Abstract
Natural killer (NK) and T lymphocytes share many properties, yet only NK cells respond rapidly to infection and cancer without pre-activation. We found that few microRNAs (miRNAs) differed significantly between human NK and T cells. Among those miRNAs, miR-181a, and miR-181b levels rose during NK cell differentiation. Prior studies indicate that miR-181a and miR-181b are critical for human NK cell development and are co-transcribed from genes on chromosome 1 (MIR181A1B1) and on chromosome 9 (MIR181A2B2). We mapped human MIR181A1B1 and MIR181A2B2 transcription start sites (TSS) to 78.3 kb and 34.0 kb upstream of the mature miRNAs, generating predominantly unspliced transcripts of 80-127 kb and ~60 kb, respectively. Unlike mouse thymocytes, human T cells expressed both MIR181A1B1 and MIR181A2B2. We tested the hypothesis that NK cells differentially transcribe the two genes during development and in response to immune regulatory cytokines. During NK cell differentiation, MIR181A2B2 expression rose dramatically and exceeded that of MIR181A1B1. TGF-β treatment increased NK cell MIR181A2B2 transcription, while IL-2, IL-15, and IL-12/IL-18 treatments upregulated MIR181A1B1. The MIR181A2B2 promoter was strongly transactivated by SMAD3 and SMAD4 transcription factors, suggesting that TGF-β signaling upregulates MIR181A2B2 expression, at least in part, through SMAD-dependent promoter activation.
INTRODUCTION
NK cell deficiency is associated with increased susceptibility to infections and cancer 1, 2. In contrast to T cells, NK cells rapidly kill infected and transformed cells without pre-activation 1, 2. They contain preformed cytotoxic granules that enclose perforin and various granzymes 1. Upon activation, NK cells rapidly exocytose these granules that kill target cells. NK cells also secrete various chemokines and cytokines, including IFN-γ, that directly affect infected and transformed cells and promote adaptive immune responses 1, 2.
Human NK cells are classified into two major subsets based on their CD56 surface expression. Most circulating blood NK cells are CD56dim, while 5-15% are CD56bright 1. CD56bright NK cells are poorly cytotoxic, but secrete high levels of cytokines and chemokines in response to inflammatory cytokines, such as IL-12 and IL-18 1. Although they respond poorly to inflammatory cytokines, CD56dim NK cells kill target cells and secrete chemokines and cytokines in response to antibody-coated cells and tumor cells 1, 3. CD56bright NK cells develop into CD56dim NK cells, and CD56dim NK cells continue to mature into subsets distinguished by CD94 expression, with CD56dimCD94hi cells being less mature than CD56dimCD94low cells 1, 4.
microRNAs (miRNA) constitute a class of ~22 nucleotide (nt) non-coding RNAs that bind to complementary sequences in the 3’ untranslated region of target mRNAs, decreasing mRNA stability and translation 5, 6. miRNAs are differentially expressed in hematopoietic tissue in which they regulate immune cell development, activation, function, and selection 5, 6. The human NK cell miRNA transcriptome has been characterized 7, 8, but to our knowledge, no one has further studied the miRNAs that are preferentially expressed in NK cells compared with the developmentally similar T cells. Although both NK cells and T cells are known to be heterogeneous, we thought that comparison of these two lymphocyte subsets would be a useful starting point.
NK cells share many developmental pathways and gene expression programs with T cells, but NK cells differ from most T cells in their gene expression programs, their ability to kill tumor cells and virus-infected cells in the absence of specific immunization, and their state of “constant readiness” 2, 9-11. Global miRNA processing defects affect NK cell maturation, function, and survival 12, 13. We found that miR-181a and miR-181b, which are important for T cell development 14, also affect NK cell maturation and activity, at least in part, by regulating the Notch signaling pathway 15.
miRNA genes are transcribed into large primary miRNAs (pri-miRNAs), which are cropped to hairpin-containing 60-110 nt precursor miRNAs (pre-miRNAs) and exported to the cytoplasm, where they are further processed to dsRNA. Then either the 5p or 3p strand of the dsRNA is retained in the silencing complex and regulates gene expression 5, 6. The 5p/3p ratio in expressed miRNAs varies considerably with tissue and developmental stage and both strands may be expressed at significant levels in the same cell 16, 17. Mature 5p miR-181a and miR-181b are both encoded by the human chromosome 1 MIR181A1B1 gene. Interestingly, identical mature miR-181a and miR-181b also are encoded by the chromosome 9 MIR181A2B2 gene. Even though the mature 5p forms are identical, the miR-181a-1 and miR-181a-2 3p strands are moderately expressed and have different seed sequences (http://www.mirbase.org/). Moreover, the pri-miRNAs and pre-miRNAs encoded by MIR181A1B1 and MIR181A2B2 are distinct. Chen and co-workers presented evidence that pre-miR-181 transcripts have direct biological activities in addition to being precursors of mature miRNAs 18. Chen proposed that both pre-miRNA and pri-miRNA regulate gene expression 19. Therefore, although the predominant mature 5p forms of miR-181a and miR-181b encoded at human chromosomes 1 and 9 are identical, the MIR181A1B1 and MIR181A2B2 loci might have distinct biological functions.
We compared miRNA expression by human blood NK and T lymphocytes. Overall, the NK and T cell miRNA transcriptomes were very similar, but a few miRNAs differed between these lymphocyte types. Focusing on two miRNAs preferentially expressed in NK cells, we explored how miRNA levels changed. Furthermore, we identified the MIR181A1B1 and MIR181A2B2 TSS and promoters and we investigated how these loci were differentially regulated during human NK cell development and in response to immune regulatory cytokines.
RESULTS
miR-181a and miR-181b are differentially expressed in NK cells and T cells and rise during NK cell development
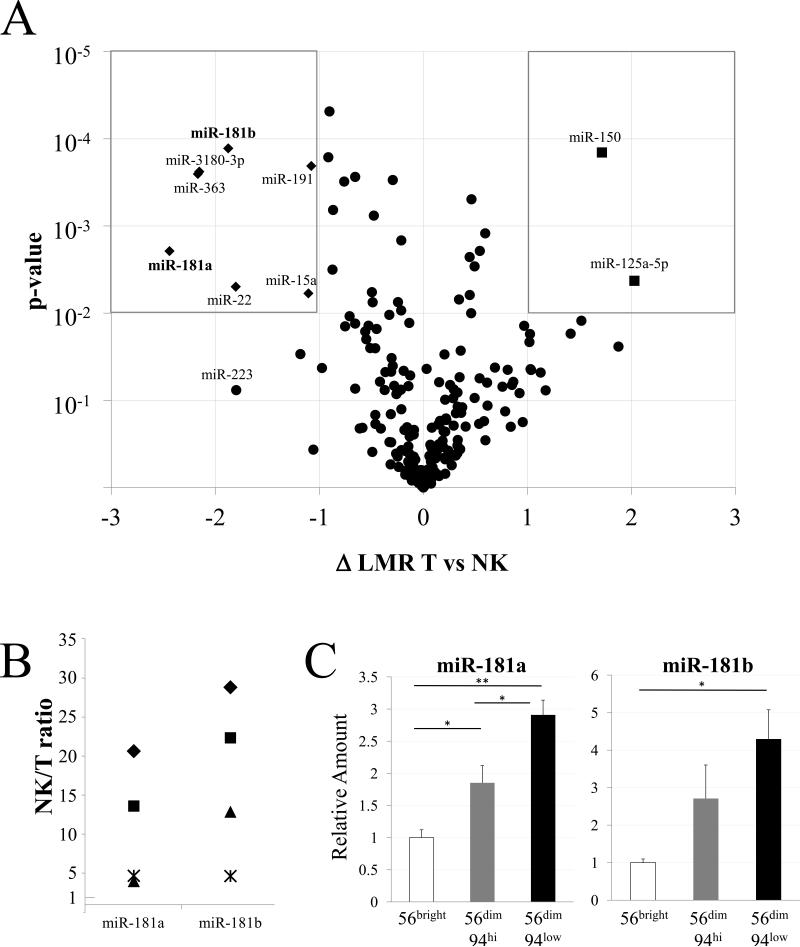
miRNA processing machinery is important for NK cell maturation, survival, and function 12, 13. We compared the NK cell miRNA transcriptome with that of extensively investigated T cells using a microarray platform. RNA extracted from NK- and T-enriched lymphocyte populations from three healthy subjects were analyzed on a miRCURY™ locked-nucleic acid array. Variations between T and NK samples were calculated as the difference between Log2 Median Ratios (ΔLMR) and were plotted against their statistical significance, shown as a “volcano” plot (Fig. 1A). miRNA expression overlapped considerably among T and NK cells, and < 4% of the miRNAs analyzed were both more than 2-fold different between T and NK cells and significant at a p-value of <0.01 (Fig. 1A boxed areas). miRNA transcriptome similarity reflects T and NK cell shared developmental pathways and functions 9-11. Among the miRNAs that are preferentially expressed in NK cells, we chose to further study miR-181a and miR-181b, which are known to regulate NK cell development 15. RT-qPCR confirmed that miR-181a, miR-181b, and other selected miRNAs were expressed at higher levels in NK cells than in T cells (Fig. 1B, Supplemental Fig. S1A). Although the ratios varied between subjects, miR-181a and miR-181b were always higher (up to 30-fold) in NK cells than in T cells (Fig. 1B).
