Abstract
A standard set of three APSY-NMR experiments has been used in daily practice to obtain polypeptide backbone NMR assignments in globular proteins with sizes up to about 150 residues, which had been identified as targets for structure determination by the Joint Center for Structural Genomics (JCSG) under the auspices of the Protein Structure Initiative (PSI). In a representative sample of 30 proteins, initial fully automated data analysis with the software UNIO-MATCH-2014 yielded complete or partial assignments for over 90% of the residues. For most proteins the APSY data acquisition was completed in less than 30 hours. The results of the automated procedure provided a basis for efficient interactive validation and extension to near-completion of the assignments by reference to the same 3D heteronuclear-resolved [1H,1H]-NOESY spectra that were subsequently used for the collection of conformational constraints. High-quality structures were obtained for all 30 proteins, using the J-UNIO protocol, which includes extensive automation of NMR structure determination.
Keywords: Automated projection spectroscopy, Automated data analysis, UNIO software, J-UNIO protocol, Protein structure determination
Introduction
Automated projection spectroscopy (APSY) was introduced nearly a decade ago (Fiorito et al. 2006; Hiller et al. 2005; 2008). In spite of a court injunction which blocked the use of APSY and related techniques for several years (Wüthrich 2011), APSY-NMR has become a standard technique for projects of the Joint Center for Structural Genomics (JCSG: www.jcsg.org). Within the J-UNIO protocol for extensive automation of protein structure determination (Serrano et al. 2012), APSY-NMR is routinely used for polypeptide backbone assignments of proteins with sizes up to about 150 amino acid residues, and significantly larger proteins have also been successfully studied (Jaudzems et al. 2014; Mohanty et al. 2014). Here we report the results obtained with a representative sample of 30 JCSG target proteins. The paper describes the characterization of the “structure-quality” protein solutions used for the APSY-NMR measurements, presents the experimental conditions for the recording of the APSY-NMR data sets, and surveys the results obtained by automated analysis of the APSY-NMR data with the use of the software UNIO-MATCH-2014 (Volk et al. 2008; T. Herrmann, to be published).
Materials and methods
Protein samples were produced using a standard cloning, expression and purification protocol (Serrano et al. 2012). Protein concentrations ranged from 0.8 to 1.5 mM in NMR buffer (20 mM sodium phosphate at pH 6.0, 50 mM sodium chloride, 5 mM NaN3 in 5% 2H2O/95% H2O (v/v); for proteins containing S−H groups, 2 mM [d10]-dithiothreitol was added). Prior to the structure determination, the targets were screened for high-quality NMR spectra, using μg amounts of [u-15N]-labeled protein and a 1.7 mm room temperature microcoil probe (Serrano et al. 2012). Protein solutions which yielded high-quality NMR-Profiles (Pedrini et al. 2013) were used for structure determination.
Backbone assignments were obtained from a standard set of three APSYNMR experiments: 4D APSY-HACANH, 5D APSY-HACACONH and 5D APSYCBCACONH (Hiller et al. 2008; Serrano et al. 2012). The APSY data were analyzed with the software GAPRO (Hiller et al. 2005; 2008), using standard parameters, except that the signal-to-noise threshold for peak identification (Herrmann et al. 2002b) was optimized for each experiment. The three GAPRO-generated listings of peak coordinates were then used as input for UNIO-MATCH-2014 (Volk et al. 2008; T. Herrmann, to be published) for automated backbone assignment. Each UNIO-MATCH calculation included 10 independent runs of optimization with the same input data in order to find a self-consistent solution; the tolerances for chemical shift matching between the different peak lists, which were used to form generic spin systems and to establish sequential connectivities, were set to 0.02 ppm and 0.2 ppm for protons and heavy atoms, respectively. The automated backbone assignments yielded by UNIO-MATCH were then validated and extended interactively following the J-UNIO protocol (Serrano et al, 2012). In this procedure, the MATCH output and the 3D 15N-resolved, 13Cali-resolved and 13Caro-resolved [1H,1H]-NOESY spectra, which are also used for automated side chain resonance assignments with the routine UNIO-ASCAN (Fiorito et al, 2008) and for the collection of conformational constraints, are loaded into CARA (Keller, 2004). Erroneous UNIO-MATCH assignments are identified and corrected, and missing assignments are added, using primarily the sequential dNN(i,i+1) and dαN(i,i+1) NOE connectivities (Wüthrich, 1986). Since the extent of the backbone chemical shift assignments yielded by UNIO-MATCH is typically about 90%, this process requires usually only a few hours of work by a spectroscopist.
