SUMMARY
Polycomb Repressive Complex 2 (PRC2) plays crucial roles in transcriptional regulation and stem cell development. However, the context-specific functions associated with alternative subunits remain largely unexplored. Here we show that the related enzymatic subunits EZH1 and EZH2 undergo an expression switch during blood cell development. An erythroid-specific enhancer mediates transcriptional activation of EZH1, and a switch from GATA2 to GATA1 controls the developmental EZH1/2 switch by differential association with EZH1 enhancers. We further examine the in vivo stoichiometry of the PRC2 complexes by quantitative proteomics and reveal the existence of an EZH1-SUZ12 sub-complex lacking EED. EZH1 together with SUZ12 form a non-canonical PRC2 complex, occupy active chromatin, and positively regulate gene expression. Loss of EZH2 expression leads to repositioning of EZH1 to EZH2 targets. Thus, the lineage- and developmental stage-specific regulation of PRC2 subunit composition leads to a switch from canonical silencing to non-canonical functions during blood stem cell specification.
INTRODUCTION
The epigenetic machinery is crucial for tissue development and cellular homeostasis, and its deregulation often drives the pathogenesis of human disorders. Polycomb Repressive Complex 2 (PRC2) represents a major class of epigenetic regulator that participates in transcriptional repression by catalyzing histone H3 lysine 27 di/tri-methylation (H3K27me2/3) (Margueron and Reinberg, 2011; Sauvageau and Sauvageau, 2010). The canonical PRC2 complex consists of EED, SUZ12, and the histone methyltransferase EZH2. While overexpression or gain-of-function of PRC2 proteins is common in many cancers (McCabe et al., 2012a; Morin et al., 2010; Varambally et al., 2002), inactivating mutations of PRC2 components have also been described in various hematopoietic malignancies (Ernst et al., 2010; Makishima et al., 2010), raising major questions regarding how this complex subserves oncogenic and tumor suppressive activities in different cellular contexts. In light of recent efforts to therapeutically target EZH2 enzymatic activities or canonical EZH2-PRC2 functions in various hematopoietic malignancies (Kim et al., 2013; Knutson et al., 2012; McCabe et al., 2012b), it will be critical to fully understand the context-dependent activity of this complex in normal developmental processes.
A confounding feature of the mammalian PRC2 complexes is the existence of two highly related enzymatic subunits EZH1 and EZH2 with near identical catalytic SET domains (Laible et al., 1997). Whereas the role of EZH2 in H3K27me3-mediated transcriptional repression has been well established (Cao et al., 2002; Czermin et al., 2002; Kuzmichev et al., 2002; Muller et al., 2002), the function of EZH1-PRC2 remains elusive and controversial. For example, in embryonic and skin stem cells, EZH1 complements EZH2 to maintain repressive chromatin and stem cell identity (Ezhkova et al., 2011; Margueron et al., 2008; Shen et al., 2008). In contrast, Ezh1 predominantly targets H3K4me3-marked active promoters and promotes RNA polymerase (Pol) II elongation in differentiating muscle cells and hippocampal neurons (Henriquez et al., 2013; Mousavi et al., 2012; Stojic et al., 2011).
Similarly, the role of PRC2 in hematopoiesis remains elusive due in part to the possible redundancy of EZH1/2 and difficulties in distinguishing effects related to canonical and non-canonical PRC2 functions that are mediated by EZH1 or EZH2 independent of the histone methyltransferase activity (Hidalgo et al., 2012; Mochizuki-Kashio et al., 2011; Xie et al., 2014). To study the role of PRC2 in hematopoiesis, we previously developed mouse models containing hematopoietic-specific genetic inactivation of Ezh2 or Eed (Shen et al., 2009; Shen et al., 2008; Xie et al., 2014). Our studies reveal complex and developmental stage-specific roles of canonical PRC2 complexes in normal hematopoietic stem cell (HSC) function (Xie et al., 2014). Therefore, to understand the context-specific functions of PRC2 in normal and malignant hematopoiesis, it is imperative to have a fuller analysis of the non-canonical PRC2 functions mediated by EZH1 independent of H3K27me2/3.
In this study, we demonstrate that the PRC2 enzymatic subunits EZH1 and EZH2 undergo an expression switch during blood cell development. We demonstrate that an erythroid-selective enhancer is indispensable for the transcriptional activation of EZH1, and a GATA2-to-GATA1 switch controls the EZH1/2 switch by developmental stage-specific association with distinct EZH1 enhancers. We determined the in vivo stoichiometry of PRC2 complexes by quantitative proteomics and uncovered the existence of an EZH1-SUZ12 sub-complex. Furthermore, through genome scale chromatin occupancy and transcriptional profiling analyses, we provide evidence that EZH1 together with SUZ12 form a non-canonical PRC2 complex, occupy active chromatin domains, and positively regulate gene expression. Importantly, loss of EZH2 expression results in repositioning of EZH1 chromatin occupancy and transcriptional activity. Thus, our study establishes a molecular link between the switch of master lineage regulators and developmental control of PRC2 subunit composition, providing a means to coordinate lineage-specific transcription and accompanying changes in the epigenetic landscape during blood stem cell specification.
RESULTS
Reciprocal Expression of EZH1 and EZH2 during Hematopoiesis and Oncogenesis
Previously we established a two-phase culture system to model the differentiation of primary human CD34+ hematopoietic stem/progenitor cells (HSPCs) ex vivo (Figure 1A). In this system, fetal liver or adult bone marrow-derived CD34+ HSPCs are expanded and differentiated into highly enriched populations of erythroid progenitor cells (proerythroblasts or ProEs). We initially determined the expression of each PRC2 core subunit during erythroid development. Whereas EED and SUZ12 mRNA and protein levels remain largely unchanged, EZH1 is progressively and significantly upregulated during differentiation (Figure S1B; Figure 1B,C). Conversely, EZH2 expression is modestly downregulated during late differentiation (day 7 to 12; Figure S1B). As a result, EZH1 and EZH2 undergo a relative switch in expression during terminal erythroid maturation (Figure 1B).
Figure 1. Reciprocal Expression of EZH1 and EZH2 during Hematopoiesis.
(A) Differentiation of fetal liver (FL) or adult bone marrow (BM) CD34+ HSPCs to proerythroblasts (ProEs) ex vivo.
(B) Expression of human EZH1 and EZH2 mRNAs in HSPCs and differentiating erythroid precursors. The mRNA expression levels relative to GAPDH are shown. Results are means ± SD of at least three independent experiments.
