Abstract
The placenta is one of the most morphologically variable mammalian organs. Four major characteristics are typically discussed when comparing the placentas of different eutherian species: placental shape, maternal–fetal interdigitation, intimacy of the maternal–fetal interface and the pattern of maternal–fetal blood flow. Here, we describe the evolution of three of these features as well as other key aspects of eutherian placentation. In addition to interspecific anatomical variation, there is also variation in placental anatomy and function within a single species. Much of this intraspecific variation occurs in response to different environmental conditions such as altitude and poor maternal nutrition. Examinations of variation in the placenta from both intra- and interspecies perspectives elucidate different aspects of placental function and dysfunction at the maternal–fetal interface. Comparisons within species identify candidate mechanisms that are activated in response to environmental stressors ultimately contributing to the aetiology of obstetric syndromes such as pre-eclampsia. Comparisons above the species level identify the evolutionary lineages on which the potential for the development of obstetric syndromes emerged.
Keywords: variation, placenta, adaptation
1. Introduction
The placenta is the conduit between the mother and fetus and directs fetal growth and development by acting as a source of nutrient exchange [1]. The placenta mediates the transfer of nutrients, including oxygen, amino acids, lipids and glucose from the mother to the fetus and provides a method for fetal waste excretion by way of the mother [2]. While these functions are conserved among placental mammals (i.e. eutherians), the morphology of the placenta is not [3]. Here we discuss interspecific placental variation in terms of how major characteristics of placental morphology differ within the eutherian mammals. Specifically, we focus on the shape of the placenta, the intimacy of the maternal–fetal interface and the interdigitation pattern of the trophoblast in relation to the maternal tissue [4]. We then discuss how these characteristics can shed light on multifactorial obstetric syndromes associated with human placental dysfunction. Lastly, we examine how environmental stressors such as hypoxia and poor nutrition can drive placental plasticity, and how adaptations to these stressors can provide insight into the aetiology of obstetric syndromes [5–7].
2. Placental variation in Eutheria
The placenta is arguably one of the most variable organs in the animal kingdom. There are many characteristics that can be used when comparing the placentas of different species. Here, we focus on three of the four major and most variable characteristics (placental shape, intimacy of maternal–fetal interface and placental interdigitation). The fourth major feature involves the arrangement of the maternal and fetal bloodstreams [4]. This feature partially determines the physiological diffusion efficiency of the various molecules exchanged between mother and fetus [2]. Unfortunately, this feature has not been described in enough mammalian species to reconstruct its evolutionary history accurately. Humans have a relatively inefficient multivillous exchange system [4]. The other three major features of the placenta are better described across mammalian diversity. The placental shape describes the pattern that makes contact with the uterine wall where nutrient exchange occurs. The maternal–fetal interface describes the type of barrier that separates the maternal and fetal tissue. Placental interdigitation refers to how the trophoblast and maternal blood supply are interwoven. These, as well as other characteristics, can be used to study different aspects of placental and fetal development.
There are five commonly described placental shapes that are found among eutherians [4]. The most common placental shape is the discoid placenta where the placental tissue makes contact with the uterine wall in the shape of a single disc. This shape was present in the most recent common ancestor of placental mammals [3,8,9]. The zonary placenta was the first shape to diverge from the ancestral discoid placenta and is present in some afrotherians such as the members of Sirenia and the African elephants as well as the carnivores of the laurasiatherian clade [3,9–12]. Another evolutionarily derived placental shape is the diffuse placenta present in some primates and most of the cetartiodactyls (i.e. cetaceans and artiodactyls) except bovines [3,4]. The diffuse placenta has a large surface area for contact with the uterine wall, allowing a high surface area for maternal–fetal nutrient exchange. Bovines have a cotyledonary placental shape which is characterized by many ‘polka dot’ like attachments to the uterine wall [4]. Among primates, there are three different placental shapes, the diffuse placenta of the strepsirrhines, the discoid placenta present in humans, other apes, some catarrhines and platyrrhines, and the bidiscoid placenta (i.e. the placenta attaches to the uterine wall as two discs that share an umbilical cord). In this case, one disc attaches on the anterior and the other disc attaches to the posterior wall of the uterus [4]. The bidiscoid placental shape is found in a subset of anthropoid primates, including some macaques and marmosets [3]. The different placental shapes among eutherians indicates varying amounts of contact between the mother and the fetus, with the diffuse placenta typically having relatively the most and the discoid having the least surface area for contact and nutrient exchange [13].
