Here, we show that ELAV-mediated 3′ UTR extension is dependent on transcription initiation. Promoters of extended genes generate 3′ UTR extension from reporter transgenes in the Drosophila nervous system. These promoters can also induce 3′ extension in non-neural tissues upon ectopic expression of ELAV. Computational analyses reveal that promoters of extended genes typically contain paused Pol II and are enriched in “pausing elements” such as the GAGA motif (Li and Gilmour, 2013). Moreover, ELAV ChIP-Seq assays suggest that ELAV associates with the promoter regions of extended genes, but is present at significantly lower levels at non-extended genes. We propose that ELAV is recruited to the promoter regions of extended genes via paused Pol II, and inhibits CPA at proximal poly(A) sites during transcription elongation.
RESULTS AND DISCUSSION
The native promoter is necessary for 3′ UTR extension
ELAV is an RNA-binding protein that has been shown to bind to U-rich regions in target mRNAs, including neuroglian (nrg) (Lisbin et al., 2001) and erect wings (ewg) (Soller and White, 2003). Recently, the Hox gene Ultrabithorax (Ubx) was shown to be bound by ELAV through similar elements to regulate alternative splicing, but ELAV was not found to bind to predicted binding sites in the Ubx 3′ UTR (Rogulja-Ortmann et al., 2014). Similarly, we also failed to identify specific ELAV recognition sequences within extended 3′ UTRs. In the present study, we investigate how ELAV is selectively recruited to appropriate targets during neurogenesis.
We examined the activities of synthetic reporter genes in transgenic embryos to determine whether extended 3′ UTRs are sufficient for the selective recruitment of ELAV in vivo. Transgenes contain the Drosophila Synthetic Core Promoter (DSCP (Pfeiffer et al., 2008)) attached to a GFP coding sequence followed by the entire extended 3′ UTR of elav, one of the targets of ELAV (Fig. 1A). If elav 3′ UTR sequences are sufficient to recruit ELAV, then this transgene should produce mRNAs containing extended 3′ UTRs.
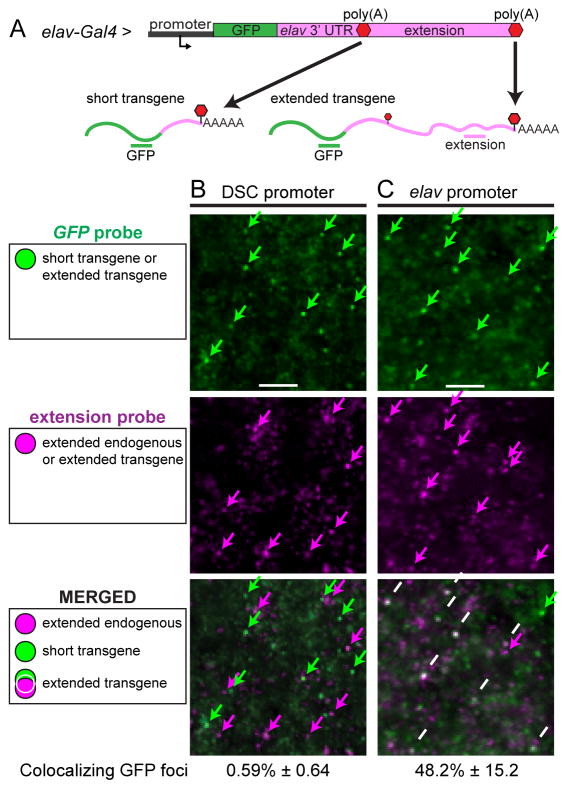
Figure 1. Native promoters are required for expression of 3′ extensions.
A. elav-Gal4 drives expression of a GFP transgene in the nervous system. Two different promoter regions were used, DSCP or the native elav promoter. The GFP coding sequence was placed upstream of the entire extended 7.2 kb elav 3′ UTR. CPA at the proximal poly(A) produces the short 3′ UTR form of the mRNA, whereas CPA at the distal-most poly(A) produces the fully extended transcript. RNA probes directed against different regions of the transcripts were used to detect mRNAs.
B,C. Double fluorescent in situ hybridization assays using probes indicated in A. Single confocal sections of a portion of the developing CNS in stage 13 embryos. Note that the extension probe detects not only the transgene, but also the endogenous elav transcript, which is expressed in the nervous system. Colocalization of the GFP and extension probes indicates expression of extended transcripts from the transgene.
B. The reporter transgene carrying the DSCP does not exhibit colocalization of GFP and extension probes. Extension signals (magenta arrows) do not colocalize with the green GFP signal, indicating that they correspond to endogenous elav mRNAs.
C. Replacing the DSCP with the native elav promoter region induces 3′ extension of the GFP transgene. There is extensive colocalization of the GFP (green arrows) and extension probes (magenta arrows), indicating expression of extended 3′ UTR sequences from the transgene (white arrows in merged image). The percentages of GFP foci that colocalized with extension foci are indicated. Numbers represent mean ± SD of six embryos for each promoter. See also Figs. S1 and S2.
Expression of 3′ UTR sequences was monitored via double labeling assays with GFP coding sequences to distinguish transgene mRNAs from endogenous elav transcripts (Fig. 1, schematics). Expression of the transgene was confirmed by colocalization of GFP with a probe directed against the short 3′ UTR (Fig. S1A). However, we did not observe colocalization of GFP with extended sequences, indicating that mRNAs produced from the transgene lack 3′ extensions (Fig. 1B). The only signals containing 3′ extensions corresponded to endogenous elav mRNAs (Fig. S1B).
Additional experiments were done to determine why the transgene fails to produce extended transcripts. We excluded the possibility that the GFP coding sequence somehow inhibits expression of extended sequences by creating GFP transgenes lacking proximal poly(A) signals (Fig. S1C–D). Such constructs no longer depended on ELAV for 3′ extension, and were found to produce mRNAs containing extended 3′ UTR sequences when expressed in ectopic tissues lacking ELAV (Fig. S1C–D).
To test whether promoter sequences play a role in ELAV recruitment, we swapped the DSCP with a 333 bp genomic DNA fragment encompassing the native elav promoter region, consisting of 92 bp upstream and 241 bp downstream of the transcription start site (TSS) (Yao and White, 1994). Strikingly, we observed colocalization of GFP and extension sequences (Fig. 1C), indicating expression of the elav 3′ UTR extension, as seen for the endogenous locus.