(C) Expression of EZH1, EZH2, EED, SUZ12 and H3K27me3 protein in fetal or adult HSPCs (day 0) and differentiating erythroid precursors (days 3 to 12). GAPDH was analyzed as a loading control.
(D) Expression of Ezh1 and Ezh2 mRNA during hematopoiesis. mRNA expression values from transcriptomic profiling (Seita et al., 2012) were shown in the indicated FACS-sorted hematopoietic stem/progenitor cells and mature lineages (Experimental Procedures).
See also Figure S1.
We next compared the expression of EZH1 and EZH2 in various hematopoietic stem/progenitor cells and mature lineages using available transcriptomic profiles (Seita et al., 2012). Notably, the expression of EZH1 and EZH2 is inversely correlated within the hematopoietic hierarchy (Figure 1D) and the developmental specification of T, B, and NK lineages (Figure S1C). Furthermore, the reciprocal expression of EZH1 and EZH2 is also observed during oncogenesis, in which increased EZH2 and concomitant decreased EZH1 expression are apparent in various cancer types (Figure S1D-G). Higher EZH2 expression is associated with lower survival in prostate cancer, whereas increased EZH1 expression is associated with higher survival (Figure S1H).
An Erythroid-selective Enhancer Controls EZH1 Activation
The expression switch between EZH2 and EZH1 during erythroid differentiation led us to ask whether EZH1 expression might be trans-activated through tissue-specific regulatory elements. We examined the epigenetic landscape surrounding the EZH1 gene in committed erythroid cells (Figure 2). Importantly, the genomic region 46-kb (or +46) downstream of the transcriptional start site (TSS) of EZH1 gene contains an enhancer signature consisting of H3K4me1, H3K27ac, and presence of DNaseI hypersensitivity (DHS). The putative enhancer signature is occupied by principal erythroid transcriptional regulators GATA1 and TAL1, present in human erythroid cell line K562 and absent in other established human cell lines (Figure 2A; Figure S2), suggesting that it functions as an erythroid-specific enhancer element. Enhancers often function through direct enhancer-promoter interaction by DNA loop formation. By chromosome conformation capture (3C) analysis (Dekker et al., 2002), we observed that the frequency of interaction between EZH1 promoter and the putative +46 enhancer is significantly higher than other tested regions (Figure 2A; bottom). Interestingly, another genomic region centered around +39 kb from TSS displays an enhancer signature in B lymphoblastoid (GM12878) cells. Of note, EZH1 expression is also progressively activated during B lymphopoiesis (Figure S1C). Thus, these results suggest that distinct tissue-specific enhancer elements are employed in the trans-activation of EZH1 expression in different cell lineages.
Figure 2. Tissue-Selective Enhancers Control EZH1 Expression.
(A) Chromatin state maps and TF occupancy within the human EZH1 gene are shown. The putative erythroid-specific enhancer is shown. The genomic regions selected for enhancer reporter assays are depicted by shaded lines. The relative interaction frequency between EZH1 promoter (anchor region) and other tested regions by 3C analysis are shown on the bottom.
(B) Transient enhancer reporter assays were performed in K562 and GM12752 cells.
(C) The putative tissue-selective enhancers activate reporter gene expression. The enhancer activity was measured by the ratio of firefly luciferase activity over renilla luciferase activity (Firefly/Renilla) and normalized to the empty vector control. Data are means ± SD of three independent experiments.
(D) Design of CRISPR/Cas9-mediated genomic engineering for enhancer deletion. The sequences of sgRNAs are shown for protospacer-adjacent motifs (PAM) highlighted in red.
(E) Protein levels of EZH1 in independent clones containing bi-allelic deletion of the +39 (lymphoid) or the +46 (erythroid) EZH1 enhancer.
(F) mRNA expression of EZH1 in independent enhancer deletion clones.
See also Figure S2.
To test this hypothesis, we first performed transient enhancer reporter assays in erythroid (K562) and lymphoid (GM12752) cell lines (Figure 2B). A genomic fragment containing the putative +46 erythroid enhancer markedly enhanced reporter expression in K562 cells, but not in GM12752 cells, compared with other tested fragments. In contrast, a fragment containing the putative +39 lymphoid enhancer significantly enhanced reporter expression in lymphoid GM12752 cells (Figure 2C). We next determined whether the identified tissue-specific enhancers are functionally relevant for EZH1 expression within their native chromatin context by using CRISPR/Cas9-mediated genomic engineering (Cong et al., 2013) (Figure 2D). We used a pair of CRISPR/Cas9 guide RNAs to create double-strand DNA breaks flanking the +39 or +46 enhancer in erythroid K562 cells. Upon non-homologous end joining (NHEJ)-mediated DNA repair, we obtained several independent clones containing bi-allelic deletion of the endogenous +39 or +46 enhancer, respectively. Removal of the +46 erythroid enhancer resulted in significant downregulation of EZH1 expression (Figure 2E,F). Deletion of the putative +39 lymphoid enhancer did not affect EZH1 expression in K562 cells, demonstrating the specificity of the erythroid regulatory element. These data provide compelling evidence that an erythroid-specific enhancer element mediates erythroid-selective transcriptional activation of EZH1.
GATA Switch Regulates EZH1 and EZH2 Switch during Erythroid Development
To gain further mechanistic insights into EZH1 trans-activation, we profiled the chromatin landscape and TF occupancy in primary human HSPCs and committed erythroid progenitor cells (ProEs). Remarkably, the erythroid enhancer signature present in ProEs is undetectable in HSPCs (Figure 3A). GATA1 and TAL1 strongly associate with the EZH1 enhancer in ProEs but occupancy is not detected in HSPCs, suggesting that the EZH1 erythroid enhancer is trans-activated by GATA1/TAL1 during erythroid differentiation. Interestingly, the genomic region centered around 38-kb from TSS is strongly associated with DNaseI hypersensitivity and occupied by GATA2 in HSPCs (Figure 3A). A switch from GATA2 to GATA1 has been previously described to regulate erythroid commitment (Bresnick et al., 2010; Dore et al., 2012; Kaneko et al., 2010; Snow et al., 2011), and expression of GATA2 and GATA1 inversely correlate during HSPC to ProE differentiation (Figure S3A,B). Therefore, our results strongly suggest that the GATA switch mediates the EZH1/2 switch during erythropoiesis.
Figure 3. GATA Switch Controls EZH1/EZH2 Switch during Erythroid Development.