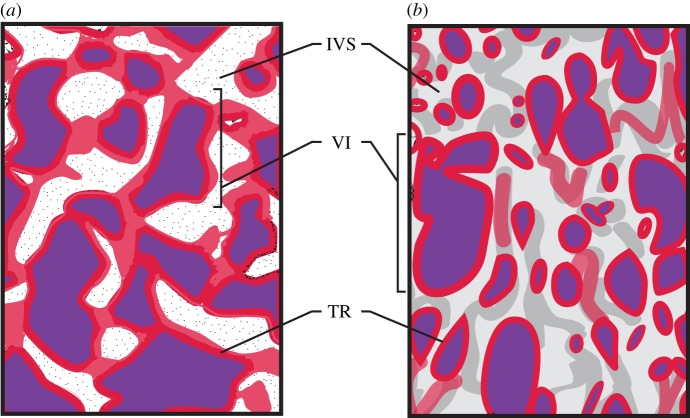
Another way to compare and classify placentas of different species is through looking at differences in their interdigitation pattern, or the way the fetal tissue interacts with the maternal tissue [14]. There are five different types of placental interdigitation observed in eutherians (labyrinthine, villous, trabecular, lamellar and folded) [4]. The most recent common ancestor of placental mammals is inferred to have had a labyrinthine pattern of interdigitation [3]. This pattern is present in most placental mammals, including all afrotherians, Glires (rodents, rabbits and allies), soricid shrews, moles, hedgehogs and bats [3,15,16]. One characteristic of the labyrinthine interdigitation pattern is web-like channels containing either maternal or fetal blood [4]. When xenarthrans (e.g. sloths, armadillos, anteaters) diverged from the other placental mammals they evolved a trabecular interdigitation pattern, in which globular folds of trophoblast terminate in branching villi-like tubes known as trabeculae [3,17]. Three different types of placental interdigitation are present in Euarchontoglires. Rodents, lagomorphs, scandentians and dermopterans have a labyrinthine interdigitiation pattern. Primates either have a trabecular interdigitation pattern, seen in tarsiers and platyrrhines, or a villous interdigitiation pattern seen in catarrhines and strepsirrhines (figure 1a). In catarrhines, fetal villi project into the maternal endometrium [18] (figure 1b). One feature that distinguishes platyrrhine trabeculae is the presence of prominent maternal channels lined with endothelium [19]. Within Laurasatheria, perissodactyls and artiodactyls have a villous interdigitation pattern, with the exception of pigs, that have a folded interdigitation pattern, i.e. fetal tissue folding in and out of maternal tissue. Some carnivores have a more complex folding pattern known as a lamellar interdigitation. Two crown laurasiatherian orders maintain the ancestral labyrinthine interdigitation, and these are Soricomorpha and Chiroptera [4].
Figure 1.
Placental interdigitation across primates. Cross-sectional view of two types of maternal–fetal interdigitation found in primates: trabecular (a) and villous (b). The trabecular interdigitation pattern is characterized as branching globular folds that terminate in villi and this is seen in tarsiers and platyrrhines. The fetal trophoblast cells (red) surround the fetal villi (purple), which contain fetal blood (purple) separating the fetal blood from the maternal blood located in the intervillous space. The villous interdigitation pattern is found in catarrhines and strepsirrhines, and this is characterized by a branching villi pattern [4]. TR, trophoblast cells; VI, villi; IVS, maternal intervillous space. Adapted from [18]. (Online version in colour.)