NMR-profiles (Pedrini et al. 2013) were generated from 2D [15N,1H]-HSQC spectra recorded on a Bruker AVANCE 700 MHz NMR instrument equipped with a 1.7 mm TXI z-gradient microcoil-probe. The APSY-NMR data sets were recorded on a Bruker AVANCE 600 MHz spectrometer equipped with a 5 mm CP2 QCI-F z-gradient cryogenic probehead. The numbers of 2D projections measured for the three different experiments are listed in Table 1. All projections were acquired with 96 × 2048 complex data points, and before Fourier transformation the spectra were multiplied in both dimension with a 45°-shifted sine bell (DeMarco and Wüthrich 1976). Since the same resolution in the indirect dimension was used throughout, the differences in the APSY NMR recording times reflect exclusively the number of acquired NMR transients, which was set individually for each protein in order to achieve sufficient peak intensity in the 2D APSY projections for analysis with the program GAPRO (Hiller et al. 2005). The 3D heteronuclear-resolved [1H,1H]-NOESY experiments were acquired on an 800 MHz Bruker AVANCE spectrometer equipped with a 5 mm room temperature TXI xyz-gradient probehead.
Table 1.
Survey of the 30 JCSG target proteins of Figure 3: GenBank accession codes, PDB accession codes, size, experimental conditions for APSY-NMR, extent of polypeptide backbone assignments
| GenBanka | PDBa | Size (aa) | Conc.b (mM) | Number of 2D APSY projections | NMR timec (h) | Percentage of residue assignment by UNIO-MATCH-2014d | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5D HACACONH | 5D CBCACONH | 4D HACANH | C | P | E | U | |||||
| BC033015 | 2m51 | 62 | 1.2 | 26 | 26 | 31 | 13 | 79.1 | 17.7 | 0.0 | 3.2 |
| NP_346341.1 | 2m7o | 70 | 1.2 | 38 | 26 | 31 | 13 | 85.7 | 10.0 | 0.0 | 4.3 |
| BC008182 | 2lo1 | 71 | 1.5 | 26 | 32 | 27 | 13 | 85.9 | 12.7 | 0.0 | 1.4 |
| NM_00109834 | 2m34 | 71 | 0.8 | 42 | 42 | 31 | 16 | 78.9 | 15.5 | 5.6 | 5.6 |
| NP_254181.1 | 2mhg | 75 | 1.2 | 42 | 44 | 23 | 25 | 78.7 | 20.0 | 9.3 | 1.3 |
| NP_390345.1 | 2lyx | 87 | 1.2 | 32 | 26 | 25 | 23 | 77.0 | 16.1 | 8.0 | 6.9 |
| BC010264 | 2m7s | 90 | 1.2 | 20 | 20 | 23 | 17 | 74.4 | 18.9 | 4.4 | 6.7 |
| BC024153 | 2lxi | 91 | 1.2 | 32 | 28 | 31 | 27 | 54.9 | 38.5 | 7.7 | 6.6 |
| NM_002139 | 2mks | 92 | 1.2 | 38 | 41 | 29 | 38 | 87.0 | 10.9 | 0.0 | 2.1 |
| NM_184234 | 2mhn | 94 | 1.2 | 42 | 42 | 29 | 22 | 83.0 | 14.9 | 6.4 | 2.1 |
| BC008071 | 2m3d | 95 | 1.2 | 20 | 20 | 23 | 9 | 70.5 | 21.1 | 3.2 | 8.4 |
| NP_809759.1 | 2m4l | 99 | 1.2 | 20 | 20 | 23 | 9 | 71.7 | 21.2 | 4.0 | 7.1 |
| ZP_02034617.1 | 2lz0 | 100 | 1.0 | 30 | 30 | 27 | 13 | 84.0 | 13.0 | 1.0 | 3.0 |
| ZP_02042476.1 | 2mct | 102 | 1.2 | 40 | 38 | 29 | 46 | 92.1 | 5.9 | 1.0 | 2.0 |