(A) Chromatin state maps and TF occupancy within the human EZH1 gene in HSPC and ProE cells are shown. The GATA2-occupied +38 HSPC-specific enhancer and the GATA1-occupied +46 erythroid enhancer are depicted by red and green shaded lines, respectively.
(B) mRNA expression of Gata1, Gata2, and PRC2 subunits upon induction of Gata1 expression in G1E/G1ER cells.
(C) mRNA expression of Ezh1 upon CRISPR/Cas9-mediated deletion of the GATA2-occupied +38 HSPC-specific regulatory element and the GATA1-occupied +46 erythroid enhancer in G1E/G1ER cells. Each dot represents a bi-allelic enhancer deletion clone. The unmodified G1E/G1ER cells were analyzed as controls. * P < 0.001; n.s. non-significant.
See also Figure S3.
To test this hypothesis, we employed G1E/G1ER cells, an established model for genetic complement of erythroid maturation upon inducible expression of Gata1 (Welch et al., 2004). Upon activation of the Gata1-ER transgene by β-estradiol treatment in G1ER cells, Gata1 mRNA was progressively elevated whereas Gata2 expression was sharply downregulated, consistent with a ‘GATA switch’ (Figure 3B). Notably, Ezh1 mRNA was progressively increased with modest downregulation of Ezh2 during later differentiation (Figure 3B). To further determine the role of GATA2 and GATA1-associated regulatory elements in EZH1 transactivation, we employed CRISPR/Cas9-mediated genomic engineering to remove the +38 or +46 regulatory region from its native chromatin context in G1E/G1ER cells (Figure 3C; Figure S3C,D). Strikingly, upon bi-allelic deletion of the +38 Gata2-associated regulatory region, the expression of Ezh1 mRNA was markedly activated in G1E progenitor cells and remained elevated upon induction of Gata1 expression. In contrast, upon bi-allelic deletion of the +46 Gata1-associated erythroid enhancer, Ezh1 expression was unchanged in G1E progenitors but failed to be activated in differentiated G1ER cells (Figure 3C). These results indicate that the Gata2-associated regulatory element negatively regulates Ezh1 expression in stem/progenitor cells, whereas the switch to Gata1 expression and the activation of Gata1-associated Ezh1 enhancer drive the transcriptional activation of Ezh1 during erythropoiesis. Thus, our findings provide evidence that the transition in expression of tissue- and stage-selective GATA factors accounts for the switch of EZH1/2 expression during lineage specification.
Differential Composition of EZH1 and EZH2-containing Polycomb Repressive Complexes
To investigate the functional similarity and difference between EZH1 and EZH2 in human erythroid development, we identified EZH1 and EZH2-containing PRC2 complexes. We generated human erythroid cell lines (K562) stably expressing subendogenous levels of PRC2 subunits (EZH1, EZH2, EED, and SUZ12) bearing a FLAG epitope and a biotin receptor site at its amino terminus, respectively. Following metabolic labeling by BirA biotin ligase in vivo, PRC2-containing multiprotein complexes were purified and identified by mass spectroscopy (MS) sequencing (Figure S4A). Although both EZH1 and EZH2 pulled down other PRC2 core components such as EED, SUZ12 and JARID2, they did not pull down each other, confirming a prior suggestion that EZH1 and EZH2 are present in mutually exclusive PRC2 complexes (Figure S4B,C) (Shen et al., 2008). Notably, more peptides corresponding to EZH2 than EZH1 were recovered in EED or SUZ12-containing complexes, indicating that a greater abundance of EZH2 than EZH1 in association with other PRC2 core subunits, consistent with previous findings in mouse embryonic stem (ES) cells (Shen et al., 2008). Furthermore, a number of nuclear factors appear to associate differentially with different PRC2 subunits, including hematopoietic-specific regulators, chromatin modifying enzymes, and the general transcription factors (Figure S4B,C).
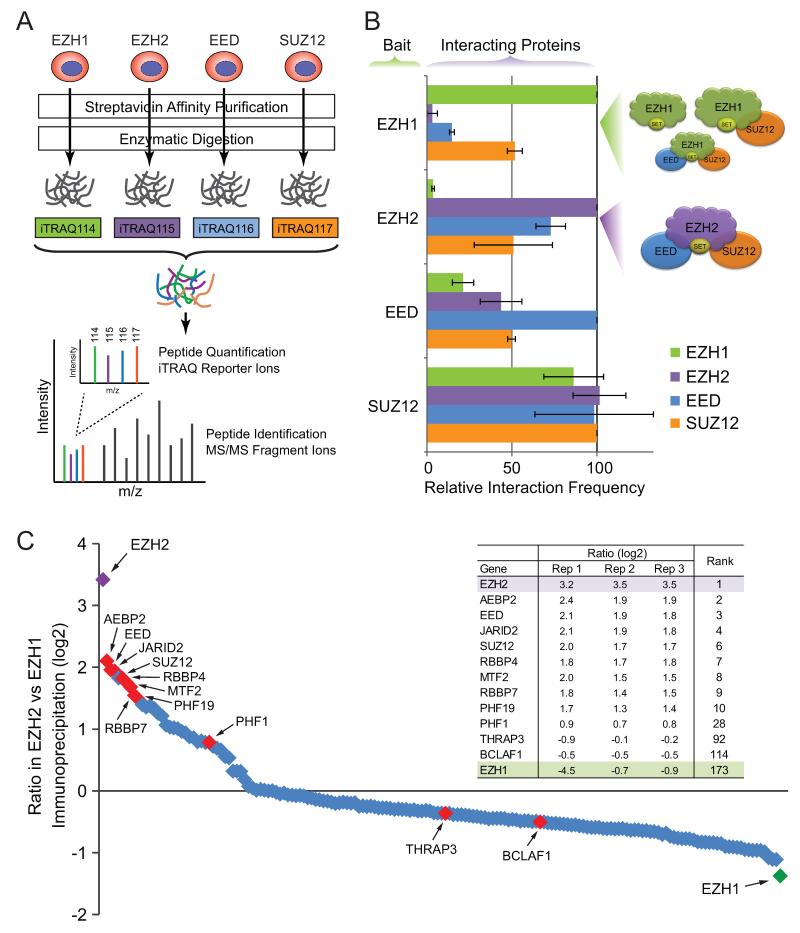
To determine further the stoichiometry of PRC2 complexes in vivo, we employed a quantitative approach by using the iTRAQ proteomics method (Figure 4A) (Ross et al., 2004). Briefly, metabolically labeled PRC2 subunits and associated multiprotein complexes were isolated by affinity purification, followed by enzymatic digestion and labeling with isobaric tags. The labeled peptides were then combined, fractionated, and analyzed by high-resolution LC-MS/MS. In EZH1 pull-down, the most abundant peptides correspond to EZH1 itself followed by SUZ12 and EED, whereas there is nearly no detectable EZH2 peptide (Figure 4B; Table S1). In contrast, in EZH2 pull-down, comparable abundances of EED and SUZ12 were observed in EZH2-containing complexes. Similarly, EED and SUZ12 were found to pull-down other core subunits in comparable frequencies (Figure 4B). These results demonstrate that EZH1 is present predominantly alone or in complex with SUZ12, but much less frequently in association with EED and SUZ12 together. In contrast, EZH2 is present predominantly in stable complexes containing both EED and SUZ12, constituting the canonical PRC2 holoenzyme (Ciferri et al., 2012).