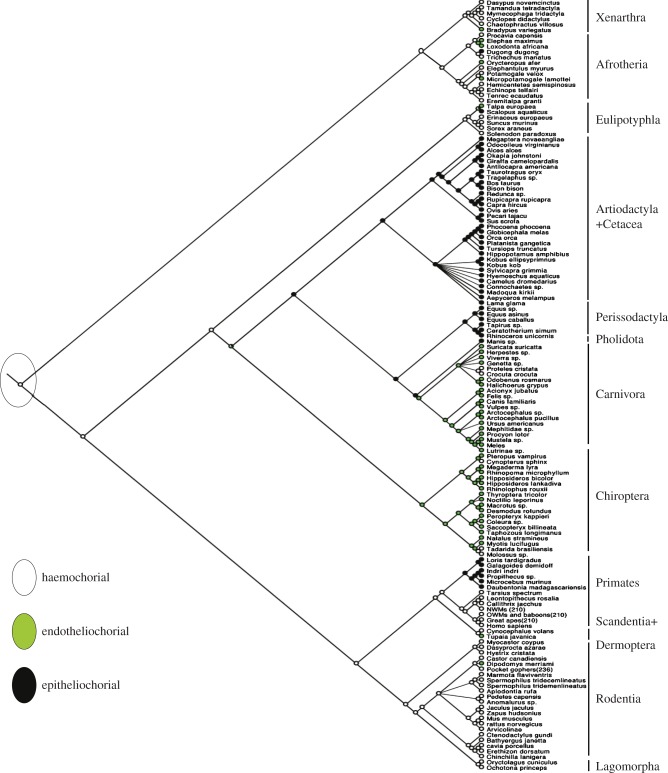
One of the most studied characteristics that can be used to classify eutherian placentas is the type of interface between the fetal tissue and the maternal blood [4]. They range in levels of intimacy, i.e. the degree of separation or number of layers separating the fetal and maternal tissues. The least intimate is the epitheliochorial placenta where there is a layer of uterine epithelial cells opposed to a layer of trophoblast cells, limiting interaction between the maternal blood and fetal tissue [4] (figure 2a). This type of barrier has evolved independently multiple times during eutherian evolution, and it is a derived character state [13] (figure 3). The second most invasive barrier is the endotheliochorial interface where, after implantation, the uterine epithelium is degraded leaving only the maternal endothelium adjacent to the trophoblast (figure 2b). The endotheliochorial interface is found in some afrotherians (elephants and aardvarks), some Euarchontoglires (tree shrews) and some laurasiatherians (moles, most carnivores and some chiropterans) [13]. The most intimate form of maternal–fetal interface is the haemochorial barrier, in which both the maternal epithelial cells and maternal endothelial cells are degraded leaving only the trophoblast cells in direct contact with the maternal blood (figure 2c). This is the most widespread type of interface present in all four of the major eutherian superordinal clades (i.e. Xenarthra, Afrotheria, Laurasiatheria and Euarchontoglires). The haemochorial interface has also been inferred to represent the ancestral state of extant placental mammals [3,8] (figure 3). The haemochorial arrangement has also been described to vary in the number of trophoblast layers that surround the syncytium [26]. The haemo-monochorial placenta has a layer of syncytium not surrounded by cytotrophoblast, haemo-dichorial placentas have a layer of cytotrophoblast on the fetal side of the syncytium; and lastly, the haemo-trichorial placenta syncytium is surrounded by a layer of cytotrophoblast on the maternal and fetal sides [4,26]. A haemochorial state requires the tolerance of the maternal immune system towards paternal antigens since the placental tissue expresses both maternal and paternal antigens [27]. If immune tolerance is disrupted, problems with placental function may occur [28]. The great variation in placenta morphology across the eutherian mammals makes it more complicated to design animal models to study human diseases associated with placental dysfunction.
Figure 2.