To confirm that 3′ extension depends on native promoter regions of extended genes, we also tested a construct bearing the fully extended brat 3′ UTR downstream of GFP, using three different promoters: the DSCP, the native promoter producing the short form of brat, and the native promoter producing the extended form of brat (Fig. S2A,A′). Only the brat promoter associated with endogenous extension mediated expression of transgenic transcripts containing 3′ UTR extensions (Fig. S2B–D). These observations suggest that the promoter regions of extended genes are essential for the ELAV-mediated expression of 3′UTR extensions.
Native promoters mediate 3′ extension in ectopic tissues
The preceding results suggest that promoter sequences are important for the synthesis of 3′ extensions in the developing nervous system. We further explored their importance by examining non-neural tissues. Ectopic ELAV can drive 3′UTR extension in ectopic tissues from endogenous loci (Hilgers et al., 2012). We sought to determine whether ectopic ELAV could also induce ectopic 3′ extensions from transgenic DNAs.
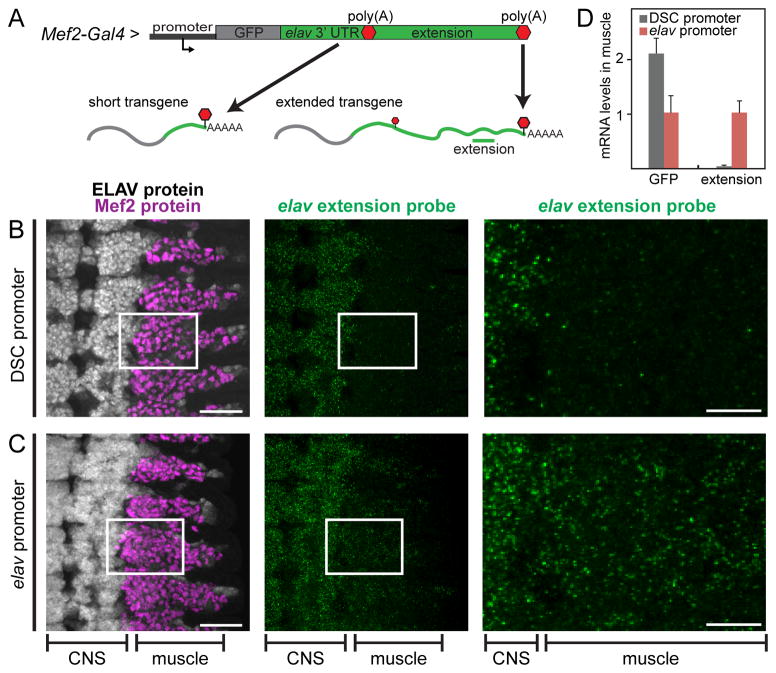
We expressed both the GFP-elav transgene and ELAV protein in muscle cells using a Mef2-Gal4 driver (Fig. 2A). In this context, mRNA expression from the reporter is easily distinguished from endogenous elav expression, which occurs only in the nervous system. The DSCP fails to generate 3′ UTR extensions (Fig. 2B, muscle), and only endogenous elav transcripts in the CNS were detected (Fig. 2B, CNS). In contrast, the GFP-elav transgene containing the native elav promoter produced transcripts with extended 3′ UTRs in muscle tissue (Fig. 2C, muscle). Quantification of transgene expression in dissected muscle tissue using qPCR shows that both promoters drive robust transgene expression (GFP signal), but only the native promoter drives expression of extension sequences (Fig. 2D). Similarly, the second brat promoter (see above), but not the DSCP, was also able to drive expression of an extended brat 3′ UTR in muscle cells (Fig. S3A–C).
Figure 2. The native elav promoter mediates 3′ extension in muscle.
A. Mef2-Gal4 drives expression of a GFP transgene in muscle cells. The promoter used for expression was either the DSCP, or the native elav promoter. The GFP coding sequence was placed upstream of the entire extended 7.2 kb elav 3′ UTR. CPA at the proximal poly(A) produces the short 3′ UTR form of the mRNA, whereas CPA at the distal-most poly(A) produces the fully extended transcript. An RNA probe directed against the elav extension was used to detect the extended transcript.
B,C. Left panels show projections of consecutive confocal sections of stage 13 embryos stained with antibodies against ELAV (white, in the nervous system) and Mef2 (magenta, in muscle). Ventral views; anterior is up. Middle panels: hybridization signals with the elav extension probe (green). Signal in the CNS corresponds to the endogenous elav mRNA. Panels on the right show enlarged views of the boxed regions in the left and middle panels. Background staining in muscle tissue is observed with the DSCP transgene (B, right panel), indicating little or no expression of the extended 3′ UTR. In contrast, there is significant expression of extended transcripts from the transgene containing the elav promoter (C, right panel).
D. mRNA quantification by qPCR using primer combinations detecting all transgene mRNAs (GFP) or specific to the extended transcript (extension). RNA was extracted from dissected muscle tissue in first instar larvae expressing the transgene depicted in A, carrying either the DSCP or the elav promoter. Levels were normalized to rp49 RNA. Both promoters foster robust transgene expression as indicated by GFP levels, but expression of extension sequences is only detected with the elav promoter. Error bars represent mean ± SD of six samples for each promoter. See also Fig. S3.
We also tested whether the promoter sequence from one extended gene could promote extension of the 3′ UTR of another such gene. Indeed, a GFP transgene containing the elav promoter and brat extended 3′ UTR exhibited ELAV-mediated APA (Fig. S3D–E). These observations suggest a link between transcription initiation and ELAV-mediated APA.
Promoters of extended genes contain GAGA and paused Pol II
To determine whether the promoter regions of extended genes share common sequence motifs, we examined 252 neural-specific transcripts produced by 219 different genes exhibiting 3′ UTR extensions (Smibert et al., 2012). The most significantly enriched motif is the GAGA element (P-value=1e-10), which occurs in nearly half of all extended genes (Fig. 3A and S4A). To investigate the functional significance of the GAGA element in promoters of extended genes, we tested whether 3′ UTR extension is diminished in animals lacking the GAGA-binding protein, Trithorax-like (Trl). For all six genes we examined, the ratio between extension sequences and coding sequences was reduced between 15 and 75% in Trl mutant flies (Fig. 3B). These observations suggest that the GAGA motifs in the promoters of extended genes are important for proper 3′ UTR extension.