Figure 4. Differential Composition of EZH1 and EZH2-containing PRC2 Complexes.
(A) iTRAQ quantitative proteomic analysis of in vivo stoichiometry of PRC2 subunit composition.
(B) Relative interaction frequency between PRC2 subunits is shown in each immunoprecipitation using EZH1, EZH2, EED, or SUZ12 as a bait. Results are means ± SD of three experiments. Schematic diagrams of EZH1 or EZH2-containing PRC2 complexes are shown on the right.
(C) PRC2-interacting partner proteins are ranked by the ratio of the frequency in EZH2 versus EZH1 immunoprecipitation using iTRAQ quantification. The ratio and rank order of the known PRC2 components are shown in the table.
To measure more precisely the frequencies of EZH1 and EZH2-interacting partner proteins in vivo, we calculated the ratio of the identified polypeptides present in EZH2 versus EZH1 immunoprecipitation by iTRAQ quantification (Figure 4C; Table S1; Experimental Procedures). Strikingly, 9 out of 10 top ranked proteins associated with EZH2 relative to EZH1 are known subunits of the PRC2 complexes, including AEBP2, EED, JARID2 and SUZ12. The recovery of all known PRC2 components demonstrates the validity of our quantitative proteomics approach. These analyses indicate that EZH2, but not EZH1, strongly favors the formation of canonical PRC2 complexes in vivo.
Differential Requirement of Polycomb Core Subunits for Global H3K27 Methylation
Since the PRC2 complex is responsible for catalyzing histone H3 lysine 27 (H3K27) methylation, we next determined the effect of depleting PRC2 subunits by shRNA or inhibitors on global H3K27 methylation levels by quantitative chromatin profiling (Jaffe et al., 2013). Upon lentiviral shRNA-mediated depletion of EZH2, EED or SUZ12 expression in primary human erythroid cells, we observed significant decreases of di- and tri-methylation of lysine 27 (H3K27me2/3) and modest increases of mono- or un-methylated lysine 27 (H3K27me0/1) and acetylated H3K27 (H3K27ac) (Figure 5; Figure S5). Similarly, interference of EZH2 enzymatic function by two small molecule inhibitors EZ5 (James Bradner et al., unpublished) and GSK126 (McCabe et al., 2012b) markedly decreased H3K27me2/3, respectively. In contrast, depletion of EZH1 led only to a slight decrease of H3K27me0/1, but no detectable effect on H3K27me2/3. These results suggest that, whereas EZH2, EED and SUZ12 are required to maintain the global level of H3K27me2/3 in vivo, EZH1 is dispensable for the activity associated with H3K27me2/3-dependent canonical PRC2 functions.
Figure 5. Global Chromatin Profiling Identifies Differential Requirement of PRC2 Subunits for H3K27 Methylation.
Primary human erythroid cells treated with lentiviral shRNA against each PRC2 subunit (shEZH1, shEZH2, shEED, or shSUZ12) or small molecule EZH2 inhibitors (EZ5 or GSK126) were subjected to molecular chromatin-signature profiling by mass spectrometry. Two independent replicates for shRNA or inhibitor treatment (0.3μM and 3μM) were analyzed. Cells transduced with non-targeting shRNA (shNT) were analyzed as controls. Each column corresponds to a H3 peptide with the indicated combination of histone marks (bottom). The value in each cell of the heatmap corresponds to the log2-fold change of the mark combination relative to value in shNT control.
See also Figure S5.
EZH1 and SUZ12 Co-occupy Non-canonical PRC2 Targets
To identify gene targets for each PRC2 core subunit, we performed RNA-seq transcriptomic analysis of differentiated erythroid progenitors (ProEs) upon shRNA-mediated silencing of each subunit (Figure 6A,B). The depletion of PRC2 subunits individually resulted in pleiotropic effects on erythroid gene expression (Figure 6C-E). Specifically, depletion of EZH2 led to upregulation of 516 genes and downregulation of 212 gene transcripts (Figure 6C; Table S2; Experimental Procedures). Similarly, depletion of EED led to upregulation of 516 genes and downregulation of 145 genes, consistent with the role of canonical PRC2 in transcriptional repression. Surprisingly, depletion of EZH1 or SUZ12 resulted in significantly more genes to be downregulated (399 and 354 genes, respectively), suggesting that these genes may be directly or indirectly dependent on EZH1 or SUZ12 for optimal expression. We therefore named these as the ‘PRC2-activated genes’, in contrast to the ‘PRC2-repressed genes’ (Figure 6C; Figure S6A-D; Table S3). Of note, the PRC2-activated genes are progressively activated during erythroid differentiation and enriched for genes important for erythroid homeostasis and functions (Figure 6D,E). In contrast, the PRC2-repressed genes are downregulated during differentiation and enriched for genes involved in developmental pathways associated with alternative lineage decisions, such as immune system process, response to stress and leukocyte development.
Figure 6. Identification of Canonical and Non-canonical PRC2 Targets in Differentiating Erythroid Cells.
(A) Transcriptomic profiling of mRNA expression changes upon shRNA-mediated depletion of each PRC2 subunit in primary erythroid cells.
(B) Expression of PRC2 subunits (EZH1, EZH2, EED, and SUZ12) upon shRNA-mediated depletion.
(C) Gene expression changes upon depletion of each PRC2 subunit. The numbers of upregulated (fold change ≥ 2, p-value ≥ 0.05; PRC2-repressed) and downregulated (PRC2-activated) genes are shown for each knockdown.
(D) Gene expression changes of PRC2-activated and PRC2-repressed genes during erythroid differentiation (day 0, HSPC; day 3-7, differentiating ProEs).