Figure 1.
Differential miRNA expression. A. miRNA expression in NK cells and T cells. The difference in Log2 Median Ratios (ΔLMR) between NK and T cell groups is shown on the x-axis against the significance of the difference (-Log10 p-value of two-tailed T-test, corrected with Bonferroni's post-hoc test) on the y-axis. miRNAs with log2 changes of >1 (higher in T cells) or <-1 (higher in NK cells) as well as high statistical significance (P<0.01) are displayed as squares when higher in T cells (upper right) and diamonds when higher in NK cells (upper left). Note that two diamonds largely overlap. Also indicated is miR-223. B. RT-qPCR data were normalized to RNU-24 levels and calculated as NK-to-T cell ratio (1 = no difference) for the miRNAs listed on the x-axis. NK and T cell RNA was analyzed from the three subjects studied above (represented as◆, ■ and▲) and from a fourth subject (represented as*). For each miRNA, T and NK cells differed significantly (p < 0.05). C. Differential miR-181a and miR-181b expression in NK cell development. The indicated NK subsets were sorted to >95% purity and the miRNA, indicated in each panel, was quantified by RT-qPCR with RNU-24 as a standard and normalized to the level in CD56bright NK cells. Differences between developmental stages are indicated: * p < 0.05, ** p < 0.01. Data shown are Avg and SEM of cells from 3 subjects, each sorted in a separate experiment.
NK cells are heterogeneous, comprising functional and maturational subsets 1, 4. We flow cytometry sorted NK cells into three groups according to their level of maturation: CD56bright, CD56dimCD94hi and CD56dimCD94low. We then examined the amount of miR-181a and miR-181b (Fig. 1C) and other NK-enriched miRNAs (Supplemental Fig. S1B) in each NK subset using RT-qPCR. miR-181a and miR-181b were lowest in the least mature CD56bright NK cells and highest in the most mature CD56dimCD94low NK cells (Fig. 1C). The rise of miR-181a and miR-181b during late NK cell maturation extends our previous finding that levels of these miRNAs increased during development from CD34+ progenitor cells into CD56+ NK cells 15. Thus, miR-181a/b levels rise during development from primitive hematopoietic precursors to late-stage NK cells.
NK and T cells differentially regulate the MIR181A1B1 and MIR181A2B2 loci
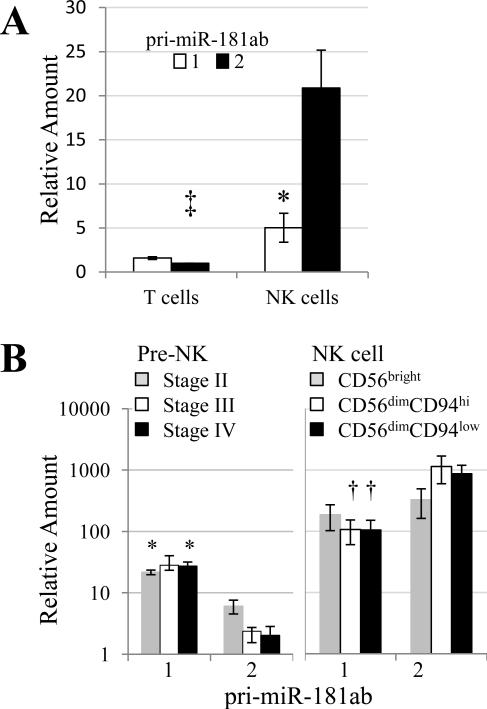
Because miR-181a and miR-181b control NK cell development 15, we asked how these miRNAs are themselves regulated. MIR181a and MIR181b are closely linked and are co-transcribed in the same pri-miRNA. miR-181 regulation is complex because human miR-181a and miR-181b are both encoded at the MIR181A1B1 locus on chromosome 1 and the MIR181A2B2 locus on chromosome 9. Mouse thymocytes express Mir181ab-1, but not Mir181ab-2 23. To explore use of MIR181ab loci in humans, we isolated blood T and NK cells and quantified pri-miR-181ab transcripts using two distinct RT-qPCR techniques, which gave virtually identical results. Unlike mouse thymocytes, freshly isolated human blood T cells expressed pri-miR-181ab-1 and primiR-181ab-2 at similar levels, with pri-miR-181ab-1 being ~1.6-fold more abundant (Fig. 2A). In contrast to both mouse thymocytes and human T cells, human NK cells expressed chromosome 9 MIR181A2B2 ~ 4-fold higher than chromosome 1 MIR181A1B1 (Fig. 2A). Compared with T cells, NK cells expressed > 20-fold more pri-miR-181ab-2 from chromosome 9 and the pri-miR-181ab-1/-2 ratio was significantly different in T cells vs. NK cells (Fig. 2A). These results show differential pri-miR-181 regulation in human blood T and NK cells and that elevated miR-181a and miR-181b levels in NK cells is due to a moderate upregulation of the chromosome 1 MIR181A1B1 locus and dramatic upregulation of the chromosome 9 MIR181A2B2 locus.
Figure 2.
MIR181A1B1 and MIR181A2B2 are regulated independently. A. Differential regulation in NK cells and T cells. RNA was assayed by RT-qPCR and quantification cycle (Cq) values were corrected with those of 18S rRNA or GAPDH, as described in Materials and Methods. The two RT-qPCR methods produced essentially identical relative amounts. Corrected Cq values were compared by t-test: pri-miR-181ab-1 vs. pri-miR-181ab-2 for NK cells (*, p < 0.03); T vs. NK cell for pri-miR-181ab-2 (‡, t < 0.02).The pri-miR-181ab-1/-2 ratio was significantly different in T cells vs. NK cells (t < 0.02). For graphic presentation, corrected Cq values were converted to numerical values and normalized to the level of T cell pri-miR-181ab-2. Avg and SEMs of 3 donors are presented. B. Differential regulation during NK cell development. Umbilical cord blood lymphocytes were sorted into Stages II-IV and adult blood lymphocytes were sorted into CD56bright, CD56dimCD94hi, and CD56dimCD94low subsets. RNA was extracted and analyzed for pri-miR-181ab-1 and pri-miR-181ab-2 levels by RT-qPCR. Shown are Avg and SEM pri-mir-181 RNA levels relative to GAPDH RNA. All values were multiplied by 10 000 for display purposes. * p < 0.05, † p < 0.01 for pri-miR-181ab-1 vs. pri-miR-181ab-2 differences.
MIR181A1B1and MIR181A2B2 expression during NK cell development and activation
We wished to trace the activity of the two MIR181ab loci during NK cell development. Caligiuri and co-workers have divided pre-NK cell development into four stages, starting with CD34+ primitive hematopoietic precursor cells and progressing through more mature stages that lose CD34 expression and eventually gain low-level CD56 expression 24. We isolated Stage II-IV cells in sufficient numbers for analysis from the umbilical cord blood of three infants, allowing us to trace pri-miR-181 levels from CD34+ precursors to CD56low/−CD94− pre-NK cells. Chromosome 1 pri-miR-181ab-1 levels did not significantly change during Stages II-IV of pre-NK cell development (Fig. 2B). In contrast, chromosome 9 pri-miR-181ab-2 transcripts were low in Stage II cells and fell to very low levels during successive pre-NK cell developmental stages. We then compared MIR181ab transcripts during mature NK cell development. To facilitate comparison with the pre-NK cells, results are presented on the same scale relative to GAPDH RNA levels. In adult subjects, the level of chromosome 1 pri-miR-181ab-1 transcript was substantially higher in CD56bright NK cells, compared with pre-NK cells (Fig. 2B). Chromosome 1 pri-miR-181ab-1 levels fell slightly during maturation into CD56dim NK cells. Chromosome 9 pri-miR-181ab-2 levels were approximately 100-fold higher in CD56bright NK cells than in pre-NK cells (Fig. 2B). The pri-miR-181ab-2 transcript appeared to rise during adult NK cell maturation and was significantly higher than the pri-miR-181ab-1 transcript in both CD56dimCD94hi and CD56dimCD94low NK cells (Fig. 2B). Although the pre-NK cell subsets isolated from cord blood and the NK cell developmental stages isolated from adult blood are not directly comparable, our results suggest that the MIR181A2B2 locus is strongly activated as cells mature from pre-NK to CD56bright.