| YP_002937094.1 | 2mca | 103 | 1.2 | 28 | 28 | 27 | 12 | 81.6 | 18.4 | 3.9 | 0.0 |
| BC043071 | 2m52 | 105 | 1.2 | 20 | 20 | 23 | 9 | 68.6 | 23.8 | 4.8 | 7.6 |
| YP_001298242.1 | 2mqc | 105 | 0.8 | 32 | 32 | 29 | 75 | 67.6 | 25.7 | 6.7 | 6.7 |
| YP_001714923.1 | 2mmb | 107 | 1.2 | 28 | 36 | 23 | 12 | 65.4 | 24.3 | 5.6 | 10.3 |
| ZP_02071672.1 | 2mhd | 110 | 1.2 | 32 | 30 | 27 | 12 | 73.6 | 16.4 | 2.7 | 10.0 |
| BC030493 | 2lq5 | 113 | 1.1 | 32 | 28 | 31 | 27 | 85.0 | 9.7 | 5.3 | 2.7 |
| ZP_02041089.1 | 2mc8 | 114 | 1.2 | 20 | 20 | 23 | 9 | 80.7 | 18.4 | 0.9 | 0.9 |
| YP_926445.1 | 2l6o | 114 | 1.2 | 42 | 42 | 27 | 15 | 84.2 | 8.8 | 7.0 | 0.9 |
| NP_809137.1 | 2mw1 | 118 | 1.2 | 38 | 38 | 29 | 28 | 79.7 | 16.9 | 3.4 | 5.1 |
| YP_001300941.1 | 2lrg | 126 | 1.2 | 42 | 38 | 23 | 55 | 92.1 | 7.1 | 0.8 | 0.8 |
| NP_390037.1 | 2lr4 | 128 | 1.2 | 32 | 32 | 27 | 37 | 89.1 | 9.4 | 1.6 | 1.6 |
| YP_001302112.1 | 2lge | 129 | 1.1 | 32 | 32 | 32 | 29 | 89.1 | 10.1 | 0.8 | 3.9 |
| NP_814968.1 | 2llg | 143 | 1.2 | 32 | 40 | 31 | 63 | 72.7 | 21.0 | 6.3 | 2.1 |
| ZP_02069618.1 | 2ml6 | 148 | 1.2 | 30 | 30 | 31 | 41 | 77.7 | 19.6 | 2.7 | 6.8 |
| YP_193882.1 | 2mwm | 151 | 1.2 | 28 | 28 | 23 | 66 | 80.8 | 9.9 | 9.3 | 1.3 |
| NP_372339.1 | 2mqb | 152 | 1.2 | 40 | 40 | 31 | 30 | 80.9 | 15.8 | 3.3 | 2.6 |
Proteins are identified by their GenBank accession codes and PDB accession codes.
Protein concentration of the NMR sample.
Total time used for acquiring the three APSY experiments (see text).
Percentage of residue assignment by UNIO-MATCH-2014. The letters C, P and E indicate the residues with complete, partial and erroneous assignments, respectively, and U represents the residues left unassigned (see also Fig. 3 and the text).
Results
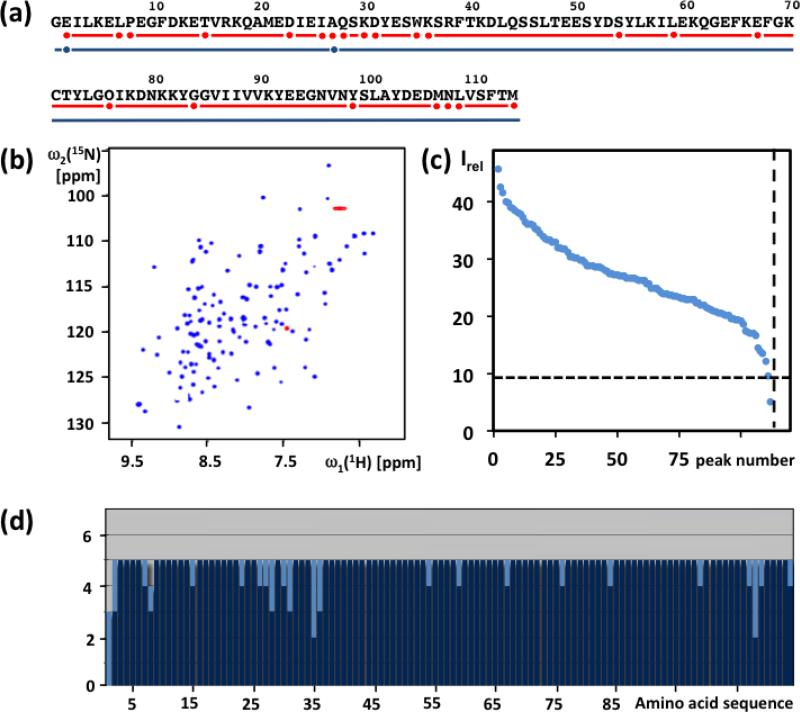
The preparation of NMR samples of the 30 proteins used in this study (Table 1) is described in Materials and methods. The proteins were selected for their biological interest in various JCSG projects, and decisions to go ahead with the NMR structure determination were based on the results from screening with 1D 1H-NMR and 2D [15N,1H]-HSQC experiments. Here, this is illustrated with the protein ZP_02041089.1 (PDB id. 2mc8) (Fig. 1,a–c). From the protein sequence, 113 15N–1H backbone amide and Trp side chain indole cross peaks were expected in the [15N,1H]-HSQC spectrum, and 112 peaks were identified in Fig. 1b after excluding the Asn, Gln and Arg side-chain signals (Pedrini et al. 2013). In the NMR-Profile (Pedrini et al. 2013), 111 of these peaks exhibited intensities above the threshold required for observation of complete APSY-NMR data sets (Figure 1c). The homogeneous peak intensity distribution in the Profile further suggested the absence of flexibly disordered polypeptide segments.