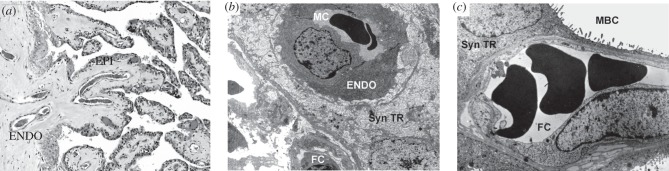
Three types for interface between the mother and the fetus. Histological examples of the three different levels maternal–fetal interface seen across the eutherians. (a) Section of placental tissue from a giraffe which has an epitheliochorial interface where the mother and the fetus are separated by layer of uterine epithelial cells (EPI) and maternal endothelial cells (ENDO). (b) An example of an endotheliochorial placenta of a three toed sloth originally from [12]. This type of placenta has one layer of maternal endothelial cells (ENDO) separating maternal blood from the trophoblast. (c) An example of a haemochorial placenta of a degu, a rodent from [20]. The haemochorial placenta is the most invasive maternal–fetal interface with fetal trophoblast in direct contact with maternal blood. ENDO, maternal endothelial cells; EPI, maternal epithelial cells; Syn TR, syncytiotrophoblast; FC, fetal capillary; MBC, maternal blood channel. With permissions from the publishers and authors.
Figure 3.
Evolution of the maternal–fetal interface. Maximum-parsimony phylogenetic reconstruction of the maternal–fetal interface of 141 mammals through parsimony analysis showing the root placental mammal having a haemochorial placenta (white) with epitheliochorial (black) and endotheliochorial (green) as divergent states [3,8,21–24]. Tree topology taken from [25]. (Online version in colour.)
3. Comparative placental transcriptomics
Sequencing technology allows us to identify genes that are differentially expressed across the different eutherian mammals. One study involving the placenta transcriptome used next-generation sequence technology to sequence the placenta transcriptome of the African elephant (Loxodonta africana) and compared it to previously published transcriptome data from humans, mice, cows, platypuses and chickens. Roughly 3000 genes were expressed in the placentas of all sampled eutherians. The list of these genes is over-represented by genes associated with many annotations, including 128 genes related to stress (e.g. corticosteroid signalling). One class of corticosteroid, glucocorticoids, has increased expression in cases of reduced fetal growth [29,30]. The conservation of corticosteroid signalling gene expression in the placenta suggests an ancient origin of the mechanism of response to stress across eutherian mammals. Other studies have used placenta transcriptomes of several equines to study parent of origin effects and genetic imprinting, where the fetus inherits genes from either the mother or the father [31,32]. One study compared placental transcriptomes of horses, donkeys and their crosses (mules and hinnys) and found that a majority of the genes that were significantly different were paternally expressed, showing that the father's size has some influence in determining the size of the fetus [31].
Placental transcriptomics has also identified novel splice variants that are processed differently in various eutharian species. Prolactin (Prl) for example, a gene involved in regulating lactation in placental mammals, has many alternative transcripts [33]. Placental mammals such as dogs and armadillos show no evidence of a myometrial Prl transcript, while humans, some New World monkeys, mice and elephants all express Prl transcripts; however, each species expresses different splice variants with alternative transcription start sites [34]. Another gene that has multiple alternative transcripts across the placental mammals is the gene encoding aromatase (CYP19). Aromatase converts androgens into oestrogens, and thus plays a role in regulating placental growth [35–37]. There is a placenta-specific CYP19 gene isoform in bovines, ovines and humans, but not rodents [35,38,39]. The human CYP19 isoform contains a long terminal repeat (LTR) that functions to localize CYP19 to the syncytiotrophoblast [35,40]. This LTR is most probably an endogenous retrovirus. This retrovirus was integrated into a promoter region, and therefore effects transcription of the gene. Endogenous retrovirus integration may have played a role in the evolution of the primate placenta [35]. Other human genes which play a role in placental development and function that contain endogenous retroviruses include the syncytins (encoded by ERVW-1 and ERVFRD-1). Syncytin-1 plays a role in cell fusion during trophoblast formation and is expressed throughout pregnancy. The main role of syncytin-2 is the suppression of the maternal immune system, but unlike syncytin-1, expression declines as pregnancy progresses [35,41]. It has been shown that down-regulation of syncytin-1 is associated with pre-eclampsia [42]. Interestingly, syncytin genes have evolved independently over the course of eutherian evolution [43]. Mice, for example, also express a pair of syncytins, syncytin-A and syncytin-B. Like syncytin-2, syncytin-B has immunosuppressive properties not seen in syncytin-1 or syncytin-A, both of which play a role in cell fusion during syncytiotrophoblast and trophoblast giant cell formation, respectively [43]. The Blind mole rat placenta expresses SynA and SynB, but these genes are not present in the Naked mole rat genome [44]. The human syncytin genes have trophoblast-specific enhancers that contain binding sites for transcription factors such as glial cell missing 1 (GCM1), that induce the expression of the syncytins [45]. GCM1 is a transcription factor that is specifically expressed in trophoblast cells and has been shown to increase syncytin-1 when overexpressed in trophoblast cell lines [46,47]. Other genes that have been shown to have placental-specific promoters or enhancers include rat placental lactogen 2 (rPLII) and AP-2 [48,49].