The GAGA element is a motif commonly found in the promoter regions of genes containing paused Pol II. Paused Pol II is a pervasive feature of gene regulation in metazoan development and at least 10–30% of all genes in Drosophila contain paused Pol II. It is thought that paused promoters are poised for rapid activation and thereby exhibit synchronous induction in the different cells of a tissue (e.g., (Boettiger and Levine, 2009)). Another function of promoter pausing might be to ensure proper recruitment of essential factors for RNA elongation and processing (Adelman and Lis, 2012).
We found that most extended genes contain paused Pol II, based on whole genome Pol II ChIP-Seq assays (Negre et al., 2011). Some extended genes express both short and long isoforms from the same promoter (for example elav, Fig. 3C), while others (e.g., brat) employ different promoters for the different isoforms. In the latter case, only the promoter driving the extended isoform contains paused Pol II (Fig. 3C′).
To determine whether paused Pol II might be associated with the formation of 3′ UTR extensions, we compared the overall Pol II pausing index (PI) of extended genes and various control genes. We found that extended transcripts are derived from significantly more paused (PI=8.58) promoters than any of the control groups, including neural-specific (but non-extended) genes (PI=5.75) (Fig. 3D and 4D). Thus, there is a clear association between Pol II pausing and 3′ UTR extension, which transcends the general pausing seen for neural-specific gene expression. Extended transcripts are also strongly paused in muscle cells (Fig. S4B; PI=7.97) where they are not actively transcribed and where ELAV is not expressed (Gaertner et al., 2012). Thus, Pol II pausing at extended genes occurs independently of ELAV.
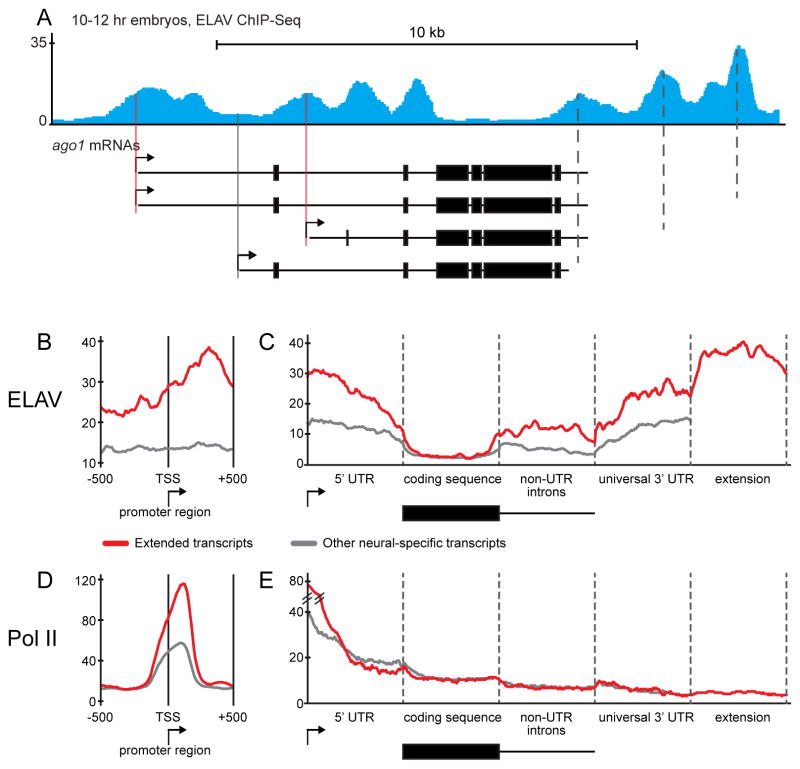
Figure 4. ELAV binds to promoter regions of extended genes.
A. Normalized ELAV ChIP-Seq reads at the ago1 locus in 10–12 hr embryos. Shown is a merged track of duplicate experiments. ELAV is found at the promoters of the extended ago1 isoforms (red lines) but not the shortest 3′ UTR form (grey line). There are peaks of ELAV binding at each proximal poly(A) site (dotted lines) where it suppresses CPA. The coding region is notably devoid of ELAV binding.
B. Meta-gene plots of ELAV ChIP-Seq datasets at the promoter region (±500 bp relative to the start site) in 10–12 hr embryos. Promoter regions of extended transcripts show significantly higher ELAV binding than other neural-specific transcripts (Wilcoxon test P-value = 1.3e-9).
C. Meta-gene analysis of ELAV binding across the entire transcription unit in 10–12 hr embryos. ELAV binding is higher in extended transcripts at the 5′ UTR, introns and the 3′ UTR compared with other neural-specific transcripts. In all genes, ELAV binding is excluded from the coding sequence.
D,E. Meta-gene analysis of Pol II binding at the promoter region (D) or across the entire transcription unit (E) in 12–16 hr embryos. Promoter regions of extended transcripts show significantly higher Pol II binding than other neural-specific transcripts. Other regions do not differ in their Pol II binding profile between the two groups. See also Tables S1 and S2 and Fig. S4.
ELAV binds to the promoter regions of extended genes
The preceding analyses raise the possibility that ELAV is selectively recruited to the promoter regions of extended genes. To test this hypothesis, we performed ChIP-Seq assays using anti-ELAV antibodies. ELAV is an RNA-binding protein that directly binds and inhibits proximal poly(A) elements of target transcripts (Hilgers et al., 2012). We therefore reasoned that it should be possible to identify the genome-wide distribution of ELAV by crosslinking ELAV/RNA complexes to associated DNA templates. ELAV ChIP-Seq assays were conducted with nuclei obtained from 6–8 hr and 10–12 hr embryos. These stages were selected based on our previous observations regarding the timing of 3′ extensions in the nervous system (Hilgers et al., 2011).