Fig. 1.
Characterization of the solution of the protein ZP_02041089.1 used for APSY-NMR experiments, and result of the automated backbone assignment with UNIO-MATCH-2014. (a) Amino acid sequence (Gly-1 results from the cloning strategy). The red line indicates residues with complete automated backbone assignment by UNIO-MATCH-2014, and red dots identify residues with incomplete assignments. The blue line and the blue dots indicate the corresponding information obtained after interactive validation of the assignments, which are complete except that for E2 and A27 only the chemical shifts of Cα, Hα and Cβ were assigned. (b) [15N,1H]-HSQC spectrum of the 15N- labeled protein recorded at 700 MHz with a 1.7 mm micro-coil probehead. Red color identifies folded peaks of arginine side chain 15N−1H moieties. (c) NMR- profile obtained from the data in (b); the dotted horizontal and vertical lines indicate, respectively, the intensity threshold for detection of APSY-NMR signals (see text) and the number of backbone amide and tryptophan indole 15N−1H correlation signals expected from the amino acid sequence. (d) UNIO-MATCH-2014 output. The dark blue and light blue bars represent the number of assigned and unassigned 1HN, 15N, Cα, Hα and Cβ atoms per residue, respectively.
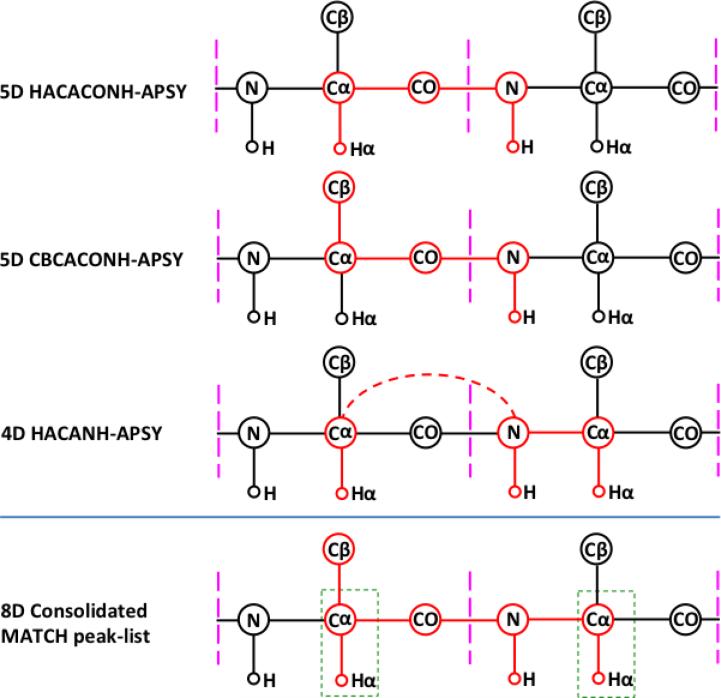
Based on the high-quality data from the micro-scale NMR experiments, we decided to produce [13C,15N]-labeled protein for the NMR structure determination. When collecting the experimental data needed for the backbone assignments, 23 projections for 4D APSY-HACANH and 20 projections for the accumulation of each of the 5D APSY-HACACONH and 5D APSY-CBCACONH experiments (Fig. 2) were recorded with 4 transients per projection. The total NMR instrument time used to measure the APSY data was 9 hours. The individual APSY experiments were processed with GAPRO (Hiller et al. 2005) to prepare the three listings of peak coordinates (Fig. 2) used as input for UNIO-MATCH-2014, which yielded complete or partial assignments of the atoms 15N, 1HN, Cα, Hα and Cβ for all residues except for the N-terminal Gly (Fig. 1d). Following the J-UNIO protocol (Serrano et al. 2012), the backbone assignments from UNIO-MATCH-2014 were interactively validated with the use of the 3D 15N-resolved and 3D 13Cali-resolved [1H,1H]-NOESY spectra. A single assignment, Cα of Asn108, was found to be erroneous, and the validation resulted in complete backbone atom assignments, with the sole exceptions of the amide moieties of Glu2 and Ala 27. This final backbone assignment was part of the input for the remainder of the structure determination, which was based on heteronuclear-resolved [1H,1H]-NOESY data analyzed with the software UNIO-ATNOS/ASCAN for automated side-chain chemical shift assignment (Fiorito et al. 2008) and UNIO-ATNOS/CANDID for automated NOE assignment (Herrmann et al. 2002a,b), in combination with the simulated annealing protocol of CYANA for structure generation (Güntert et al. 1997). The resulting structure of ZP_02041089.1 is precisely defined (PDB id. 2mc8), with backbone and heavy atoms RMSD values of 0.60 ± 0.10 Å and 1.04 ± 0.10 Å, respectively.