There are other larger gene families that have placenta-specific members [35]. One example is the human leucocyte antigen (HLA) gene family. HLA-g is a placenta-specific immune system gene that plays a role in trophoblast invasion [50]. Another example of a eutherian-specific gene expressed in placenta that acts differently in various eutherian species is placenta-specific 1 (PLAC1) [51]. This gene is mainly expressed in the placenta but may also be expressed in the testis [35,52]. Orthologues of PLAC1 are found only in placental mammals [35,51,52]. PLAC1 is expressed throughout gestation in humans but only during e7.5–e14.5 in rodents [53,54]. Galectins are another gene family with placenta-specific members that play a role in the maternal–fetal immune tolerance and implantation [55]. All members of this family are expressed at the maternal–fetal interface [56]. The placenta-specific galectins are located in a cluster on chromosome 19. This chromosome contains many placenta-specific genes including the pregnancy-specific glycoproteins and a cluster of placenta-specific microRNAs [57,58].
4. Animal models for placental dysfunction
Owing to the wide variation among placental mammals, it is challenging to develop animal models of obstetric syndromes associated with placental dysfunction [59,60]. While some mouse models have been used to study obstetric syndromes associated with placental dysfunction, such as implantation failure, miscarriages and pre-eclampsia, the results may not be directly relevant to the human condition [60–63]. One difficulty to overcome with mouse models of obstetric syndromes is the relatively superficial invasion of the fetal tissue into the maternal decidua [27]. Human placentas typically have a deeper invasion into the maternal myometrium than mice, and since pre-eclampsia is characterized by relatively shallow trophoblast invasion (i.e. the typical condition in mice), studies of reduced placental invasiveness may be difficult to model using mice [64]. Also, the cellular make-up of the murine placenta is different than the human placenta. Mice have polyploid trophoblast giant cells. Like human cytotrophoblast cells, these giant cells regulate uterine implantation. Mice also have a layer of spongiotrophoblast similar to the human extravillous trophoblast [65]. Sheep have also been used to study placental dysfunction [66]. Sheep pregnancy has been used to model human intrauterine growth restriction by inducing maternal hypothermia, removal of endometrial caruncles to limit placental growth, limiting blood supply through the restriction of uterine blood flow and maternal overnutrition [66]. All these methods result in altered oxygen and nutrient transfer leading to a decrease in fetal growth [66]. Guinea pigs have also been used to model human placentation, because they share features with humans [60]. Guinea pigs have smaller litter sizes, longer gestation and deeper trophoblast invasion than other rodents [60,67]. Guinea pig models have been developed to study placental transfer and intrauterine growth restriction [60,68,69].