We identified 6879 genomic regions bound by ELAV in 6–8 hr embryos (Table S1) and 8076 regions in 10–12 hr embryos (Table S2). There is a striking enrichment of ELAV in the promoter regions of extended genes. For example, argonaute1 (ago1) produces multiple APA isoforms driven from three different promoters. The two promoters that produce extended transcripts display ELAV peaks, whereas the promoter that expresses the short (ubiquitous) isoform does not (Fig. 4A and S4C, filled lines). High levels of ELAV are also found at 3′ poly(A) sites (Fig. 4A and S4C, dotted lines), consistent with previous RIP assays (Hilgers et al., 2012).
We combined the ChIP-Seq data into a ‘meta-gene’ plot that provides simple visualization of key sites of ELAV binding (Fig. 4B–C and S4D–E, see Experimental Procedures). There is a significant enrichment of ELAV at the promoter regions of extended genes as compared with neural-specific non-extended genes (Fig. 4B, Wilcoxon test P-value = 1.3e-9). A distinct ELAV peak is seen near the transcription start site, although ELAV binding continually increases across the 5′ UTR and peaks at ~300 bp downstream of the start site.
ELAV not only binds to promoter regions, but also to 3′ UTRs and introns of extended genes. ELAV is strikingly depleted from coding sequences. As expected, binding markedly increases in the vicinity of proximal poly(A) sites and remains high across extended regions where there are additional poly(A) elements (Fig. 4C and S4E).
We also performed a meta-gene analysis of previously published Pol II ChIP-Seq data (Negre et al., 2011). Pol II binding is highly enriched in the promoter regions of extended genes, which is consistent with our earlier evidence that such genes tend to contain paused Pol II (Fig. 4D and S4F). The Pol II binding profile did not otherwise differ from non-extended neural-specific genes (Fig. 4E and S4G). It is possible that ELAV binds to both nascent transcripts and associated DNA templates since ELAV is usually detected at distal poly(A) sites of extended genes prior to full transcriptional extension (e.g., Fig. S4C).
We have presented evidence that paused Pol II fosters selective recruitment of ELAV and coordinates expression of extended 3′ UTR sequences during neurogenesis. The basis for selective recruitment of ELAV is a bit of a mystery since it has been shown to interact with broadly distributed low-complexity RNA sequences (e.g., U-rich). Increased interaction between paused promoters and termination regions might help promote 3′ extension, for example by bringing ELAV to the promoter via gene looping (Henriques et al., 2012; O’Sullivan et al., 2004; Tan-Wong et al., 2012). The observed association of ELAV with the paused promoter regions of extended genes provides a foundation for selectivity and also strengthens the link between transcription initiation and 3′ cleavage (Hsin and Manley, 2012). It is improbable that paused Pol II is sufficient for recruitment of ELAV since not all paused genes exhibit APA. It is therefore likely that additional sequence elements, for example in extended 3′ UTRs, are essential for recruitment. ELAV proteins are highly conserved and it is easy to imagine that the regulation of 3′ extension in the vertebrate CNS depends on selective promoter recruitment as seen in Drosophila.
EXPERIMENTAL PROCEDURES
Plasmids and fly strains
Flies were cultured on standard medium and crosses were performed at 25°C. Trl mutants had the genotype Trl62/Trl67. Mef2-Gal4 and elav-Gal4 strains were obtained from the Bloomington Stock Center. Trl62 and Trl67 flies were provided by Paul Schedl. GFP reporter plasmids were constructed by inserting the eGFP coding sequence BglII/NotI into pBID-UASc (Wang et al., 2012). Native promoter sequences (300–350 bp surrounding the TSS) were amplified from fly genomic DNA and cloned into pBID-UASc-eGFP SacI/BglII, thus removing the DSC promoter and maintaining the UAS repeats. Extended 3′ UTR sequences were amplified from genomic DNA and cloned into the modified pBID-UASc-eGFP NotI/XbaI. Extension sequences lacking the short 3′ UTR including the proximal poly(A) were cloned in the same way. In those constructs, additional proximal poly(A) signals present in the extension sequences were mutated from AATAAA into AACAAA. Constructs were injected and transgenic flies were generated using targeted integration. Primer sequences are available in the Supplemental Information.
In situ hybridization and immunocytochemistry
Embryos were collected, fixed and hybridized with riboprobes according to standard protocols. Detection of RNA probes was carried out with anti-digoxigenin and anti-biotin primary antibodies (Roche) and fluorescent secondary antibodies (Molecular Probes). Rat anti-ELAV-7E8A10 was obtained from the Developmental Studies Hybridoma Bank (DSHB), and rabbit anti-DMef2 was a gift from Bruce Paterson. Confocal imaging was performed on a Zeiss LSM 700 microscope. Colocalizing GFP foci were manually counted in confocal images. Approximately 150 GFP foci were assessed per embryo for at least six embryos per experiment.
RNA quantification
Total RNA was extracted from dissected first instar larval muscle tissue (Fig. 2D) or dissected third instar larval brains (Fig. 3B) using TRIzol (Invitrogen). DNase treatment and reverse transcription used the QuantiTect Reverse Transcription Kit (Qiagen). qPCR was performed on a 1:20 dilution of the samples and monitored in a Viia7 real-time PCR system using SYBR Green reagents (Applied Biosystems). Primer sequences are available on request.
Chromatin Immunoprecipitation and sequencing
ChIP-Seq from Drosophila embryos was performed essentially as described in (Oktaba et al., 2008) with modifications as described in the Supplemental Information. ChIP-Seq libraries were constructed with the NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (NEB) using NEBNext Multiplex Oligos for Illumina (NEB). ChIP and input DNA libraries were single-end sequenced with 50 bp reads using an Illumina HiSeq2000 instrument by the Functional Genomics Laboratory at the University of California at Berkeley. Data were processed as described in the Supplemental Information.
Computational analysis of promoters of extended transcripts
Known nervous system specific extended transcripts and control groups of transcripts were defined and filtered as described in the Supplemental Information. Enriched sequence motifs in the promoters of 3′ extended genes were identified using HOMER software (Heinz et al., 2010). A region ±200 bp relative to the TSS was searched and all other annotated gene promoters were used as the background set. Pausing indexes were determined as described in the Supplemental Information.