(E) Gene ontology (GO) analysis of PRC2 activated or repressed genes.
(F) Unsupervised hierarchical clustering of ChIP-seq datasets within the proximal promoter regions (−2 to +1 kb of TSS). Heatmap depicting the Pearson correlation coefficient of ChIP-seq read densities is shown for the indicated PRC2 subunits and histone marks. (G) ChIP-seq density heatmaps are shown for H3K4me3, H3K27me3, and PRC2 subunits within each promoter category (left). K-means clustering of all PRC2-associated promoters identifies canonical and non-canonical PRC2 targets (right).
(H) mRNA expression values are shown for canonical, non-canonical PRC2 targets, and each promoter category in ProEs. Boxes show median line and quartiles. Whiskers show the boundary to define outliers (red dots).
(I) Gene ontology (GO) analysis of canonical and non-canonical PRC2 target genes.
(J) Gene expression correlation analysis of PRC2 subunit composition and transcriptional activities. See also Figure S6, Tables S2 and S3.
To relate gene expression changes directly with occupancy of the PRC2 complexes, we next determined the chromatin targets of each PRC2 core subunit by ChIP-seq analysis. At the genomic scale, EED and EZH2 highly colocalize with H3K27me3, and inversely correlate with H3K4me3 and H3K27me1, histone marks associated with active transcription (Bernstein et al., 2006; Cui et al., 2009). In contrast, the occupancy of EZH1 and SUZ12 positively associates with H3K4me3 and H3K27me1 at a global scale (Figure 6F).
We then categorized all human genes into bivalent, repressed, active or null state based on the presence of H3K4me3 and H3K27me3 (Experimental Procedures). By this analysis, it is apparent that EZH2 and EED are almost exclusively enriched at bivalent and repressed genes, whereas EZH1 is predominantly enriched at active genes. SUZ12 is enriched at both bivalent/repressed and active genes (Figure 6G). We have extensively validated the EZH1 and EZH2 ChIP-seq analyses by three independent ChIP-seq experiments using different antibodies (Figure S6E,F; Experimental Procedure). These analyses reveal largely consistent chromatin occupancy confirming the reproducibility of these observations.
We further extracted the promoters that are occupied by at least one subunit, and performed k-means clustering analysis. Importantly, PRC2 targeted genes can be separated into two distinct categories: one group is predominantly occupied by SUZ12, EED, and EZH2, highly enriched for H3K27me3, and consists mostly of repressed and bivalent genes (Figure 6G). We therefore named this category ‘Canonical PRC2 targets’. In contrast, the ‘Non-Canonical PRC2 targets’ are predominantly occupied by SUZ12 and EZH1, and are enriched for H3K4me3, DHS, and H3K27me1, in addition to many TFs known to activate erythroid gene expression (Figure 6G; Figure S6G; Table S3). Of note, canonical PRC2 targets primarily consist of repressed and bivalent genes that display lower mRNA expression, whereas non-canonical PRC2 targets mostly consist of active genes that are highly expressed (Figure 6H). By gene ontology analysis, we observed that canonical and non-canonical PRC2 targets are enriched for distinct biological processes (Figure 6I), suggesting that they participate in different cellular functions.
We then integrated the chromatin binding data with the gene expression changes (Experimental Procedure), and correlated combinations of PRC2 subunit binding with PRC2-mediated repression or activation of target gene expression. Importantly, EZH2 together with SUZ12 are the most repressive, such that the genes occupied by EZH2 and SUZ12 are more likely to be repressed compared with other combinations (Figure 6J). Similarly, EED+SUZ12, EZH2+EED+SUZ12, or EED alone strongly correlate with transcriptional repression. In striking contrast, EZH1+SUZ12 strongly correlate with gene activation. EZH1 or SUZ12 alone also associates with activation. Therefore, EZH2 or EZH1, through differential association with SUZ12, mediates distinct transcriptional outputs: EZH2+SUZ12 is predominantly repressive, whereas EZH1+SUZ12 is predominantly activating.
EZH1 Complements EZH2 Loss within Canonical PRC2 Targets
The PRC2 catalytic subunits EZH1 and EZH2 undergo a relative switch in expression during erythroid differentiation (Figure 1). EZH1 and EZH2 form mutually exclusive PRC2 complexes (Figure 4), and are differentially required for the maintenance of global H3K27me2/3 (Figure 5). Furthermore, EZH1 and EZH2 occupy largely non-overlapping chromatin domains and, together with SUZ12, associate with opposing transcriptional changes (Figure 6). Based on these findings, we hypothesized that the expression switch of EZH2 and EZH1 may lead to a functional switch of canonical versus non-canonical PRC2 functions during development. To test this hypothesis, we examined the chromatin occupancy of EZH2 and EZH1 in the presence or absence of EZH2 knockdown. In control knockdown cells, the global occupancy of EZH2 highly overlaps with H3K27me3, whereas EZH1 largely colocalizes with H3K4me3-enriched promoters (Figure 7A). These results are consistent with our previous findings (Figure 6F-J), indicating that EZH2-containing canonical PRC2 complexes and EZH1-SUZ12-containing non-canonical complexes occupy distinct chromatin targets. Upon shRNA-mediated depletion, EZH2 occupancy is eliminated due to the depletion of EZH2 protein (Figure 7A; Figure 6B). Remarkably, in the absence of EZH2, there is a global repositioning of EZH1 to H3K27me3-marked promoters that were previously occupied by EZH2 (Figure 7A), such as the known PRC2 targets HOXA and HOXD gene clusters (Figure 7B; Figure S7). Importantly, 979 (or 68%) of EZH2 targets are ‘recovered’ by EZH1 upon EZH2 loss (Figure 7C; Table S4). Furthermore, EZH1 selectively recovers genes previously targeted by canonical PRC2 complexes containing EZH2, such as EZH2+EED+SUZ12 and EZH2+SUZ12 (Figure 7D,E). Of note, the genes recovered by EZH1 display intermediate mRNA expression level compared to genes remained bound by EZH2 or not recovered by EZH1 (Figure 7F). Furthermore, EZH1-recovered EZH2 targets still display substantial loss of H3K27me3 at their promoter regions (Figure S7), suggesting that the enzymatic function of EZH1 in catalyzing H3K27me3 may be much weaker when recruited to EZH2 target genes, consistent with previous findings (Margueron et al., 2008; Shen et al., 2008). These results demonstrate that, although EZH1 may be repositioned to chromatin targets of EZH2, it is insufficient to execute full repression of EZH2 targets. We conclude that lineage-specifying transcription factors modulate the tissue-specific expression of the core subunits of PRC2 complexes, resulting in differential composition of the chromatin modifying complexes associated with distinct transcriptional outputs.