Pre-eclampsia is an example of an obstetric syndrome related to shallow trophoblast invasion [70,71]. In pre-eclampsia, the spiral arteries that supply maternal blood to the developing fetus are poorly remodelled due to decreased trophoblast invasion; therefore blood supply to the fetus may be limited [72–74]. Some studies have identified non-human primate models that can be used to study placental invasiveness especially in those with a bidiscoid placenta such as the rhesus monkey [75,76]. Pre-eclampsia is typically thought to be a human-specific syndrome [75,77]. However, recent work has challenged this view, primarily due to the fact that placental invasiveness is also quite deep in the closest relatives of humans, chimpanzees and gorillas [78–80]. Thus, it is likely that the potential for pre-eclampsia due to dysregulation of placental invasiveness predated the most recent common ancestor of humans, chimpanzees and gorillas during the Miocene. A recent study supporting this hypothesis examined the evolution of orthologous protein coding gene sequences and found evidence for adaptive evolution (i.e. positive selection as measured through comparison of non-synonymous and synonymous substitution rates) in genes associated with the more invasive phenotype seen on the evolutionary lineage leading to the most recent common ancestor of these three species [81]. In particular, genes associated with immune function, regulation of blood vessel size and hypertension were enriched among the adaptively evolving genes. These findings suggest that the molecular mechanisms for the induction of pre-eclampsia were in place long before the evolution of modern humans.
5. Natural human models for studying placental dysfunction
While animal models can be useful for studying specific symptoms associated with the complex diseases involving placental dysfunction, some researchers have begun using environmentally variable, natural human models to study the genetic and morphological differences associated with placental dysfunction [6,82,83]. The human placenta brings practical advantages to biomedical studies as it is a temporary organ that, like the fetus (i.e. the child), is delivered [4]. Some human populations that have been used to study placental morphology in regard to placental dysfunction are high altitude natives as well as multigenerational populations who experienced periods of poor nutrition in at least one generation [5,6].
Populations native to high altitude, such as native Andeans and Tibetans, are candidates to study how hypoxic conditions affect the placenta and contribute to complex obstetric syndromes such as intrauterine growth restriction and pre-eclampsia [5,83]. These populations also allow for a case–control-type experimental design in which high altitude native populations are used as the case populations and natives in low altitude regions can be used as the control populations [82]. The natives of the Tibetan Plateau and Andes Mountains have been exposed to hypoxic conditions for roughly 20 000 and 10 000 years, respectively [84,85]. Infant cohorts exposed to the hypoxic conditions of high altitude during gestation experience lower than average birthweights when compared with their sea-level counterparts [86]. The exceptions to this pattern are infant cohorts born to mothers who have an ancestral pattern of high altitude hypoxia. These infants exhibit birth-weights comparable to those at sea-level, suggesting some level of adaptation to the hypoxic environment [85]. Morphological studies of placentas of high altitude dwelling individuals show an increased capillary surface area and greater placental mass. These features combine to increase the total area for oxygen exchange between the mother and fetus [87]. Physiological studies have shown that Andeans and Tibetans are different at high altitude with Tibetans being more adapted to high altitude with attributes more similar to sea-level populations [84]. Genetic studies have shown that differences in the genetics between Andean and Tibetans resulted in convergent phenotypic evolution between high and low altitude populations [85,88,89]. Recently, a genome-wide study compared two populations native to high altitude for several millennia: (i) the native inhabitants of the Tibetan Plateau and (ii) the native inhabitants of the Andean Altiplano of South America. These populations have been exposed to chronic hypoxia, using low altitude individuals as control populations. In 2010, Bigham et al. were able to identify gene regions and single nucleotide polymorphisms (SNPs) that show evidence of positive selection (i.e. adaptive evolution). Of the dozens of genes identified, one deserves special attention. EGLN1 encodes hypoxia-inducible factor prolyl hydroxylase 2, a protein that regulates the expression of HIF-1a, the transcription factor responsible for the hypoxic response [90]. EGLN1 shows evidence for positive selection at 25 SNPs in Andeans and 28 SNPs in Tibetans [89]. This finding suggests there is a genetic basis for the adaptation to hypoxia in these populations, and future work should test how these positively selected SNPs affect the oxygen exchange between the mother and fetus via the placenta. This is important since recent migrants to high altitude tend to have offspring with relatively low birthweights compared with their native counterparts who have evolved adaptions to hypoxia [91].