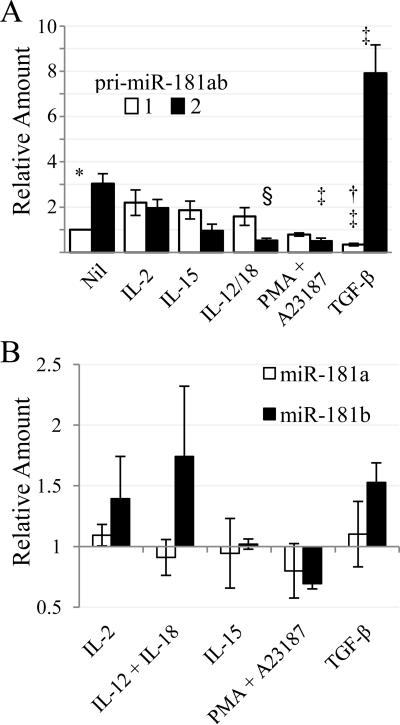
We next tested how immune regulatory cytokines affect the two MIR181ab loci. NK cells expressed relatively more of the chromosome 9 pri-miR-181ab-2 than the chromosome 1 transcript after overnight culture in culture medium alone. IL-2, IL-15, and IL-12/18 treatment tended to increase the chromosome 1 pri-miR-181ab-1 and tended to decrease the chromosome 9 pri-miR-181ab-2 so that the ratios were significantly different than those in culture medium alone (Fig. 3A). In contrast, TGF-β decreased the chromosome 1 transcript 3-fold and increased the chromosome 9 transcript 2.6-fold (Fig. 3A). TGF-β and the other stimuli did not significantly alter mature miR-181a and miR-181b levels, reflecting the opposing changes in pri-miR-181ab-1 and -2 levels (Fig. 3B). Our results show that NK cells differentially regulate pri-miR-181ab in response to cytokines. Because pri-miRNA is upstream of Drosha and other RNA processing events, we propose that pri-miR-181 levels reflect differences in MIR181A1B1 and MIR181A2B2 transcription.
Figure 3.
Primary miR-181ab-1 and miR-181ab-2 levels change in response to immune regulatory cytokines. Blood NK cells were cultured overnight in the presence of media alone (Nil) or the indicated stimulants. Avg and SEMs of 3 donors are presented. A. Cq values were corrected with those of 18S rRNA. Statistical significance is indicated: pri-miR-181ab-1 and -2 differ under the indicated conditions (*, p < 0.05; †, p < 0.01); the pri-miR level differs between media alone and the indicated stimulation (‡, p < 0.05; §, p < 0.01). Compared to media alone, the ratio of pri-miR-181ab-1 vs -2 was significantly different for all stimulations (IL-12/IL-18, p < 0.05; all others, p < 0.01). For graphic presentation, corrected Cq values were converted to numerical values and normalized to the level of pri-miR-181ab-1 (Nil). B. miRNA levels were assessed by RT-qPCR and were normalized to RNU-24 and to the unstimulated controls.
Mapping MIR181A1B1 and MIR181A2B2 transcriptional units
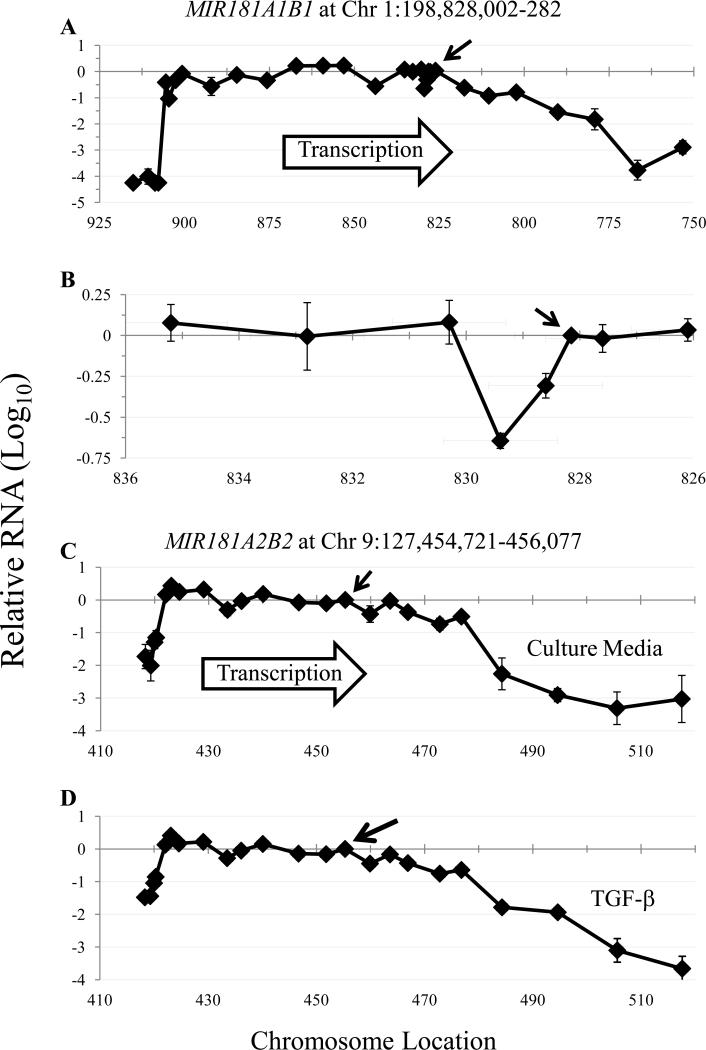
The MIR181A1B1 and MIR181A2B2 transcriptional units and their promoters have not been established, so we first performed RT-qPCR at intervals upstream and downstream of the mature miR-181 sequences to measure the extent of the MIR181ab transcriptional units. Fig. 4 shows that RT-qPCR signals were relatively constant for several kb upstream and downstream of the mature miR-181a/b sequences on both chromosomes 1 and 9 in human NK cells. Variations in RT-qPCR signal strength may be due to unequal RNA degradation rates, differences in priming efficiency, or RNA secondary structure formation affecting reverse transcription efficiency. Some unspliced pri-miRNAs are present at higher levels than their spliced counterparts 25 and the relative constancy of pri-miR-181 levels across the transcriptional units suggests that the major forms are unspliced transcripts. Levels of pri-miR-181ab-1 gradually fell 67-fold in the interval 2-49 kb downstream of the mature miRNA sequences (Fig. 4A). This gradual decline suggests the absence of a single dominant RNA polymerase II termination site in MIR181A1B1. In contrast, pri-miR-181ab-2 levels fell > 50-fold somewhere in the interval 21.5-29 kb downstream of the mature miR-181a/b-2 coding sequences (Fig. 4C), consistent with a dominant MIR181A2B2 RNA polymerase II termination site.
Figure 4.
Primary miR-181ab-1 and -2 transcript mapping. Purified NK cells of three subjects were cultured overnight with culture media (A-C) or with TGF-β (D). Following reverse transcription, qPCR was performed using primers with sequences homologous to genomic DNA at the location shown by figure data points. For each subject, determination of relative RNA levels compared to the “middle primers” located between the miR-181a and miR-181b coding sequences (arrows) were calculated as: fold difference = 2 (Cq middle primer - Cq specific primer). When no amplification was detected, the Cq was set to 40.0. The Avg and SEM are presented on a log10 scale; some error bars are too small to be seen. The numbers in each graph indicate genomic position. In A, for example, “825” indicates Chromosome 1:198 825 000. In C, “450” represents Chromosome 9:127 450 000. A, B. MIR181A1B1. B. An expanded view of Chromosome 1:198 826 100-198 835 200, which highlights the region near pre-miR-181a. C, D. MIR181A2B2. The amount of pri-miR-181ab-2 RNA in NK cells incubated in culture media (C) or with TGF-β (20 ng/mL, D).