Figure 7. Loss of EZH2 Leads to Repositioning of EZH1 to Canonical PRC2 Targets.
(A) ChIP-seq density heatmaps are shown for H3K27me3, H3K4me3, EZH2 and EZH1 in the presence or absence of shEZH2, ranked by H3K27me3 read intensity within ±5 kb of TSS in ProEs.
(B) Several representative EZH1-recovered EZH2 target gene loci are shown.
(C) Overlap analysis of EZH1 and EZH2 chromatin targets defined by ChIP-seq in control (shNT) and EZH2-depleted (shEZH2) ProEs.
(D) The distribution of EZH1-recovered chromatin targets within various PRC2 subunit combinations as defined in Figure 6J.
(E) The number and percentage of canonical PRC2 target genes upon EZH2 knockdown are shown.
(F) mRNA expression levels of PRC2 target genes are shown for the indicated categories. Boxplots are constructed as described in Figure 6H.
(G) Model of developmental context-dependent alternative PRC2 subunit composition in transcriptional regulation.
DISCUSSION
Developmental Control of Epigenetic Pathways by Lineage Master Regulators
The epigenetic machinery is critical for tissue homeostasis, and its deregulation underlies many human disorders. While the roles of epigenetic pathways in lineage-specific gene expression have been extensively studied in various model systems, little is known about how lineage master transcriptional regulators contribute to the expression and/or function of epigenetic regulators. By characterizing the regulatory mechanisms controlling erythroid specification from primary human hematopoietic stem/progenitor cells, we uncovered a novel molecular link between lineage-specifying master regulators (GATA2 and GATA1) and the Polycomb regulators. Specifically, by differential association with the distal regulatory elements of the EZH1 gene in stem/progenitor and committed erythroid cells, GATA factors differentially modulate the lineage-specific transactivation of EZH1 expression. The switch from GATA2 to GATA1 is essential for proper erythroid lineage commitment from hematopoietic stem/progenitor cells (Bresnick et al., 2010; Snow et al., 2011). Although the molecular mechanisms controlling the GATA switch and their respective gene targets remain elusive, increasing evidence suggests that GATA2 and GATA1 may regulate a set of shared gene targets in a highly context-specific manner (Dore et al., 2012; Snow et al., 2011). Hence, our study provides one of the first examples that lineage-specifying regulators modulate the context-specific transcriptional activities of a major epigenetic pathway during lineage commitment. Furthermore, these findings close the loop in the transcriptional network in which the epigenetic regulators cooperate with lineage factors in setting an epigenetic ‘landscape’, whereas the lineage master regulators also modify epigenetic complexes to coordinate differentiation.
Differential Composition of Multi-Subunit Chromatin Modifying Complexes Regulates Chromatin Targeting and Transcriptional Activity
Like other multi-subunit chromatin modifying complexes, PRC2 is composed of a set of central subunits providing the core enzymatic activity, along with auxiliary subunits such as JARID2 that serve specialized roles in modulating PRC2 activity and/or recruitment of additional regulatory machinery (Di Croce and Helin, 2013; Margueron and Reinberg, 2011; Shen et al., 2009). The canonical PRC2 complex consists of EED, SUZ12, and the histone methyltransferase EZH2. Here we demonstrate that the alternative PRC2 enzyme subunit EZH1 together with SUZ12 form a non-canonical PRC2 complex, occupy active chromatin domains independent of H3K27me3, and positively regulate gene transcription. The expression of EZH1 and EZH2 inversely correlates during normal hematopoiesis. Loss of EZH2 expression leads to global repositioning of EZH1 to EZH2 targets. Thus, the differential assembly of PRC2 core subunits contributes to the non-canonical PRC2 functions in development and disease.
By demonstrating a functional subunit swap in the PRC2 complexes during hematopoiesis, our study demonstrates that PRC2 composition may regulate differential chromatin targeting and transcriptional activity. These results are analogous to previously characterized cofactor swaps in the SWI/SNF complex during neurogenesis (Lessard et al., 2007) and tumor transformation (Kadoch and Crabtree, 2013). However, in contrast to the developmental subunit swaps in the SWI/SNF complex, the differential subunit composition that we describe here is associated with a ‘qualitative’ change in PRC2 composition consisting of an EZH1/2 swap and the loss of the H3K27me3-binding core component EED. Thus, the resulting EZH1-SUZ12 non-canonical PRC2 composition confers drastically different transcriptional outcomes compared to EED-containing canonical PRC2 complexes. Therefore, these findings support the concept that the differential combination of subunits, rather than the mere presence of a specific subunit, provides functional specificity required for cell-type and developmental context-specific roles of these ubiquitously expressed chromatin modifying complexes.
These findings also raise the possibility that subunit swap and complex composition may be reversed by activating stem/progenitor subunits or inactivating pro-differentiation subunits, thereby allowing mature lineages to acquire stem/progenitor cell features. For example, EZH2 plays predominant roles in embryonic and adult stem cells, and is downregulated in many adult tissues, whereas EZH1 is highly expressed in adult differentiated tissues. Hence, reversal of the EZH2 to EZH1 switch may account for the aberrant EZH2 functions in various human cancers (Figure S1D-H) by directing cells toward a stem/progenitor cell-like state. Further work is needed to elucidate the regulation of EZH1 and EZH2 expression during development and oncogenesis, and to develop therapeutic approaches to target oncogenic EZH2 for cancer intervention.
Non-Canonical PRC2 Functions in Development and Disease
While most studies have focused on PRC2-mediated repression through trimethylation of H3 lysine 27 (H3K27me3), increasing evidence suggests that individual PRC2 subunits may regulate gene expression independent of H3K27me3. Specifically, the catalytic subunit EZH2 acts as a transcriptional coactivator for critical transcriptional factors in breast and prostate cancers, in which its histone methyltransferase activity may not be required (Lee et al., 2011; Xu et al., 2012b). Similarly, conflicting evidence has been reported pointing to compensatory or divergent functions of the PRC2 enzymatic subunits EZH1 and EZH2. For example, EZH1 appears to be a backup enzyme for EZH2 in embryonic and skin stem cells, and participate in transcriptional repression (Ezhkova et al., 2011; Margueron et al., 2008; Shen et al., 2008). In contrast, EZH1 and EZH2 display opposing functions in other cellular contexts, such as myogenesis and neurogenesis, in which EZH1 is associated with transcriptional activation by promoting RNA Polymerase II elongation (Henriquez et al., 2013; Mousavi et al., 2012; Stojic et al., 2011).