Meta-gene analysis
ELAV ChIP-Seq data from two biological replicates in 10–12 hr embryos and Pol II ChIP-Seq data in 12–16 hr embryos (Negre et al., 2011) were used for this analysis. Enriched ELAV and Pol II binding regions were identified as described in the Supplemental Information. All the reads outside the ELAV or Pol II binding regions were filtered out, respectively. Each of the following 6 gene body regions was divided into 100 windows: (1) promoter (TSS ±500 bp), (2) 5′ UTR, (3) coding sequence, (4) non-UTR introns, (5) universal 3′ UTR, and (6) 3′ UTR extension. The filtered reads were mapped to these regions and reads per kb per million mapped reads (RPKM) were calculated for each window. Meta-gene plots were smoothened by using the moving average of 7 windows.
Analysis of ELAV binding at promoter regions
ELAV binding at promoter regions was calculated as the ELAV enrichment over background (input DNA), averaged between two biological replicates, within ±500 bp relative to the TSS. ELAV binding at 252 promoters of 3′ UTR extended transcripts was compared to 1219 promoters of non-extended neural-specific transcripts using the Wilcoxon rank sum test.
Supplementary Material
Figure S1. Transgene properties that promote expression of 3′ extensions. Related to Figure 1.
A-B. Native promoters are required for expression of 3′ extensions.
A. elav-Gal4 drives expression of a GFP transgene in the nervous system. The promoter used for expression was the DSCP. The GFP coding sequence was placed upstream of the entire extended 7.2 kb elav 3′ UTR. CPA at the proximal poly(A) produces the short 3′ UTR form of the mRNA, whereas CPA at the distal-most poly(A) produces the fully extended transcript. RNA probes directed against different regions of the transcripts were used to detect mRNAs. Shown are double fluorescent in situ hybridization assays. Single confocal sections of a portion of the developing CNS in stage 13 embryos. Colocalization of the GFP and 3′ UTR probes indicates expression of the short transcript from the transgene.
B. Virtually all foci from the elav extension probe colocalize with the probe directed against the endogenous elav coding sequence, which indicates that the extension signal originates from the endogenous elav transcript. Numbers represent mean ± SD of six embryos for each sample.
C–D. Bypassing the requirement for ELAV recruitment allows for transcription of extension sequences from the DSCP.
Mef2-Gal4 drives expression of GFP transgenes in muscle cells. The promoter used for expression was the DSCP. The GFP coding sequence was placed upstream of the extended portion of the elav 3′ UTR (C) or the extended portion of the brat 3′ UTR (D), thereby excluding the respective short 3′ UTRs and proximal poly(A) signals. CPA at the indicated poly(A) produces a transcript that was detected using RNA probes directed against the GFP coding sequence (C) as well as a distal region of the elav (C) or brat (D) 3′ UTR extension. Shown are double fluorescent in situ hybridization assays combined with antibody staining against Mef2 protein as a muscle marker. Projections of consecutive confocal sections of stage 13 embryos. Ventral views; anterior is up.
C. The GFP mRNA signal in muscle cells shows muscle-specific expression of the transgene. Signal from the elav extension probe in the central nervous system (CNS) corresponds exclusively to the endogenous extended elav transcript. Detection of extension sequences in muscle cells indicates expression of extended transcripts from the transgene.
D. Signal from the brat 3′ UTR (that is not present in the transgene mRNA) and brat extension probes in the CNS corresponds exclusively to endogenous transcripts. Detection of extension sequences in muscle cells indicates expression of extended transcripts from the transgene.
Figure S2. Native promoters are required for expression of 3′ extensions. Related to Figure 1.
A. elav-Gal4 drives expression of a GFP transgene in the nervous system. Three different promoter regions were used: DSCP (B), the native promoter producing the short form of brat (C), and the native promoter producing the extended form of brat (D). A′ depicts the configuration of endogenous extended and short brat mRNAs with their respective promoters. The GFP coding sequence was placed upstream of the entire extended 8.5 kb brat 3′ UTR. CPA at the proximal poly(A) produces the short 3′ UTR form of the mRNA, whereas CPA at the distal-most brat poly(A) produces the fully extended transcript. RNA probes directed against different regions of the transcripts were used to detect mRNAs.
B–D. Double fluorescent in situ hybridization assays using probes indicated in A. Single confocal sections of a portion of the developing CNS in stage 13 embryos. Note that the extension probe detects not only the transgene, but also the endogenous brat transcript, which is expressed in the nervous system. Colocalization of the GFP and extension probes indicates expression of extended transcripts from the transgene.
B,C. The reporter transgenes carrying the DSCP (B) or the native promoter of the short form of brat (C) do not exhibit colocalization of GFP and extension probes. Extension signals (magenta arrows in merged image) do not colocalize with the green GFP signals, indicating that they correspond to endogenous brat mRNAs.
D. Replacing the DSCP with the native promoter producing the extended form of brat induces 3′ extension of the GFP transgene. There is extensive colocalization of the GFP (green arrows) and extension probes (magenta arrows), indicating expression of extension sequences from the transgene (white arrows in merged image). Non-colocalizing GFP signal (e.g., green arrow in merged image) corresponds to the short transgene, and non-colocalizing signal from the extension probe (e.g., magenta arrow in merged image) corresponds to the endogenously expressed extended brat mRNA. Numbers represent mean ± SD of six embryos for each promoter (except C: three embryos).
Figure S3. The native brat and elav promoters mediate brat 3′ UTR extension. Related to Figure 2.
A-C: The native brat promoter mediates 3′ UTR extension in ectopic tissues.
A: Mef2-Gal4 drives expression of a GFP transgene in muscle cells. The promoter used for expression was either the DSCP, or the native brat promoter. The GFP coding sequence was placed upstream of the entire extended 8.5 kb brat 3′ UTR. CPA at the proximal poly(A) produces the short 3′ UTR form of the mRNA, whereas CPA at the distal-most poly(A) produces the fully extended transcript. RNA probes directed against different regions of the transcripts were used to detect mRNAs.