We “walked upstream” until the RT-qPCR signal precipitously declined, consistent with a dominant TSS. Chromosome 1 pri-miR-181ab-1 RT-qPCR signal dropped > 10 000-fold, ~ 78 kb upstream of the miR-181a/b-1 coding sequences (Fig. 4A). Likewise, ~ 35 kb upstream of the mature miR-181a/b-2 sequences, RT-qPCR signal dropped ~ 100-fold (Fig. 4C). Overlapping with the putative MIR181A2B2 transcriptional unit on chromosome 9, the NR6A1 gene is expressed at low level in many adult somatic tissues 26, and is transcribed anti-sense relative to MIR181A2B2. Thus the relatively small 100-fold drop-off upstream of the putative MIR181A2B2 TSS is likely due to background NR6A1 RNA rather than weak MIR181A2B2 promoter strength. Interestingly, the putative promoter sites indicated by our analysis are consistent with bioinformatic data. For both MIR181ab loci, the putative promoters localized with sites of DNase I hypersensitivity, FAIRE clusters 27, chromHMM hot spots 28 and chromatin immunoprecipitation of transcription factors, acetylated histone H3K27, and trimethylated histone H3K4, all of which are associated with TSS (data not shown). No other high-likelihood promoter elements were located in the interval between the putative TSS and the miR-181a/b coding sequences (data not shown). We more finely mapped the region just upstream of the mature miR-181a sequence on chromosome 1 because another group had cloned a putative promoter based on the assumption that “the core promoter of miRNA is known to locate close to its pre-miRNA” 29. RT-qPCR signal was 2.0- to 4.4-fold lower immediately upstream of the pre miR-181a sequence, but this was within the range of variation that we saw elsewhere in the transcript (Fig. 4B). Although we cannot exclude the possibility that a weak promoter lies immediately upstream of the pre-miR-181a-1 sequence, our data indicate that the major MIR181A1B1 promoter lies much further upstream.
We tested whether TGF-β stimulation induced transcription at alternative sites. Across the ~60 kb pri-miR-181ab-2 transcript, levels were 3- to 4-fold higher after TGF-β stimulation than after incubation of NK cells in culture media alone (Fig. 4C, D and data not shown). In TGF-β-treated NK cells, there was a precipitous decrease in pri-miR-181ab-2 level precisely where the TSS was located in unstimulated NK cells (Fig. 4C, D). Therefore, TGF-β appears to be acting at the same chromosome 9 promoter that drives constitutive pri-miR-181ab-2 expression in NK cells. Likewise, MIR181A1B1 appeared to use the same chromosome 1 TSS in both resting and TGF-β-stimulated NK cells (Fig. 4A and data not shown). Upon TGF-β treatment, the chromosome 1 pri-miR-181ab-1 RNA level slightly fell across the transcript (data not shown).
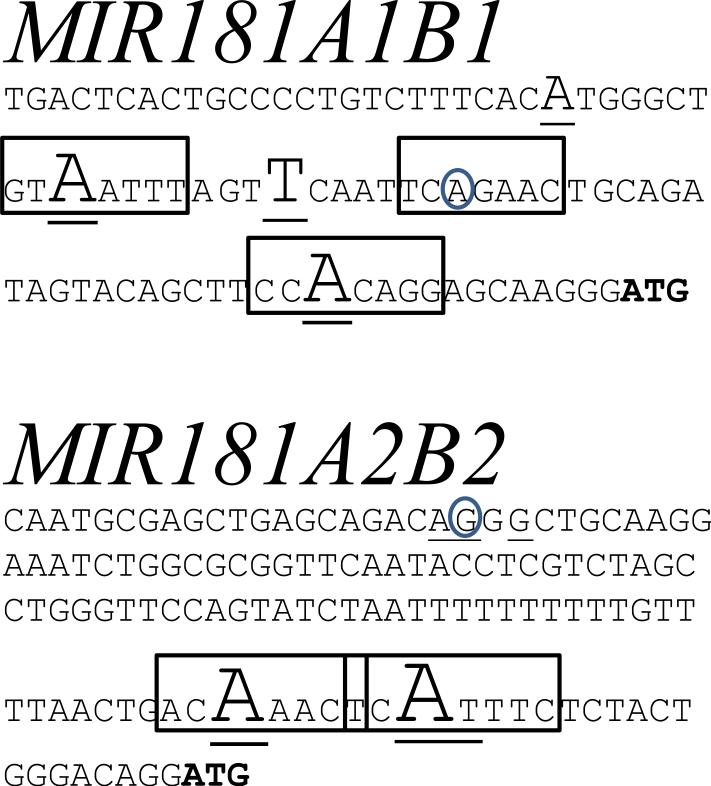
To confirm and more precisely map the TSS locations, we carried out 5’RACE on RNA from freshly isolated blood NK cells using RT and PCR primers homologous to regions just downstream of the putative MIR181ab TSS (Supplemental Table 1, Fig. 4). For MIR181A1B1, we identified several TSS near where a previously cloned cDNA had been mapped (Fig. 5). These TSS are located 78.3 kb upstream of the mature miR-181a/b coding sequences. For MIR181A2B2 (Fig. 5), two major TSS localized to consensus sites for Initiators, core promoter elements that are often required for efficient gene transcription and are found in miRNA gene promoters 30, 31. We identified additional weaker MIR181A2B2 TSS, one of which corresponds to the 5’ end of a previously cloned cDNA. These TSS are located 34.0 kb upstream of the mature miR-181a/b coding sequences.
Figure 5.
MIR181A1B1 and MIR181A2B2 TSS. RNA was harvested from the NK cells of two (MIR181A2B2) or three (MIR181A1B1) subjects and 5’RACE was performed. Underlined nt denote the 5’ end of RACE clones, with major TSS indicated by larger font size. Boxes enclose potential Initiator elements (consensus sequence, YYANWYY). Transcription direction is left to right. A. Chr1:198 906 606 - 519 depicting MIR181A1B1 TSS. The number of 5’RACE clones found (underlined letters from left to right) were 2 (A), 16 (A), 9 (T), 15 (A). The circled “A” shows the 5’ end of the previously reported NR_040073 transcript. B. Chr9:127 420 695 – 824 depicting MIR181A2B2 TSS. The number of 5’RACE clones found (underlined letters from left to right) were 1 (A), 1 (G), 1 (G), 11 (A), 17 (A), and 1 (T). The underlined and circled “G” refers to the beginning of the previously reported NR_038975 transcript, which coincides with the start of a 5’RACE clone. For each sequence, an “ATG” that was modified to “TTG” in the luciferase reporter plasmids is shown as bold letters.
Previous work suggested the identification of a MIR181A1B1 promoter just proximal of the pre-miR-181a sequence. Therefore, we performed 5’RACE using primers homologous to regions between or immediately upstream of the mature miR-181a/b-1 sequences using two different strategies (primers in Supplemental Table 1). Yet, no TSS were identified near the premiR-181a coding sequence (data not shown). We conclude that the major MIR181A1B1 TSS in human NK cells is located 78.3 kb upstream of the miR-181a/b-1 coding sequences.