Fig. 2.
Scalar connectivities detected by the three APSY-NMR experiments used and consolidated peak list generated by UNIO-MATCH-2014. The three upper panels show the correlations obtained from the individual APSY experiments. The higher-dimensional correlation generated by UNIO-MATCH-2014 is indicated at the bottom. Magenta vertical bars delimit the individual residues. Correlated atoms are highlighted in red and connected by red lines. The 4D HACANH experiment yields interresidue (broken line) and intraresidue correlations. Overlaps of the resonances from the atoms in the green rectangles are used by UNIO-MATCH-2014 to identify sequential connectivities between neighboring residues.
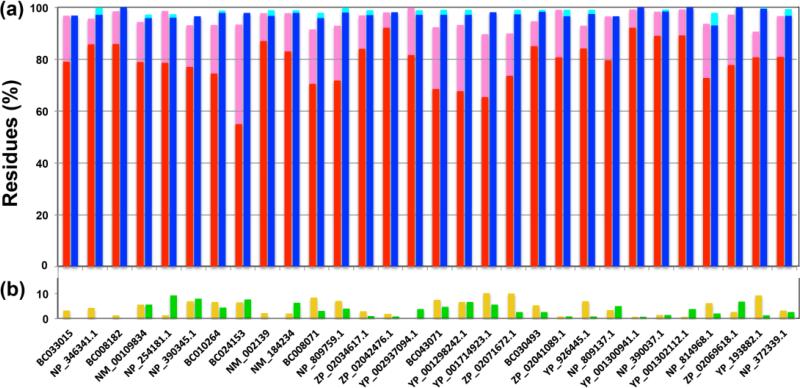
Analogous procedures to those illustrated in Fig. 1 for ZP.02041089.1 were applied with all the other proteins in Table 1. The table shows that the protein size varied from 62 to 152 residues, and that the protein concentration in the NMR samples varied between 0.8 and 1.5 mM, with most solutions containing 1.2 mM protein. For the 4D APSY-HACANH experiments, 23 to 31 projections were acquired, and 20 to 44 projections were recorded for each of the two 5D APSY experiments (Table 1). The recording time was adjusted so as to obtain sufficient signal intensity for reliable data processing with GAPRO (Hiller et al. 2005; 2008). For YP_001298242.1, 24 transients were acquired, and for the other proteins, 4 to 16 transients were accumulated per projection. The NMR time thus used to measure the APSY data ranged from 9 to 75 hours, and for most of the proteins it was less than 30 hours. The extent of the assignments obtained is presented in Fig 3. For all proteins, at least 89.7% of all the residues were completely or partially assigned automatically with UNIO-MATCH-2014, and for most proteins the extent of the automated assignments was above 95% (Table 1; red and pink bars in Fig. 3a). The listed measurement times were chosen based on the intensities observed in the NMR-Profiles, the decisions being largely at the discretion of the individual spectroscopists. The extent of automated assignments obtained depends obviously largely on the performance of UNIO-MATCH-14, which will be discussed elsewhere (Herrmann et al. private communication), and can be affected by conformational exchange line broadening in discrete polypeptide segments, and by peak overlap. Important features of the automated assignment procedures are of course the percentages of missed and erroneously assigned residues (Fig. 3b). The number of residues with erroneous partial or complete assignments, which are contained in the pink and red bars of Fig. 3a, was below 10% for all proteins (green bars in Fig. 3b), with an average of 3.4 ± 2.7 % over all 30 data sets (a residue is counted as erroneously assigned if at least one of the five backbone atoms of interest is wrongly assigned). For all 30 proteins, at most 10% of the residues remained unassigned (yellow bars in Fig. 3b), with an average of 4.5 ± 3.0 %. After validation and extension with the use of the NOESY experiments recorded for the collection of conformational constraints (see Materials and Methods), the polypeptide backbone assignments obtained automatically from the APSY-NMR experiments (blue bars in Fig. 3) provided the basis for high-quality NMR structure determinations with the J-UNIO protocol, with RMSD values among the 30 proteins of Table 1 ranging from 0.42 to 0.74 Å for the backbone atoms, and from 0.77 to 1.21 Å for all heavy atoms (see the PDB deposits listed in Table 1).