B,C. Double fluorescent in situ hybridization assays using probes indicated in A, combined with antibody staining against ELAV protein. Projections of consecutive confocal sections of stage 13 embryos. Lateral views; anterior is up. The weak ELAV signal in muscle cells corresponds to ectopic expression driven by Mef2-Gal4, whereas the strong signal in the CNS corresponds to endogenous ELAV. Arrowheads indicate neurons of the PNS (strong ELAV signal). Ectopic expression of the short 3′ UTR GFP transgene can be achieved from both DSC and brat promoters, as shown by detection of GFP probe signal in muscle cells in B and C (middle panels, magenta). Right panels exhibit hybridization signals with the brat extension probe (green). Signal in the CNS corresponds exclusively to the endogenous extended brat transcript, whereas expression in the muscle corresponds exclusively to reporter expression. Background staining in muscle is observed with the DSCP transgene (B), indicating little or no expression of the extended 3′ UTR from the GFP transgene. In contrast, there is significant expression of extended transcripts from the transgene containing the brat promoter in muscle (C).
D,E. The native elav promoter mediates brat 3′ UTR extension.
D. elav-Gal4 drives expression of a GFP transgene in the nervous system. The promoter used for expression was the native elav promoter. The GFP coding sequence was placed upstream of the entire extended 8.5 kb brat 3′ UTR. CPA at the proximal poly(A) produces the short 3′ UTR form of the mRNA, whereas CPA at the distal-most poly(A) produces the fully extended transcript. RNA probes directed against different regions of the transcripts were used to detect mRNAs.
E. Double fluorescent in situ hybridization assays using probes indicated in D. Single confocal sections of a portion of the developing CNS in a stage 13 embryo. Note that the extension probe detects not only the transgene, but also the endogenous brat transcript, which is expressed in the nervous system. There is extensive colocalization of the GFP (green arrows) and extension probes (magenta arrows), indicating expression of extended 3′ UTR sequences from the transgene (white arrows in merged image). Non-colocalizing GFP signal (e.g., green arrow in merged image) corresponds to the short transgene, and non-colocalizing signal from the extension probe (e.g., magenta arrow in merged image) corresponds to the endogenously expressed extended brat mRNA. Numbers represent mean ± SD of six embryos.
Figure S4. Promoters of extended genes contain the GAGA motif and paused Pol II and are bound by ELAV. Related to Figure 3 and Figure 4.
A. Frequency of occurrence and distribution of identified GAGA motifs in promoters of extended or control genes relative to the TSS. GAGA motifs are most often located between -100 bp and the TSS in both groups of promoters and occur significantly more frequently in promoters of extended genes.
B. Pausing index (PI) distribution and median pausing index values of the promoters of the indicated groups of transcripts in muscle tissues (see Supplemental Experimental Procedures), where ELAV is absent. The numbers in parentheses denote the number of transcripts in each group. Promoters of extended transcripts are significantly more paused than promoters of any control group. Wilcoxon rank sum test P-values were calculated by comparing the pausing index of extended transcripts with each group of controls.
C. Normalized ELAV ChIP-Seq reads at the ago1 locus in 6–8 hr and 10–12 hr embryos. Shown are merged tracks of duplicate experiments. ELAV peaks at each proximal poly(A) site (dotted lines) are found in both 6–8 hr and 10–12 hr embryos.
D,E. Meta-gene plots of ELAV ChIP-Seq datasets at the promoter region (D) (±500 bp relative to the TSS) or across the entire transcription unit (E) in 10–12 hr embryos. Each line (meta-gene) averages the ChIP-Seq data of all indicated transcripts. ELAV binding is higher in extended transcripts compared to other transcripts (see exception below) at the promoter region, 5′ UTR, introns and the 3′ UTR. In all genes, ELAV binding is excluded from the coding sequence. Differences in ELAV binding between extended transcripts and ‘other isoforms of extended genes’ are not significant. We think the reason is that transcripts from these two groups share many gene regions including sequences as close as ±100 bp relative to the TSS, introns and the universal 3′ UTR. Moreover, both groups of transcripts are relatively small (252 and 187 transcripts, respectively).
F,G. Meta-gene analysis of Pol II binding at promoter region (F) or across the entire transcription unit (G) in 12–16 hr embryos. Promoter regions of extended transcripts show significantly higher Pol II binding than other control groups of transcripts. Other regions downstream of the TSS do not differ in their Pol II binding profile between the four groups, except the “other neuronal active transcripts” that show higher Pol II binding at the 5′ UTR and the coding sequence due to their high level of expression. See also Table S1 and Table S2 for ELAV peak coordinates.
Listing of chromosomal coordinates (UCSC dm3 release) of 6879 ELAV binding peaks in 6–8 hr embryos identified by ChIP-Seq.
Listing of chromosomal coordinates (UCSC dm3 release) of 8076 ELAV binding peaks in 10–12 hr embryos identified by ChIP-Seq.
Acknowledgments
We thank James Manley for critical reading of the manuscript. V.H. is supported by a fellowship from the German Research Foundation (DFG HI 1552/3–1). K.O. is supported by a fellowship from the European Molecular Biology Organization (EMBO ALTF 492–2011). This study was funded by a grant from the NIH (GM34431). This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303.
Footnotes
AUTHOR CONTRIBUTIONS
V.H. designed, performed and analyzed experiments and wrote the paper. K.O. performed and analyzed experiments. W.Z. analyzed data. T.S.L., D.J.J., S.B.L., S.P.N. and E.E. performed experiments. M.L. designed experiments and wrote the paper.