MIR181A1B1 and MIR181A2B2 promoters
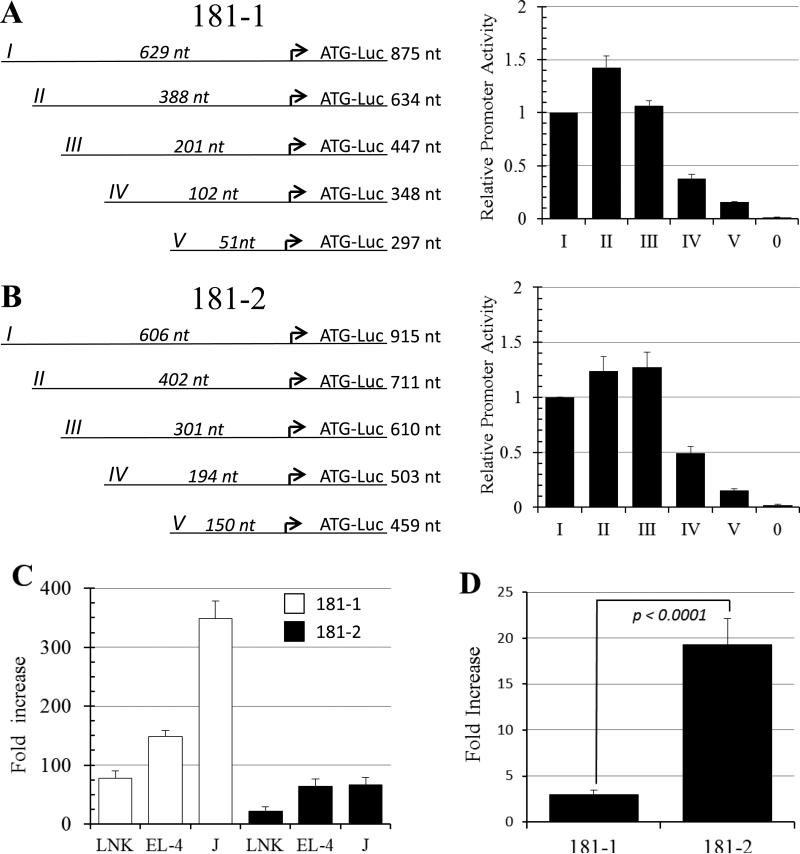
Next, we tested promoter function by cloning regions surrounding the MIR181A1B1 and MIR181A2B2 TSS. Sequences were placed into a Luciferase reporter vector and tested for promoter activity in NK and T cell lines by transient transfection. Both full length promoters (containing ~900 nt of MIR181A1B1 and MIR181A2B2 sequence, constructs 181-1 I and 181-2 I) produced strong transcriptional activity in YT NK cells and in other NK and T cell lines (Fig. 6A-C). To further characterize the promoters, we created a series of 5’ truncations (constructs II-V) and tested them for activity in YT NK cells. For both promoters, constructs I-III directed similar transcriptional activity, suggesting that the minimal promoter was contained within 201 nt and 301 nt regions upstream of the MIR181A1B1 and MIR181A2B2 major TSS, respectively. Further deletion of 5’ sequences decreased transcriptional activity, suggesting that the regions 51 to 201 nt and 150 to 301 nt upstream of the major TSS for the MIR181A1B1 and MIR181A2B2 genes, respectively, contained important transcription factor binding sites (Fig. 6A, B).
Figure 6.
Characterizing the MIR181A1B1 and MIR181A2B2 promoters. A, B. Shown are schematic views of promoter truncation experiments. The arrow indicates the major TSS. Full length (I) and truncation fragments (II-V) of the 181-1 (A) and 181-2 (B) promoter were transfected into YT-HY NK cells and promoter activity was measured as described in Materials and Methods. The length of the promoter fragment 5’ of the major TSS and the length of the total promoter fragment are shown. pXPG-181-1-I-Luc or pXPG-181-2-I-Luc produced luciferase values that were 70-fold and 45-fold above empty vector background. Values represent averages from tests of 2-4 different plasmid preparations over at least two different experiments. “0” indicates background level produced by the empty pXPG-Luc basic vector. C. MIR181 promoters are active in T and NK cell lines. The white and back bars indicate the 181-1 and 181-2 promoters, respectively. Fold increase refers to normalized luciferase activity compared to the empty pXPGLuc basic vector. The experiment was done 3 times with two different plasmid preps each time; all Avg were significant above those of the empty vector (*p < 0.002). D. SMAD3/4 strongly transactivate the 181-2 promoter. HepG2 cells were transfected with pXPG-181-1-I-Luc and pXPG-181-2-I-Luc reporter plasmids, along with expression constructs for SMAD3 and 4, or empty vector. Fold increase refers to transactivation activity of SMAD3/4 divided transactivation activity by empty vector. Shown is Avg and SEM of 5 experiments.
Because MIR181A2B2 transcription was activated by TGF-β (Fig. 3A), we asked if TGF-β acted on the promoter. TGF-β activation is often mediated through SMAD3 and SMAD4, which after ligand binding are recruited by the TGF-β receptor, are phosphorylated, and then enter the nucleus and bind to specific promoter sites 32. We therefore tested whether 181-1 and 181-2 promoters were transactivated by SMAD3 and SMAD4 transcription factor expression plasmids. The 181-2 promoter-reporter Luciferase construct was strongly transactivated by SMAD3 and SMAD4 transcription factors, with relatively weaker activation of the 181-1 promoter-reporter Luciferase construct (Fig. 6D). SMAD3/4 transactivated the 181-2 promoter 6.6-fold more than the 181-1 promoter (Fig. 6D). This finding is consistent with the hypothesis that TGF-β signaling upregulates MIR181A2B2 expression, at least in part, through SMAD-dependent promoter activation.
DISCUSSION
We showed that human blood NK cells and T cells share largely overlapping miRNA transcriptomes with <4% of the miRNAs significantly differing between the two lymphocyte types. It is likely that a few more miRNAs would have reached statistical significance had more subjects been analyzed and it is likely that expression of other miRNAs differs between specific T cell subsets and NK cells as a whole or specific NK cell subsets. Even with this consideration, a relatively small number of miRNAs differed between NK cells and T cells, which was anticipated because NK cells and T cells share developmental precursors, receptors, and functions 9, 10. Because of the importance of miR-181 in NK cell development and activation 15, we investigated how miR-181a and miR-181b are regulated. miR-181a and miR-181b expression rose in concert with NK maturation from the least mature CD56bright NK cells through CD56dimCD94hi cells to the most mature CD56dimCD94low NK cells. Combined with our prior results, this shows that miR-181a and miR-181b expression levels rise from primitive hematopoietic progenitor cells through the most mature NK cells 15. Other members of the miR-181 family, miR-181c and miR-181d, were below the limit of microarray detection in both NK cells and T cells. miR-181a and miR-181b are differentially expressed in hematopoietic cells and help to commit primitive cells to lineage differentiation 33. Specifically, we showed that miR-181a and miR-181b are required for efficient NK development from primitive progenitor cells 15. Notch signaling is needed for proper NK cell development and is suppressed by Nemo-like kinase, which in turn is regulated by miR-181a and miR-181b 15, 34.
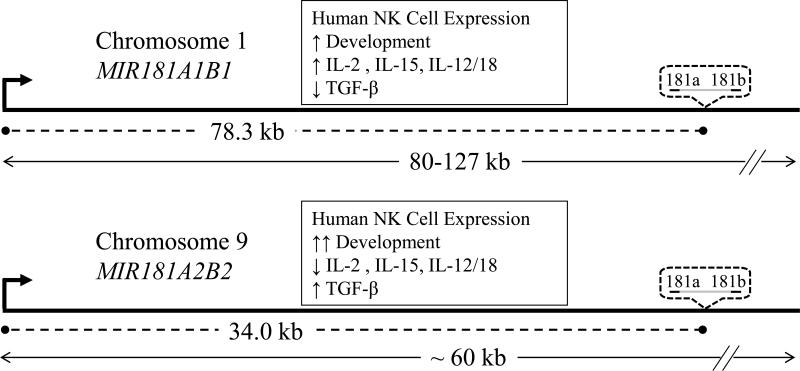
Mature miR-181a and miR-181b each are encoded by loci on chromosomes 1 and 9. Gene knockout of the MIR181A1B1 locus in mice abolished mature miR-181a and miR-181b in the thymus, whereas knockout of MIR181A2B2 did not change mature miR-181a and miR-181b levels 23. This indicated that the MIR181A2B2 locus is not significantly expressed by mouse thymocytes. Examining pri-miR-181ab by two different RT-qPCR strategies, we found that the chromosome 1 MIR181A1B1 locus was expressed at levels similar to those of the chromosome 9 MIR181A2B2 locus in human blood T cells. Therefore, the MIR181ab loci appear to be regulated much differently in mouse and human T lymphocytes. In human blood NK cells, pri-miR-181ab-2 levels were higher than pri-miR-181ab-1 levels and both were higher than in T cells. This finding is consistent with our microarray and RT-qPCR data showing that mature miR-181a and miR-181b levels were significantly higher in NK cells than in T cells. Treatment with immune regulatory cytokines showed that the two pri-miR-181ab loci were independently regulated in NK cells. In resting NK cells, pri-miR-181ab-2 was more highly expressed than pri-miR-181ab-1. However, the ratio was reversed after culture with several cytokines known to stimulate NK cells: IL-2, IL-15, and IL-12/IL-18. Interestingly, the immunosuppressive cytokine, TGF-β, accentuated the ratio by suppressing pri-miR-181ab-1 expression while boosting pri-miR-181ab-2 expression (summarized in Fig. 7). TGF-β and other TGF family members increase miR-181 expression in a variety of cancer cell lines and contribute to metastasis 35, 36. Taylor et al showed that TGF-β treatment upregulated pre-miR-181a1 slightly and pre-miR-181a2 extensively in the MDA-MB-231 human breast cancer cell line 36.