Fig. 3.
Survey of the assignment results for 30 JCSG target proteins. Backbone assignments obtained automatically by UNIO-MATCH-2014, and final assignments after interactive validation using a 3D 15N-resolved [1H,1H]-NOESY spectrum are shown. (a) Histograms showing the percentages of residues assigned. Residues assigned completely and partially by UNIO-MATCH-2014 are represented by red and pink bars, respectively. Blue and cyan bars represent the corresponding results after interactive validation and extension of the backbone assignment with the use of 3D 15N-resolved and 13Cali-resolved [1H, 1H]- NOESY data. (b) Percentages of residues that were erroneously assigned by UNIO-MATCH-2014 are indicated with green bars (these are contained in the red and/or pink bars), and those that were left unassigned are represented by yellow bars. From left to right, the proteins are arranged according to molecular weights, showing that the assignment results are not correlated with the protein size over the range from 62 to 152 residues, but rather with the quality of the NMR spectra (see also the text).
Discussion and conclusions
The JCSG is a PSI:Biology high-throughput protein structure determination center. Work in this environment confirmed the previously discussed advantages of APSY-NMR (Fiorito et al. 2006; Hiller et al. 2005; 2008) and its use with the JUNIO protocol (Serrano et al. 2012) in daily practice. Specifically, the use of APSY-NMR ensures important savings of instrument time when compared with the use of conventional triple resonance experiments for obtaining corresponding information. Table 1 shows that the total measuring time for the presently used combination of three APSY-NMR experiments was shorter than 15 h for 12 proteins, and shorter than 30 h for 22 of the 30 proteins studied. Furthermore, the high digital resolution of the 2D APSY projections facilitates automated analysis of the data, as illustrated with the presently used software UNIO-MATCH-2014. Another significant advantage is that the APSY-based automated assignments can efficiently be validated with the use of 3D heteronuclear-resolved [1H,1H]-NOESY spectra, due to the fact that most of the backbone chemical shifts are precisely known and correctly assigned from the APSY-NMR measurements. In the overall structure determination procedure, validation against the NOESY data sets is efficient because no data need to be recorded in addition to the measurements required for the collection of conformational constraints (Serrano et al. 2012). Besides the identification of residues with erroneous assignments generated by the automated procedure (Fig. 3b), the NOESY data also enable to determine the correct chemical shifts for these residues, and to close gaps in the sequential assignments that may be left by the automated data analysis (Fig. 3).
In conclusion, considering the aforementioned unique assets, the use of APSY-NMR experiments in combination with a suitable automated assignment routine, as exemplified here with UNIO-MATCH-2014, is a valid alternative to the many previously proposed approaches for automated polypeptide backbone NMR assignment in proteins (e.g., Atreya et al. 2000; Bartels et al. 1997; Crippen et al. 2010; Fredriksson et al. 2012; Lee et al. 2014; Lemak et al. 2008; Lescop and Brutscher 2009; Moseley et al. 2001; Schmidt and Güntert 2012; 2013; Schmucki et al. 2009; Staykova et al. 2008; Tikole et al. 2012; Zawadzka-Kazimierczuk et al. 2012; Zimmermann et al. 1997). The Figure 3 illustrates that within the range covered by the 30 proteins of Table 1, the result of polypeptide backbone assignments based on the present protocol of using APSY-NMR and UNIO-MATCH-2014 does not depend critically on the protein size. Based on these results, it is not surprising, that the same approach has successfully been applied for proteins with about 200 amino acid residues (Jaudzems et al. 2014; Mohanty et al. 2014).