ACCESSION NUMBER
ChIP-Seq data has been deposited in the GEO database under GSE63323.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen PK, Jensen TH, Lykke-Andersen S. Making ends meet: coordination between RNA 3′-end processing and transcription initiation. Wiley interdisciplinary reviews. RNA. 2013;4:233–246. doi: 10.1002/wrna.1156. [DOI] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, Booth BW, Wen J, Park S, Suzuki AM, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo O, Manley JL. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Molecular cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- Elkon R, Drost J, van Haaften G, Jenal M, Schrier M, Vrielink JA, Agami R. E2F mediates enhanced alternative polyadenylation in proliferation. Genome biology. 2012;13:R59. doi: 10.1186/gb-2012-13-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nature reviews Genetics. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- Gaertner B, Johnston J, Chen K, Wallaschek N, Paulson A, Garruss AS, Gaudenz K, De Kumar B, Krumlauf R, Zeitlinger J. Poised RNA polymerase II changes over developmental time and prepares genes for future expression. Cell reports. 2012;2:1670–1683. doi: 10.1016/j.celrep.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nature structural & molecular biology. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T, Ji Z, Tan-Wong SM, Carmo AM, Tian B, Proudfoot NJ, Moreira A. Transcription termination between polo and snap, two closely spaced tandem genes of D. melanogaster. Transcription. 2012;3:198–212. doi: 10.4161/trns.21967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Lemke SB, Levine M. ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes & development. 2012;26:2259–2264. doi: 10.1101/gad.199653.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B. Neural-specific elongation of 3′ UTRs during Drosophila development. Proceedings of the National Academy of Sciences. 2011;108:15864–15869. doi: 10.1073/pnas.1112672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes & development. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PloS one. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gilmour DS. Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. The EMBO journal. 2013;32:1829–1841. doi: 10.1038/emboj.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisbin MJ, Qiu J, White K. The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes & development. 2001;15:2546–2561. doi: 10.1101/gad.903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield KD, Keene JD. Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nucleic acids research. 2012;40:2734–2746. doi: 10.1093/nar/gkr1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3′ end processing and transcription initiation. Molecular cell. 2010;40:410–422. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome research. 2013;23:812–825. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. Molecular cell. 2011;41:409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nature genetics. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- Oktaba K, Gutierrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EE, Muller J. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Developmental cell. 2008;15:877–889. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Picao-Osorio J, Villava C, Patraquim P, Lafuente E, Aspden J, Thomsen S, Technau GM, Alonso CR. The RNA-binding protein ELAV regulates Hox RNA processing, expression and function within the Drosophila nervous system. Development. 2014;141:2046–2056. doi: 10.1242/dev.101519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Alternative polyadenylation: new insights from global analyses. Rna. 2012;18:2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, et al. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell reports. 2012;1:277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M, White K. ELAV inhibits 3′-end processing to promote neural splicing of ewg pre-mRNA. Genes & development. 2003;17:2526–2538. doi: 10.1101/gad.1106703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ. Gene loops enhance transcriptional directionality. Science. 2012;338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic acids research. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, Sive H, Bartel DP. Extensive alternative polyadenylation during zebrafish development. Genome research. 2012;22:2054–2066. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Beck ES, McCabe BD. A modular toolset for recombination transgenesis and neurogenetic analysis of Drosophila. PloS one. 2012;7:e42102. doi: 10.1371/journal.pone.0042102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao KM, White K. Neural specificity of elav expression: defining a Drosophila promoter for directing expression to the nervous system. Journal of neurochemistry. 1994;63:41–51. doi: 10.1046/j.1471-4159.1994.63010041.x. [DOI] [PubMed] [Google Scholar]
- Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, Ansari AZ. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. The Journal of biological chemistry. 2012;287:8541–8551. doi: 10.1074/jbc.M111.335687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome biology. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Transgene properties that promote expression of 3′ extensions. Related to Figure 1.
A-B. Native promoters are required for expression of 3′ extensions.
A. elav-Gal4 drives expression of a GFP transgene in the nervous system. The promoter used for expression was the DSCP. The GFP coding sequence was placed upstream of the entire extended 7.2 kb elav 3′ UTR. CPA at the proximal poly(A) produces the short 3′ UTR form of the mRNA, whereas CPA at the distal-most poly(A) produces the fully extended transcript. RNA probes directed against different regions of the transcripts were used to detect mRNAs. Shown are double fluorescent in situ hybridization assays. Single confocal sections of a portion of the developing CNS in stage 13 embryos. Colocalization of the GFP and 3′ UTR probes indicates expression of the short transcript from the transgene.
B. Virtually all foci from the elav extension probe colocalize with the probe directed against the endogenous elav coding sequence, which indicates that the extension signal originates from the endogenous elav transcript. Numbers represent mean ± SD of six embryos for each sample.
C–D. Bypassing the requirement for ELAV recruitment allows for transcription of extension sequences from the DSCP.
Mef2-Gal4 drives expression of GFP transgenes in muscle cells. The promoter used for expression was the DSCP. The GFP coding sequence was placed upstream of the extended portion of the elav 3′ UTR (C) or the extended portion of the brat 3′ UTR (D), thereby excluding the respective short 3′ UTRs and proximal poly(A) signals. CPA at the indicated poly(A) produces a transcript that was detected using RNA probes directed against the GFP coding sequence (C) as well as a distal region of the elav (C) or brat (D) 3′ UTR extension. Shown are double fluorescent in situ hybridization assays combined with antibody staining against Mef2 protein as a muscle marker. Projections of consecutive confocal sections of stage 13 embryos. Ventral views; anterior is up.
C. The GFP mRNA signal in muscle cells shows muscle-specific expression of the transgene. Signal from the elav extension probe in the central nervous system (CNS) corresponds exclusively to the endogenous extended elav transcript. Detection of extension sequences in muscle cells indicates expression of extended transcripts from the transgene.
D. Signal from the brat 3′ UTR (that is not present in the transgene mRNA) and brat extension probes in the CNS corresponds exclusively to endogenous transcripts. Detection of extension sequences in muscle cells indicates expression of extended transcripts from the transgene.
Figure S2. Native promoters are required for expression of 3′ extensions. Related to Figure 1.
A. elav-Gal4 drives expression of a GFP transgene in the nervous system. Three different promoter regions were used: DSCP (B), the native promoter producing the short form of brat (C), and the native promoter producing the extended form of brat (D). A′ depicts the configuration of endogenous extended and short brat mRNAs with their respective promoters. The GFP coding sequence was placed upstream of the entire extended 8.5 kb brat 3′ UTR. CPA at the proximal poly(A) produces the short 3′ UTR form of the mRNA, whereas CPA at the distal-most brat poly(A) produces the fully extended transcript. RNA probes directed against different regions of the transcripts were used to detect mRNAs.
B–D. Double fluorescent in situ hybridization assays using probes indicated in A. Single confocal sections of a portion of the developing CNS in stage 13 embryos. Note that the extension probe detects not only the transgene, but also the endogenous brat transcript, which is expressed in the nervous system. Colocalization of the GFP and extension probes indicates expression of extended transcripts from the transgene.
B,C. The reporter transgenes carrying the DSCP (B) or the native promoter of the short form of brat (C) do not exhibit colocalization of GFP and extension probes. Extension signals (magenta arrows in merged image) do not colocalize with the green GFP signals, indicating that they correspond to endogenous brat mRNAs.