Figure 7.
Schematic diagram showing the distances (dashed lines) between human MIR181A1B1 and MIR181A2B2 TSS (bent arrows) and mature miRNA coding sequences (dotted bubbles) and length (double arrow lines) of the pri-miRNAs. Distances are not drawn to scale. Summarized is human NK cell differential regulation of MIR181A1B1 and MIR181A2B2 gene expression in response to developmental signals and immune regulatory cytokines (boxes).
We had previously shown that miR-181a and miR-181b levels rise as cells develop through pre-NK cell stages into CD56+ NK cells 15. In the current work, we extend this finding to show that miR-181a and miR-181b levels continue to rise during NK cell differentiation, from CD56bright NK cells through more mature CD56dimCD94hi and CD56dimCD94low stages. The relatively smooth rise of mature miR-181a/b levels belies the marked changes in MIR181A2B2 gene expression. The chromosome 9 gene was expressed at very low levels in pre-NK cell development and then rose dramatically by the time cells entered the CD56bright NK cell stage. During pre-NK cell development, pri-miR-181ab1 was higher than pri-miR-181ab2. Although pri-miR-181ab1 levels rose as cells transitioned from Stage IV pre-NK cells into CD56bright NK cells, pri-miR-181ab2 levels rose even more, and were statistically higher than pri-miR-181ab1 levels in CD56dim mature NK cells. MIR181 gene expression reflects the dramatic changes in phenotype that cells undergo as they transition from pre-NK cells into mature CD56+ NK cells. miR-181a and miR-181b help cause this change by regulating the expression of nemo-like kinase and Notch signaling 15, 34. It will be of interest to investigate which transcription factors are responsible for the dramatic increase in MIR181A2B2 gene expression.
We located TSS of the pri-miR-181ab loci by “walking” upstream of the miR-181 mature sequences with a series of RT-qPCR primers. Although others have used Drosha knockdown or knockout to block pri-miRNA processing and thereby increase detection of pri-miRNAs 37, we analyzed human NK cells without changing Drosha expression. This avoided artifacts that might be associated with manipulation of miRNA processing. miRNA can be the products of spliced or unspliced primary transcripts 6, 25. For both MIR181A1B1 and MIR181A2B2, pri-miRNA levels were relatively constant over many kb. This strongly suggests that the dominant forms of primiR-181ab1 and pri-miR-181ab2 are unspliced transcripts, as has been reported for other primiRNAs 25. We confirmed and precisely mapped the MIR181A1B1 and MIR181A2B2 TSS using 5’RACE on freshly isolated NK cells with normal intact miRNA processing. DNA regions just upstream of the TSS cloned into luciferase reporter vectors demonstrated strong promoter activity in both NK cells and in T cells, consistent with these regions being promoters for the transcriptional units in these cell types (Fig. 6A-C). In co-transfection experiments, the MIR181A2B2 promoter was strongly transactivated by SMAD3 and SMAD4 transcription factors, suggesting that TGF-β treatment upregulated MIR181A2B2 transcription by direct or indirect SMAD-dependent activation of the MIR181A2B2 promoter.
Other investigators started with the assumption that miRNA core promoter elements are located close to the presumptive 110 nt pre-miR-181a-1 DNA sequence. A 600 nt segment immediately upstream of the pre-miR-181a-1 DNA sequence had significant promoter activity in squamous cell carcinoma cells and in embryonic kidney cells and bound C/EBPα isoforms in vitro 29, 38. However, these authors did not provide any independent evidence that pri-miR-181ab-1 transcription initiated in this region. We cannot rule out the possibility that this site or other sites can be used as alternative promoters in NK cells or other cells, but our data indicate that the major promoters in NK cells are 78.3 kb and 34.0 kb upstream of the mature miR-181a/b sequences on chromosomes 1 and 9, respectively (shown schematically in Fig. 7). It is remarkable that 22-23 nt gene products are produced from such large primary transcripts, 80-127 kb for pri-miR-181ab-1 and ~60 kb for pri-miR-181ab-2. However, large sizes have been reported for other pri-miRNAs 31.
Several bioinformatic algorithms have been used to predict MIR TSS 31. For example, a recent described algorithm predicted five potential TSS for MIR181A2B2, distanced 7.8, 33.0, 33.5, and 34.0 kb upstream of the miR-181a coding sequence 39 and an earlier algorithm predicted a TSS 33.7 kb upstream of the coding sequence 40. Several of these predicted sites approximate the 34.0 kb distance that we identified by “chromosome walking” and 5’RACE. In contrast, forecast of the MIR181A1B1 TSS was less successful. One algorithm 39 listed possible TSS at 2.4, 28.6, 44.8, 47.1, and 47.4 kb upstream of the miR-181a coding sequence, all very different than the TSS that we identified 78.3 kb upstream of the coding sequence; the other algorithm did not predict a MIR181A1B1 TSS 40. Our results show that although bioinformatic information can be quite useful, it does not replace experimental verification. In our view, this requires “chromosome walking” to approximate the TSS, which is then precisely located with 5’RACE or comparable technique. Confirmation of the same TSS under different stimulatory or developmental conditions further strengthens its identification.
Given that the chromosome 1 and 9 sites produce identical 23 nt miR-181a and miR-181b mature 5p species, is there biological significance to the differential regulation of MIR181A1B1 and MIR181A2B2? There are at least two ways in which pri-miR-181ab-1 and -2 might have different effects. First, the 5p/3p ratio in expressed miRNAs varies considerably with tissue and developmental stage and both strands may be expressed at significant levels in the same cell 16, 17. RNA processing machinery favors the 5p form of both miR-181a and miR-181b, but a significant amount of the 3p strand is selected from both pre-miR-181a-1 and pre-miR-181a-2 in NK cells (data not shown). Unlike the 5p strands, the 3p strands differ between miR-181a-1 and miR-181a-2, including the nt 2-8 seed sequences that dictate target mRNA selection: 5’ ACCA(UC/CU)GACCGUUGA(U/C)UGUACC. Second, there is increasing evidence that regions outside the mature miRNA 22-23 nt sequence regulate mRNA 19. Mutation and swapping miR-181a-1and miR-181c sequences suggested that loop nt and other pre-miRNA sequences influence miRNA biological action unrelated to mature miRNA production 18. Interestingly, knockout of the mouse MIR181A2B2 locus, but not of MIR181A1B1 locus, affected the rate of embryonic stem cell self renewal, despite expression of MIR181A1B1, MIR181A2B2, and MIR181CD loci in WT cells 19. Therefore, the differential expression of pri-miR-181ab-1 and -2 may influence cell physiology other than simply by dictating levels of the predominant 5p form of the mature miRNA.
We have demonstrated differential control of two genes that both encode mature 5p forms of miR-181a and miR-181b. Given that differential gene expression is found in both mice and humans, this begs the question of what use the dual gene system has for cells, including lymphocytes. Future research should investigate gene regulatory roles of pri-miRNA and premiRNA. In addition, we plan to investigate how MIR181A1B1 and MIR181A1B1 promoters are controlled by developmental signals and immune regulatory cytokines.
MATERIALS AND METHODS
Cell isolation and stimulation
Tissues and informed consent were obtained in accordance with the guidelines of the Human Subjects Institution Review Board. NK cells and T cells were isolated from heparinized adult blood by negative selection using RosetteSep® Human NK Cell Enrichment and Human T Cell Enrichment cocktails, respectively (Stemcell Technologies, Vancouver, British Columbia, Canada), as per manufacturer's instructions. The purity of the isolated NK and T cell populations were consistently > 90% and > 98%, respectively. Umbilical cord blood was obtained from the Memorial Blood Bank (Minneapolis, MN), Placental Blood Program of the New York Blood Center (New York, NY), St. Louis Cord Blood Bank (St. Louis, MO), or local obstetric units. Mononuclear cells were isolated by density gradient centrifugation, and CD34+ hematopoietic cells were purified using the MACS magnetic bead selection system (Miltenyi Biotech). Primary NK cells were cultured in NK complete medium (RPMI 1640, 10% fetal bovine serum, amino acids, antibiotics and other additives, Lonza Life Sciences, Ann Arbor, MI and HyClone, South Logan, UT).