Acknowledgements
This work was supported by the Joint Center for Structural Genomics through the NIH Protein Structure Initiative (PSI) grant U540-GM074898 from the National Institute of General Medical Sciences (www.nigms.nih.gov). Kurt Wüthrich is the Cecil H. and Ida M. Green Professor of Structural Biology at The Scripps Research Institute. We thank our TSRI graduate students Bryan Martin, Sergey Shnitkind, Lukas Susac and Arndt Wallmann, who each provided the APSY-based assignments for one of the proteins in Table 1. T.H. thanks his graduate student Viet Dung Duong for contributions to the software UNIO-MATCH-2014.
Contributor Information
Samit Kumar Dutta, Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA, and Joint Center for Structural Genomics (http://www.jcsg.org.), La Jolla, CA 92037, USA.
Pedro Serrano, Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA, and Joint Center for Structural Genomics (http://www.jcsg.org.), La Jolla, CA 92037, USA.
Andrew Proudfoot, Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA, and Joint Center for Structural Genomics (http://www.jcsg.org.), La Jolla, CA 92037, USA.
Michael Geralt, Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA, and Joint Center for Structural Genomics (http://www.jcsg.org.), La Jolla, CA 92037, USA.
Bill Pedrini, Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA, and Joint Center for Structural Genomics (http://www.jcsg.org.), La Jolla, CA 92037, USA; Institute of Molecular Biology and Biophysics, ETH Zürich, CH-8093 Zürich, Switzerland.
Torsten Herrmann, Institut des Sciences Analytiques, Centre de RMN à Très Hauts Champs, Université de Lyon, UMR 5280 CNRS, ENS Lyon, UCB Lyon 1, 5 rue de la Doua, 69100 Villeurbanne, France.
Kurt Wüthrich, Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA, and Joint Center for Structural Genomics (http://www.jcsg.org.), La Jolla, CA 92037, USA; Skaggs Institute for Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA; Institute of Molecular Biology and Biophysics, ETH Zürich, CH-8093 Zürich, Switzerland.
References
- Atreya HS, Sahu SC, Chary KVR, Govil G. A tracked approach for automated NMR assignments in proteins (TATAPRO). J Biomol NMR. 2000;17:125–136. doi: 10.1023/a:1008315111278. [DOI] [PubMed] [Google Scholar]
- Bartels C, Güntert P, Billeter M, Wüthrich K. GARANT-a general algorithm for resonance assignment in multidimensional nuclear magnetic resonance spectra. J Comput Chem. 1997;18:139–149. [Google Scholar]
- Crippen GM, Rousaki A, Revington M, Zhang Y, Zuiderweg ERP. SAGA: rapid automatic mainchain NMR assignment for large proteins. J Biomol NMR. 2010;46:281–298. doi: 10.1007/s10858-010-9403-2. [DOI] [PubMed] [Google Scholar]
- DeMarco A, Wüthrich K. Digital filtering with a sinusoidal window function: An alternative technique for resolution enhancement in FT NMR. J Magn Reson. 1976;24:201–204. [Google Scholar]
- Fiorito F, Hiller S, Wider G, Wüthrich K. Automated resonance assignment of proteins: 6D APSY-NMR. J Biomol NMR. 2006;35:27–37. doi: 10.1007/s10858-006-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorito F, Herrmann T, Damberger F F, Wüthrich K. Automated amino acid side-chain NMR assignment of proteins using 13C- and 15N-resolved 3D [1H,1H]-NOESY. J Biomol NMR. 2008;42:23–33. doi: 10.1007/s10858-008-9259-x. [DOI] [PubMed] [Google Scholar]
- Fredriksson J, Bermel W, Staykova DK, Billeter M. Automated protein backbone assignment using the projection-decomposition approach. J Biomol NMR. 2012;54:43–51. doi: 10.1007/s10858-012-9649-y. [DOI] [PubMed] [Google Scholar]
- Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002a;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR. 2002b;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- Hiller S, Fiorito F, Wüthrich K, Wider G. Automated projection spectroscopy (APSY). Proc Natl Acad Sci USA. 2005;102:10876–10881. doi: 10.1073/pnas.0504818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller S, Wider G, Wüthrich K. APSY-NMR with proteins: practical aspects and backbone assignment. J Biomol NMR. 2008;42:179–195. doi: 10.1007/s10858-008-9266-y. [DOI] [PubMed] [Google Scholar]