In this study, we demonstrate that the alternative composition of PRC2 complexes also contributes to the non-canonical functions of PRC2 in developing erythroid cells. In contrast to the canonical PRC2 complex comprised of EZH2, EED and SUZ12, the non-canonical PRC2 complex assembled by EZH1 and SUZ12 is associated with actively transcribed genes and positive regulation of gene expression. The role of non-canonical PRC2 complexes in the context of canonical EZH2-PRC2 functions remains unclear. Several possible mechanisms may account for the functional switch between EZH2- and EZH1-PRC2 complexes. In one model, EZH1 may function ‘passively’ to antagonize the activity of EZH2 by blocking its access to non-canonical target genes. Thus, by assembling EZH1 and SUZ12 in the absence of EED and H3K27me3, the non-canonical PRC2 complex may prevent transcriptional silencing of its target genes by the canonical PRC2 complex. Alternatively, EZH1 may ‘actively’ regulate gene expression by modulating the transcriptional machinery at non-canonical targets. This mechanism is supported by previous studies of EZH1 in skeletal muscle development, in which EZH1 was noted to interact physically with RNA PolII complex and promote transcriptional elongation (Mousavi et al., 2012). Further investigation is needed to elucidate the molecular mechanisms by which canonical and non-canonical PRC2 complexes attain their target specificity. In addition, it has been suggested that EZH1 may recruit PRC2-EZH2 to its chromatin targets through dimerization between EZH1 and EZH2 (Son et al., 2013). In this study, we were unable to identify evidence of EZH1/2 dimerization using a variety of assays (Figures 4, 6 and S4). In contrast, we noted that EZH1 and EZH2 form mutually exclusive PRC2 complexes in vivo (Figures 4 and S4), consistent with previous studies in ES cells (Shen et al., 2008). Thus, it remains unclear whether these discrepancies are due to different cell models used (e.g. ES, myoblasts, and erythroid cells) or other variables, and will need to be addressed in future studies.
Implications for Therapeutic Targeting of PRC2 Functions in Hematopoietic Malignancies
Owing to recent findings that aberrant expression or gain-of-function mutations of EZH2, as well as other PRC2 core subunits, are common in various malignancies including lymphoma, breast and prostate cancers, intensive efforts have been devoted to developing therapeutic approaches to target EZH2 function in cancers (McCabe et al., 2012a; Morin et al., 2010; Varambally et al., 2002). The discovery of small molecules or peptides that specifically inhibit oncogenic EZH2 raises the exciting possibility of therapeutic targeting of this epigenetic regulator (Kim et al., 2013; Knutson et al., 2012; McCabe et al., 2012b). Major considerations, however, will need to be addressed in further considering EZH2 as a potential therapeutic target. Importantly, evidence for inactivating EZH2 mutations in myeloid disease suggests a tumor suppressor function for EZH2 in other cellular contexts (Ernst et al., 2010; Makishima et al., 2010). Thus, intervention of the oncogenic functions of EZH2 should avoid or limit interference with the tumor-suppressor role of wild-type EZH2. Furthermore, loss of EZH2 expression results in the repositioning of EZH1 to canonical gene targets (Figure 7), suggesting that the relative expression levels of EZH1 and EZH2 may also have an effect on the activities of the canonical versus non-canonical PRC2 functions. Given that the available agents are designed to preferentially target EZH2 enzymatic activities or canonical EZH2-PRC2 functions, their effects on non-canonical PRC2 activity need to be tested carefully to ensure a beneficial on-target effect. The ‘histone code’ has been suggested to decipher how diverse combinations of histone modifications control gene activity (Strahl and Allis, 2000). Our study suggests that the combinatorial assembly of multi-protein chromatin modifying complexes could act in an equally complex manner to specify the context-dependent transcriptional activities in development and diseases.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture
Primary human fetal liver CD34+ HSPCs were isolated from second-trimester fetal livers as described (Van Handel et al., 2010). Primary erythroblasts were generated ex vivo as described (Xu et al., 2010). The K562-BirA, K562-FLAG-Bio-EZH1, EZH2, EED, and SUZ12 stable cell lines were generated as described (Xu et al., 2012a).
Multiprotein Complex Purification and Proteomics Analysis
PRC2-interacting multiprotein complexes were purified and characterized as described (Kim et al., 2009). K562 cells expressing only biotin ligase BirA were used as negative control. For quantitative proteomics by isobaric tags for relative and absolute quantification (iTRAQ), multiprotein complexes from K562 stable cell lines expressing FLAG-Bio-tagged PRC2 subunits were purified and digested to peptides, labelled with iTRAQ reagents (AB SCIEX LLC), combined, and analyzed by LC-MS/MS (Ross et al., 2004). See also Supplemental Experimental Procedures.
Western Blot
Western blot was performed as described (Xu et al., 2012a) using the following antibodies: EZH1 (ab13665, Abcam), EZH2 (612666, BD Biosciences), EED (17-663, Millipore), SUZ12 (39357, Active Motif), H3K27me3 (07-449, Millipore), and GAPDH (sc-26778, Santa Cruz Biotechnology).
Chromatin Profiling
Histone extraction and chromatin profiling were performed as described (Jaffe et al., 2013). 25 μg histones were used for each sample. All rations were normalized to the heavy/light ratio of the H3 41-49 peptide and log2 transformed. Unsupervised hierarchical clustering was performed in Gene-E.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described (Xu et al., 2012a). See also Supplemental Experimental Procedures.
ChIP-seq Data Analysis
For ChIP-seq using the Illumina HiSeq2000 or HiSeq2500, 10 ng of ChIP DNA was processed for library generation using the NEBNext ChIP-seq Library Prep Master Mix following the manufacturer’s protocol (New England Biolabs). Raw sequencing reads were processed using the Illumina software pipeline, and aligned to the reference human genome (UCSC, hg18). See also Supplemental Experimental Procedures for more details on ChIP-seq data analysis.
Gene Expression Analysis
Total RNA was isolated using RNeasy Plus Mini Kit (Qiagen) following manufacturer’s protocol. RNA-seq library was prepared using the Truseq v2 LT Sample Prep Kit (Illumina). The sequencing reads from all RNA-seq experiments were aligned to human reference genome hg18 by TopHat (Trapnell et al., 2009). Differential gene expression analyses were performed using DEseq (Anders and Huber, 2010). See also Supplemental Experimental Procedures.