For flow cytometry analysis and cell sorting of adult blood, fluorescently-labeled antibodies to the following antigens were used, CD3 (UCHT1), CD16 (3G8) CD56 (HCD56) and CD94 (DX22; BioLegend, San Diego, CA); for cord blood, CD34 (581), CD117 (104D2), CD94 (HP-3D9), CD3 (UCHT1), and CD56 (NCAM16.2) (BD Biosciences, Franklin Lake, NJ). NK cells (1 × 106) were incubated in 1 ml of NK complete medium for 24 h at 37°C in 5% CO2 with IL-2 (100 U/ml, Biological Resources Branch, National Cancer Institute, Frederick, MD), IL-12 (10 U/ml, Peprotech, Rocky Hill, NJ) plus IL-18 (100 ng/ml, R&D Systems, Minneapolis, MN), IL-15 (100 ng/ml, BioLegend), TGF-β (10 ng/ml, R&D Systems), or PMA (10 ng/ml, Enzo) plus the calcium ionophore, A23187 (250 ng/ml, Sigma-Aldrich, St. Louis, MO).
Mononuclear cells or purified NK cells were incubated with human IgG (Sigma-Aldrich) to block Fc receptors, and then stained with antibody to CD3, CD16 and CD56. Stained samples were read on an LSR-II cytometer (BD Biosciences) and data were analyzed using FlowJo software (Treestar). NK-enriched samples were sorted into CD3–CD56brightCD16–, CD3– CD56dimCD16+CD94hi and CD3–CD56dimCD16+CD94low populations using MoFlo Legacy® High-Speed Cell Sorter (Beckman Coulter). For cord blood, antibody-stained NK cell precursors were sorted using a FACS Aria (BD Biosciences) based on their developmental stage: Stage I (CD3−CD34+CD117-CD56−CD94−), Stage II (CD3−CD34+CD117+CD56−CD94−), Stage III (CD3-CD34−CD117+CD56−CD94−), and Stage IV (CD3−CD34−CD117+CD56low/−CD94−). The numbers of Stage I cells recovered were insufficient for miRNA analysis.
miRNA analysis
RNA was extracted from NK- and T-enriched lymphocyte populations from three subjects using standard TRIzol® reagent (Invitrogen, Carlsbad, CA) and analyzed on a miRCURY™ locked-nucleic acid microarray (Version 11.0; Exiqon; Vedbaek, Denmark) consisting of control probes, mismatch probes, and 1769 capture probes complementary to human, mouse, rat, and viral sequences from the miRBase v.14.0 release. miRNA levels were calculated based on normalized Hy3 labeling signals (log2 transformed) from all hybridizations using image analysis software. Only miRNAs with six valid measurements were analyzed. Microarray data was deposited with GEO, Accession # GSE61215. Mature miRNA levels were quantified in reverse transcription-quantitative PCR (RT-qPCR) using the appropriate TaqMan primer/probe sets and the TaqMan PCR Mastermix (Applied Biosystems, Foster City, CA) in a modification of the manufacturer's instructions. Briefly, RNA (25 ng) was reverse transcribed in a 10 μL reaction containing 1× RT buffer, dNTPs (each at 0.375 mM), 5 U/μL MultiScribe reverse transcriptase, 50 nM stem-loop RT primers and 0.38 U/μL RNase inhibitor (Applied Biosystems) at 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. Real-time PCR amplifications were performed in triplicate in 10 μL volumes. Quantitative miRNA expression data were acquired and analyzed on Applied Biosystems 7000 and 7500 instruments, using RNU-24 small nucleolar RNA as an endogenous control. Control and target miRNA standard curves were generated in each experiment.
pri-miR-181 assay
RNA was harvested with TRIzol, DNase I-treated (1 U/μg, Promega), and reverse transcribed with random octamers and oligo(dT) primers (RNA to cDNA High Capacity Kit, Invitrogen) at 37 °C for 60 min. pri-miR-181 was assayed with two methods, which produced virtually identical results. In the first method, qPCR was carried out using primers (Supplemental Table I) at 500 nM in 1x SsoAdvanced SYBR Green Mix (Bio-Rad) on a CFX96 Real-Time qPCR instrument (Bio-Rad), using GAPDH mRNA as an endogenous control, with initial melting at 95 °C (3 min), then 95 °C (5 s), 64.5 °C (30 s) for 40 cycles. The second method used hsa-mir-181a-1 and -2 TaqMan® Pri-miRNA Assays (Life Technologies) including proprietary primers and TaqMan probes, as per the manufacturer's instructions, using 18S rRNA as an endogenous control. Thermal cycling conditions were: initial melting at 95°C (10 min), then 95°C (15 s), 60°C (60 s) for 40 cycles.
TSS mapping
TSS were identified using the RLM-RACE (Ambion) kit followed by cloning 5’rapid amplification of cDNA ends (5’RACE) products into the pCR4-TOPO vector. Briefly, DNase I- treated RNA was phenol:chloroform:isoamyl alcohol (25:24:1, Invitrogen) extracted and treated with calf alkaline phosphatase (Ambion). Reverse transcription of the RNA adapter-tagged sample was primed using gene specific primers (sequences in Supplemental Table S1) using Superscipt III (Invitrogen ) at 50 °C, following manufacturer instructions. A first PCR was carried as described above for 32 cycles using the “outer” Ambion primer with the 1st nested gene-specific primer (Supplemental Table I). A second (nested) PCR was carried out as above using the “inner” Ambion primer with the 2nd nested gene-specific primer, at one of several annealing/extension temperatures (54 – 68 °C, 60 s) for 40 cycles. Products were purified using a Qiagen spin column and cloned into the pCR4-TOPO vector using the TOPO-TA cloning kit (Invitrogen). Sequencing was carried out by the Northwestern University sequencing facility (Chicago, IL).
Promoter-Luciferase constructs
Regions 879 and 915 nt in length located at 78.3 and 34.0 kb upstream of the 181-a/b-1 and 181-a/b-2 sequences, respectively, were cloned into the Kpn I/Hind III and KpnI/NcoI sites of the pXPG-luciferase vector 20. ATGs near the TSS were mutated to TTG (indicated in Fig. 5) to eliminate potential translation start sites upstream of the luciferase sequence. Inserts were confirmed by DNA sequencing. Promoter regions were 5’ truncated using PCR producing pXPG-181-1-Luc constructs of 634 nt, 447 nt, 348 nt, and 297 nt (constucts II-V). Similarly, pXPG-181-2-Luc constructs of 711 nt, 610 nt, 503nt, and 459 nt were produced (constructs II-V, Supplemental Table 1).
YT-HY, Jurkat, and LNK were cultured and transfected with plasmid mini-preps (Qiagen) using DEAE-dextran or electroporation, as previously described 21, 22. EL-4 (a kind gift from F. Marti) was cultured and transfected in the same manner as Jurkat 22, but using 950 uF and 280 V electroporator settings. HepG2 (a kind gift of B. Spear) was cultured in 1:1 Ham's F-12: DMEM (Sigma) supplemented with 10% Cosmic Calf Serum (Hyclone), glutamine (2 mg/mL, Invitrogen), and insulin (4 ug/mL, Invitrogen). HepG2 cells (~2 × 106) in individual 35-mm wells were transfected using Lipofectamine and Plus reagent (Invitrogen) with 1.0 μg of pXPG-181-I-Luc or pXPG-181-2-I-Luc promoter plasmid, 5 ng of control SV40-renella plasmid (Promega), and 250 ng of CS2-Myc-SMAD3 plus 250 ng CS2-HASMAD4 expression constructs (a kind gift of T. Woodruff) or 500 ng of an empty vector in place of the SMAD expression constructs. Manufacturer's instructions were followed, except that cells received fresh media at 3 and 24 h after transfection. We determined that SMAD3/4 variably decreased control renilla activity, and so we report SMAD3/4-induced MIR181 promoter activity without normalization. When results were normalized to renilla luciferase activity, SMAD3/4 caused a 12.0-fold increase in pXPG-181-2-I-Luc promoter activity, compared with that of pXPG-181-I-Luc (data not shown). Preferential action of SMAD3/4 on the MIR181A2B2 promoter was confirmed in transfections carried out without renella plasmid control (data not shown).