D. Replacing the DSCP with the native promoter producing the extended form of brat induces 3′ extension of the GFP transgene. There is extensive colocalization of the GFP (green arrows) and extension probes (magenta arrows), indicating expression of extension sequences from the transgene (white arrows in merged image). Non-colocalizing GFP signal (e.g., green arrow in merged image) corresponds to the short transgene, and non-colocalizing signal from the extension probe (e.g., magenta arrow in merged image) corresponds to the endogenously expressed extended brat mRNA. Numbers represent mean ± SD of six embryos for each promoter (except C: three embryos).
Figure S3. The native brat and elav promoters mediate brat 3′ UTR extension. Related to Figure 2.
A-C: The native brat promoter mediates 3′ UTR extension in ectopic tissues.
A: Mef2-Gal4 drives expression of a GFP transgene in muscle cells. The promoter used for expression was either the DSCP, or the native brat promoter. The GFP coding sequence was placed upstream of the entire extended 8.5 kb brat 3′ UTR. CPA at the proximal poly(A) produces the short 3′ UTR form of the mRNA, whereas CPA at the distal-most poly(A) produces the fully extended transcript. RNA probes directed against different regions of the transcripts were used to detect mRNAs.
B,C. Double fluorescent in situ hybridization assays using probes indicated in A, combined with antibody staining against ELAV protein. Projections of consecutive confocal sections of stage 13 embryos. Lateral views; anterior is up. The weak ELAV signal in muscle cells corresponds to ectopic expression driven by Mef2-Gal4, whereas the strong signal in the CNS corresponds to endogenous ELAV. Arrowheads indicate neurons of the PNS (strong ELAV signal). Ectopic expression of the short 3′ UTR GFP transgene can be achieved from both DSC and brat promoters, as shown by detection of GFP probe signal in muscle cells in B and C (middle panels, magenta). Right panels exhibit hybridization signals with the brat extension probe (green). Signal in the CNS corresponds exclusively to the endogenous extended brat transcript, whereas expression in the muscle corresponds exclusively to reporter expression. Background staining in muscle is observed with the DSCP transgene (B), indicating little or no expression of the extended 3′ UTR from the GFP transgene. In contrast, there is significant expression of extended transcripts from the transgene containing the brat promoter in muscle (C).
D,E. The native elav promoter mediates brat 3′ UTR extension.
D. elav-Gal4 drives expression of a GFP transgene in the nervous system. The promoter used for expression was the native elav promoter. The GFP coding sequence was placed upstream of the entire extended 8.5 kb brat 3′ UTR. CPA at the proximal poly(A) produces the short 3′ UTR form of the mRNA, whereas CPA at the distal-most poly(A) produces the fully extended transcript. RNA probes directed against different regions of the transcripts were used to detect mRNAs.
E. Double fluorescent in situ hybridization assays using probes indicated in D. Single confocal sections of a portion of the developing CNS in a stage 13 embryo. Note that the extension probe detects not only the transgene, but also the endogenous brat transcript, which is expressed in the nervous system. There is extensive colocalization of the GFP (green arrows) and extension probes (magenta arrows), indicating expression of extended 3′ UTR sequences from the transgene (white arrows in merged image). Non-colocalizing GFP signal (e.g., green arrow in merged image) corresponds to the short transgene, and non-colocalizing signal from the extension probe (e.g., magenta arrow in merged image) corresponds to the endogenously expressed extended brat mRNA. Numbers represent mean ± SD of six embryos.
Figure S4. Promoters of extended genes contain the GAGA motif and paused Pol II and are bound by ELAV. Related to Figure 3 and Figure 4.
A. Frequency of occurrence and distribution of identified GAGA motifs in promoters of extended or control genes relative to the TSS. GAGA motifs are most often located between -100 bp and the TSS in both groups of promoters and occur significantly more frequently in promoters of extended genes.
B. Pausing index (PI) distribution and median pausing index values of the promoters of the indicated groups of transcripts in muscle tissues (see Supplemental Experimental Procedures), where ELAV is absent. The numbers in parentheses denote the number of transcripts in each group. Promoters of extended transcripts are significantly more paused than promoters of any control group. Wilcoxon rank sum test P-values were calculated by comparing the pausing index of extended transcripts with each group of controls.
C. Normalized ELAV ChIP-Seq reads at the ago1 locus in 6–8 hr and 10–12 hr embryos. Shown are merged tracks of duplicate experiments. ELAV peaks at each proximal poly(A) site (dotted lines) are found in both 6–8 hr and 10–12 hr embryos.
D,E. Meta-gene plots of ELAV ChIP-Seq datasets at the promoter region (D) (±500 bp relative to the TSS) or across the entire transcription unit (E) in 10–12 hr embryos. Each line (meta-gene) averages the ChIP-Seq data of all indicated transcripts. ELAV binding is higher in extended transcripts compared to other transcripts (see exception below) at the promoter region, 5′ UTR, introns and the 3′ UTR. In all genes, ELAV binding is excluded from the coding sequence. Differences in ELAV binding between extended transcripts and ‘other isoforms of extended genes’ are not significant. We think the reason is that transcripts from these two groups share many gene regions including sequences as close as ±100 bp relative to the TSS, introns and the universal 3′ UTR. Moreover, both groups of transcripts are relatively small (252 and 187 transcripts, respectively).
F,G. Meta-gene analysis of Pol II binding at promoter region (F) or across the entire transcription unit (G) in 12–16 hr embryos. Promoter regions of extended transcripts show significantly higher Pol II binding than other control groups of transcripts. Other regions downstream of the TSS do not differ in their Pol II binding profile between the four groups, except the “other neuronal active transcripts” that show higher Pol II binding at the 5′ UTR and the coding sequence due to their high level of expression. See also Table S1 and Table S2 for ELAV peak coordinates.
Listing of chromosomal coordinates (UCSC dm3 release) of 6879 ELAV binding peaks in 6–8 hr embryos identified by ChIP-Seq.
Listing of chromosomal coordinates (UCSC dm3 release) of 8076 ELAV binding peaks in 10–12 hr embryos identified by ChIP-Seq.