- Jaudzems K, Pedrini B, Geralt M, Serrano P, Wüthrich K. Automated J UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4. J Biomol NMR. 2014 doi: 10.1007/s10858-014-9886-3. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. Computer-aided resonance assignment. Cantina: 2004. http://cara.nmr.ch/ [Google Scholar]
- Lee W, Stark JL, Markley JL. PONDEROSA-C/S: client–server based software package for automated protein 3D structure determination. J Biomol NMR. 2014;60:73–75. doi: 10.1007/s10858-014-9855-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W, Hu K, Tonelli M, Bahrami A, Neuhardt E, Glass KC, Markley JL. Fast automated protein NMR data collection and assignment by ADAPT- NMR on Bruker spectrometers. J Magn Reson. 2013;236:83–88. doi: 10.1016/j.jmr.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemak A, Steren CA, Arrowsmith CH, Llinás M. Sequence specific resonance assignment via multicanonical Monte Carlo search using an ABACUS approach. J Biomol NMR. 2008;41:29–41. doi: 10.1007/s10858-008-9238-2. [DOI] [PubMed] [Google Scholar]
- Lescop E, Brutscher B. Highly automated protein backbone resonance assignment within a few hours: the <<BATCH>> strategy and software package. J Biomol NMR. 2009;44:43–57. doi: 10.1007/s10858-009-9314-2. [DOI] [PubMed] [Google Scholar]
- Mohanty B, Serrano P, Geralt M, Wüthrich K. NMR structure determination of the protein NP_344798.1 as the first representative of the Pfam PF06042. J Biomol NMR. 2014 doi: 10.1007/s10858-014-9878-3. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley HN, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Meth Enzym. 2001;399:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- Pedrini B, Serrano P, Mohanty B, Geralt M, Wüthrich K. NMR-Profiles of protein solutions. Biopolymers. 2013;99:825–831. doi: 10.1002/bip.22348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E, Güntert P. A new algorithm for reliable and general NMR resonance assignment. J Am Chem Soc. 2012;134:12817–12829. doi: 10.1021/ja305091n. [DOI] [PubMed] [Google Scholar]
- Schmidt E, Güntert P. Reliability of exclusively NOESY-based automated resonance assignment and structure determination of proteins. J Biomol NMR. 2013;57:193–204. doi: 10.1007/s10858-013-9779-x. [DOI] [PubMed] [Google Scholar]
- Schmucki R, Yokohama S, Güntert P. Automated assignment of NMR chemical shifts using peak-particle dynamics simulation with the DYNASSIGN algorithm. J Biomol NMR. 2009;43:97–109. doi: 10.1007/s10858-008-9291-x. [DOI] [PubMed] [Google Scholar]
- Serrano P, Pedrini B, Mohanty B, Geralt M, Herrmann T, Wüthrich K. The J-UNIO protocol for automated protein structure determination by NMR in solution. J Biomol NMR. 2012;53:341–354. doi: 10.1007/s10858-012-9645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staykova DK, Fredriksson J, Bermel W, Billeter M. Assignment of protein NMR spectra based on projections, multi-way decomposition and a fast correlation approach. J Biomol NMR. 2008;42:87–97. doi: 10.1007/s10858-008-9265-z. [DOI] [PubMed] [Google Scholar]
- Tikole S, Jaravine V, Rogov VV, Rozenknop A, Schmöe K, Löhr F, Dötsch V, Güntert P. Fast automated NMR spectroscopy of short-lived biological samples. ChemBioChem. 2012;13:964–967. doi: 10.1002/cbic.201200044. [DOI] [PubMed] [Google Scholar]
- Volk J, Herrmann T, Wüthrich K. Automated sequence-specific protein NMR assignment using the memetic algorithm MATCH. J Biomol NMR. 2008;41:127–138. doi: 10.1007/s10858-008-9243-5. [DOI] [PubMed] [Google Scholar]
- Wüthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
- Wüthrich K. Celebrating its 20th anniversary in 2011, the Journal of Biomolecular NMR looks forward to the second decade of the 21st century. J Biomol NMR. 2011;49:1–2. doi: 10.1007/s10858-010-9466-0. [DOI] [PubMed] [Google Scholar]
- Zawadzka-Kazimierczuk A, Koźmiński W, Billeter M. TSAR: a program for automatic resonance assignment using 2D cross-sections of high dimensionality, high-resolution spectra. J Biomol NMR. 2012;54:81–95. doi: 10.1007/s10858-012-9652-3. [DOI] [PubMed] [Google Scholar]
- Zimmermann DE, Kulikowski CA, Huang Y, Feng W, Tashiro M, Shimotakahara S, Chien C, Powers R, Montelione GT. Automated analysis of protein NMR assignments using methods from artificial intelligence. J Mol Biol. 1997;269:592–610. doi: 10.1006/jmbi.1997.1052. [DOI] [PubMed] [Google Scholar]