Statistics and Analysis
The Student T-test with and without Bonferroni correction was used to evaluate microarray data and other data, respectively. ANOVA test with Bonferroni correction was used to analyze miRNA responses to cytokine stimulation. F-statistics indicated that luciferase data showed equivalent variances after logarithmic transformation, and these were tested using paired, two-tailed t tests. The p values are listed in the figures and figure legends. All error bars represent SEM. Chromosome sites were accessed on the UCSC Genome Browser (https://genome.ucsc.edu/), based on assembly Feb. 2009 (GRCh37/hg19).
Supplementary Material
ACKNOWLEDGEMENT
We thank Jeffrey Ebersole, Yelena Alimova, Peter Nelson and Luke Bradley for use of equipment, Martha L. Peterson, Peter T. Nelson, Wangxia Wang, and Francesc Marti for advice, Brett Spear and Francesc Marti for cell lines, Teresa Woodruff for plasmids, Dennis Williams and the Kentucky Blood Center for help with blood products, and the National Cancer Institute for IL-2. This work was supported by grants from the National Institutes of Health R01 AI56506 to CTL, R01 55417 to JSM.
Footnotes
Conflict of Interest: The authors declare no actual or potential conflicts of interest.
Supplementary information is available at Genes and Immunity's website
REFERENCES
- 1.Caligiuri MA. Human natural killer cells. Blood. 2008;112(3):461–9. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fauriat C, Long EO, Ljunggren H-G, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115(11):2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu J, Mao HC, Wei M, Hughes T, Zhang J, Park IK, et al. CD94 surface density identifies a functional intermediary between the CD56bright and CD56dim human NK-cell subsets. Blood. 2010;115(2):274–81. doi: 10.1182/blood-2009-04-215491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 6.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 7.Wang P, Gu Y, Zhang Q, Han Y, Hou J, Lin L, et al. Identification of resting and type I IFN-activated human NK cell miRNomes reveals microRNA-378 and microRNA-30e as negative regulators of NK cell cytotoxicity. J Immunol. 2012;189(1):211–21. doi: 10.4049/jimmunol.1200609. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Wang Y, Sun Q, Yan J, Huang J, Zhu S, et al. Identification of microRNA transcriptome involved in human natural killer cell activation. Immunol Lett. 2012;143(2):208–17. doi: 10.1016/j.imlet.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Obata-Onai A, Hashimoto S, Onai N, Kurachi M, Nagai S, Shizuno K, et al. Comprehensive gene expression analysis of human NK cells and CD8(+) T lymphocytes. Int Immunol. 2002;14(10):1085–98. doi: 10.1093/intimm/dxf086. [DOI] [PubMed] [Google Scholar]
- 10.Blom B, Spits H. Development of human lymphoid cells. Annu Rev Immunol. 2006;24(1):287–320. doi: 10.1146/annurev.immunol.24.021605.090612. [DOI] [PubMed] [Google Scholar]
- 11.Bezman NA, Kim CC, Sun JC, Min-Oo G, Hendricks DW, Kamimura Y, et al. Molecular definition of the identity and activation of natural killer cells. Nat Immunol. 2012;13(10):1000–9. doi: 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bezman NA, Cedars E, Steiner DF, Blelloch R, Hesslein DGT, Lanier LL. Distinct requirements of microRNAs in NK cell activation, survival, and function. J Immunol. 2010;185(7):3835–3846. doi: 10.4049/jimmunol.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan RP, Leong JW, Schneider SE, Keppel CR, Germino E, French AR, et al. MicroRNA-deficient NK cells exhibit decreased survival but enhanced function. J Immunol. 2012;188(7):3019–30. doi: 10.4049/jimmunol.1102294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q-J, Chau J, Ebert PJR, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–61. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, et al. Cutting Edge: MicroRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187(12):6171–5. doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15(4):354–63. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24(10):992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu G, Min H, Yue S, Chen CZ. Pre-miRNA loop nucleotides control the distinct activities of mir-181a-1 and mir-181c in early T cell development. PloS one. 2008;3(10):e3592. doi: 10.1371/journal.pone.0003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CZ. An unsolved mystery: the target-recognizing RNA species of microRNA genes. Biochimie. 2013;95(9):1663–76. doi: 10.1016/j.biochi.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bert AG, Burrows J, Osborne CS, Cockerill PN. Generation of an improved luciferase reporter gene plasmid that employs a novel mechanism for high-copy replication. Plasmid. 2000;44(2):173–82. doi: 10.1006/plas.2000.1474. [DOI] [PubMed] [Google Scholar]
- 21.Presnell SR, Zhang L, Ramilo CA, Chan H-W, Lutz CT. Functional redundancy of transcription factor-binding sites in the killer cell Ig-like receptor (KIR) gene promoter. Int. Immunol. 2006;18(8):1221–1232. doi: 10.1093/intimm/dxl043. [DOI] [PubMed] [Google Scholar]
- 22.Presnell SR, Zhang L, Chlebowy CN, Al-Attar A, Lutz CT. Differential transcription factor use by the KIR2DL4 promoter under constitutive and IL-2/15-treated conditions. J Immunol. 2012;188(9):4394–4404. doi: 10.4049/jimmunol.1103352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fragoso R, Mao T, Wang S, Schaffert S, Gong X, Yue S, et al. Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1. PLoS Genet. 2012;8(8):e1002855. doi: 10.1371/journal.pgen.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, et al. Evidence for discrete stages of human natural killer cell differentiation in vivo. J. Exp. Med. 2006;203(4):1033–1043. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slezak-Prochazka I, Kluiver J, de Jong D, Kortman G, Halsema N, Poppema S, et al. Cellular localization and processing of primary transcripts of exonic microRNAs. PloS one. 2013;8(9):e76647. doi: 10.1371/journal.pone.0076647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan ZJ, Xu X, Chung AC, Cooney AJ. Extra-germ cell expression of mouse nuclear receptor subfamily 6, group A, member 1 (NR6A1). Biol Reprod. 2009;80(5):905–12. doi: 10.1095/biolreprod.107.067322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17(6):877–85. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9(3):215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin KH, Bae SD, Hong HS, Kim RH, Kang MK, Park NH. miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem Biophys Res Commun. 2011;404(4):896–902. doi: 10.1016/j.bbrc.2010.12.055. [DOI] [PubMed] [Google Scholar]
- 30.Lo K, Smale ST. Generality of a functional initiator consensus sequence. Gene. 1996;182:13–22. doi: 10.1016/s0378-1119(96)00438-6. [DOI] [PubMed] [Google Scholar]
- 31.Liang R, Bates DJ, Wang E. Epigenetic control of microRNA expression and aging. Current genomics. 2009;10(3):184–93. doi: 10.2174/138920209788185225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massague J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C-Z, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 34.Bachanova V, McCullar V, Lenvik T, Wangen R, Peterson KA, Ankarlo DE, et al. Activated notch supports development of cytokine producing NK cells which are hyporesponsive and fail to acquire NK cell effector functions. Biol Blood Marrow Transplant. 2009;15(2):183–94. doi: 10.1016/j.bbmt.2008.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neel JC, Lebrun JJ. Activin and TGFβ regulate expression of the microRNA-181 family to promote cell migration and invasion in breast cancer cells. Cellular signalling. 2013;25(7):1556–66. doi: 10.1016/j.cellsig.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP. TGF-beta upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123(1):150–63. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirigin FF, Lindstedt K, Sellars M, Ciofani M, Low SL, Jones L, et al. Dynamic microRNA gene transcription and processing during T cell development. J Immunol. 2012;188(7):3257–67. doi: 10.4049/jimmunol.1103175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hickey CJ, Schwind S, Radomska HS, Dorrance AM, Santhanam R, Mishra A, et al. Lenalidomide-mediated enhanced translation of C/EBPα-p30 protein up-regulates expression of the antileukemic microRNA-181a in acute myeloid leukemia. Blood. 2013;121(1):159–69. doi: 10.1182/blood-2012-05-428573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsico A, Huska MR, Lasserre J, Hu H, Vucicevic D, Musahl A, et al. PROmiRNA: a new miRNA promoter recognition method uncovers the complex regulation of intronic miRNAs. Genome Biol. 2013;14(8):R84. doi: 10.1186/gb-2013-14-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE, et al. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19(10):1742–51. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.