Chromatin Conformation Capture (3C)
Chromatin conformation capture (3C) assay was performed as described (Xu et al., 2012a). See also Supplemental Experimental Procedures.
Lentiviral RNAi
Lentiviral RNAi was performed in primary human erythroid precursors as previously described (Xu et al., 2012a). The following shRNA clones in the pLKO.1-puro vector were used for EZH1 (sh1: TRCN0000002489; sh3: TRCN0000002441), EZH2 (sh1: TRCN0000286227; sh3: TRCN0000040075), EED (sh2: TRCN0000021205; sh3: TRCN0000021206), and SUZ12 (sh2: TRCN0000038725; sh5: TRCN0000038728). All shRNA clones were obtained from Sigma-Aldrich. Cells transduced with the vector containing non-targeting shRNA (shNT) were used as control.
Enhancer Reporter Assay
Dual luciferase enhancer reporter assay was performed in K562 and GM12752 cells using the Dual-Luciferase Reporter Assay System (Promega). See also Supplemental Experimental Procedures.
Genomic Engineering by CRISPR/Cas9
The clustered regularly interspersed palindromic repeats (CRISPR)/CRISPR associated (Cas) 9 nuclease system was used to introduce enhancer deletion mutations in K562 and G1E/G1ER cells following recently published protocols (Cong et al., 2013; Mali et al., 2013). See also Supplemental Experimental Procedures.
Supplementary Material
Acknowledgment
We thank Sidinh Luc, Cong Peng, Matthew Cancer, Daniel Bauer and members of the Orkin laboratory for assistance and discussion, Ross Tomaino and Steven Gygi for assistance with proteomics analysis, and John Stamatoyannopoulos for assistance with DNase-seq. We thank Kambiz Mousavi and Vittorio Sartorelli for providing the EZH1 antibody, and Ben van Handel and Hanna Mikkola for providing the fetal liver CD34+ cells. This work was supported by funding from the National Institutes of Health (NIH) (G.C.Y., and S.H.O.). S.H.O. is an Investigator of the Howard Hughes Medical Institute (HHMI). J.X. is supported by a NIDDK Career Development Award K01DK093543 and R03DK101665. The authors declare no conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The ChIP-seq, RNA-seq and microarray data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE59090 and GSE36994.
Author Contributions
J.X., D.L., H.X., W.K., and J.E.T. performed experiments and analyzed the data. J.X., Z.S., J.H., L.P., K.G., and G.C.Y. performed bioinformatic analyses. J.E.T. and J.D.J. performed the chromatin profiling experiments and analyzed the data. J.X. and S.H.O. designed the project and interpreted the results. J.X., Z.S., and S.H.O. wrote the manuscript.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R, Nogales E. Molecular architecture of human polycomb repressive complex 2. eLife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nature structural & molecular biology. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- Dore LC, Chlon TM, Brown CD, White KP, Crispino JD. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012;119:3724–3733. doi: 10.1182/blood-2011-09-380634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez B, Bustos FJ, Aguilar R, Becerra A, Simon F, Montecino M, van Zundert B. Ezh1 and Ezh2 differentially regulate PSD-95 gene transcription in developing hippocampal neurons. Molecular and cellular neurosciences. 2013;57:130–143. doi: 10.1016/j.mcn.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Hidalgo I, Herrera-Merchan A, Ligos JM, Carramolino L, Nunez J, Martinez F, Dominguez O, Torres M, Gonzalez S. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;11:649–662. doi: 10.1016/j.stem.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Jaffe JD, Wang Y, Chan HM, Zhang J, Huether R, Kryukov GV, Bhang HE, Taylor JE, Hu M, Englund NP, et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet. 2013;45:1386–1391. doi: 10.1038/ng.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat Protoc. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, Orkin SH. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nature chemical biology. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nature chemical biology. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, Liou YC, Yu Q. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H, Jankowska AM, Tiu RV, Szpurka H, Sugimoto Y, Hu Z, Saunthararajah Y, Guinta K, Keddache MA, Putnam P, et al. Novel homo- and hemizygous mutations in EZH2 in myeloid malignancies. Leukemia. 2010;24:1799–1804. doi: 10.1038/leu.2010.167. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci U S A. 2012a;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012b;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Mochizuki-Kashio M, Mishima Y, Miyagi S, Negishi M, Saraya A, Konuma T, Shinga J, Koseki H, Iwama A. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 2011;118:6553–6561. doi: 10.1182/blood-2011-03-340554. [DOI] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol Cell. 2012;45:255–262. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & cellular proteomics: MCP. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita J, Sahoo D, Rossi DJ, Bhattacharya D, Serwold T, Inlay MA, Ehrlich LI, Fathman JW, Dill DL, Weissman IL. Gene Expression Commons: an open platform for absolute gene expression profiling. PloS one. 2012;7:e40321. doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow JW, Trowbridge JJ, Johnson KD, Fujiwara T, Emambokus NE, Grass JA, Orkin SH, Bresnick EH. Context-dependent function of “GATA switch” sites in vivo. Blood. 2011;117:4769–4772. doi: 10.1182/blood-2010-10-313031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Shen SS, Margueron R, Reinberg D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013;27:2663–2677. doi: 10.1101/gad.225888.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojic L, Jasencakova Z, Prezioso C, Stutzer A, Bodega B, Pasini D, Klingberg R, Mozzetta C, Margueron R, Puri PL, et al. Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenetics & chromatin. 2011;4:16. doi: 10.1186/1756-8935-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Handel B, Prashad SL, Hassanzadeh-Kiabi N, Huang A, Magnusson M, Atanassova B, Chen A, Hamalainen EI, Mikkola HK. The first trimester human placenta is a site for terminal maturation of primitive erythroid cells. Blood. 2010;116:3321–3330. doi: 10.1182/blood-2010-04-279489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Xie H, Xu J, Hsu JH, Nguyen M, Fujiwara Y, Peng C, Orkin SH. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell. 2014;14:68–80. doi: 10.1016/j.stem.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010;24:783–798. doi: 10.1101/gad.1897310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Shao Z, Glass K, Bauer DE, Pinello L, Van Handel B, Hou S, Stamatoyannopoulos JA, Mikkola HK, Yuan GC, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Developmental cell. 2012a;23:796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012b;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.