Another class of human populations that have shed light on placental plasticity are those in which pregnant woman were exposed to periods of low nutrient availability during all or part of their pregnancy. One of the first studies using famine as a mediator of fetal growth and development examined samples from a Dutch population who had suffered through the famine during the Second World War [6]. Famine studies have shown that those women who were exposed to famine conditions during their third trimester had offspring with low fetal birthweights as they experienced limited nutrient availability [6,92]. Those who experienced famine conditions during their first trimester produced offspring with heavier placentas. This was thought to be an adaptive mechanism to allow for increased surface area for nutrient exchange [7].
Epigenetic studies have also examined placental gene expression and placental methylation differences in famine populations and those individuals with low birthweights. They found differential expression in genes associated with fetal growth as well as differential methylation in those growth-related genes [93–95]. One study examined differential methylation along insulin-growth factor 2 (IGF2) [95], a gene known to promote fetal growth [96]. This study used whole blood from 60 individuals conceived during the Dutch famine and compared them with their control, same sex, siblings born before or conceived after the famine [95]. The study describes five methylation sites along the promoter region of IGF2 and found decreased methylation across all sites in the famine-exposed individuals when compared with the control siblings. This finding suggests that during times of low nutrition, genes such as IGF2 can be differentially methylated and potentially provide a link between maternal nutrition and fetal gene expression [95]. These results add another layer of placental adaptation in which during times of low nutritional availability, the placenta responds by either expanding in size to increase surface area of nutrient exchange or repressing genes associated with fetal growth in order to protect the mother.
Low birthweight as a result of poor maternal nutrition not only affects the conceptus during fetal development but also impacts their adult life [97]. According to the Developmental Origins of Health and Disease (DOHaD) paradigm, low fetal birthweight can lead to complications such as diabetes, obesity and renal malfunction later in life [98]. This has been hypothesized to be an adaptive mechanism, as the fetus has become accustomed to the low nutrient environment in the womb. Later, if the child is exposed to a healthy, more enriched diet after birth, their body reacts differently than would children who had sufficient nutrients during fetal development. This reaction to a more enriched diet after birth can result in poor outcomes later in life including obesity and obesity-related conditions [99].
6. Parent–offspring conflict
Robert Trivers pioneered investigations on the conflict between the parents and their offspring. Trivers defined parental investment as, ‘any investment by the parent in an individual offspring that increases the offspring's chance of surviving (and hence reproductive success) at the cost of the parent's ability to invest in other offspring’ [100, p. 139]. In this manner, the survivability of future offspring is affected if the current offspring takes an excess amount of maternal resources, limiting the amount of resources available to the offspring's future siblings [101].
David Haig extended the idea of parent–offspring conflict by adding that there is also conflict between the mother and the fetus. In this view, the fetus aims to extract the maximum amount of maternal nutrients (thus engendering conflict), but the mother and the fetus must also cooperate during a normal pregnancy [102,103]. Trophoblast invasion is an example of a balance between maternal–fetal conflict and cooperation. During a normal human pregnancy, the trophoblast invades into the maternal myometrium and extracts nutrients from maternal blood [104]. Conflict arises because while the mother wants to limit trophoblast invasion, the placenta invades deeply thereby extracting more nutrients [104,105]. Cooperation is necessary to balance invasion. Optimal placenta invasion is deep enough into the maternal tissue to ensure the fetus can absorb enough nutrients but shallow enough to protect the mother from syndromes such as placenta accreta [104]. One obstetric syndrome that is related to the improper balance of conflict and cooperation is pre-eclampsia. Conflict theory suggests that fetal actions need to be distinguished from maternal responses in pre-eclampsia [104,105].
General conflict between the mother and the fetus can be mediated by genetic processes such as gene imprinting. Imprinted genes are those genes in which only one parental allele is expressed, whereas the other allele is silenced through mechanisms such as DNA methylation [106,107]. Conflict theory predicts that maternally expressed imprinted genes restrict fetal growth by retaining maternal resources. By contrast, paternally expressed imprinted genes promote fetal growth and maximize resource extraction [108,109]. Despite the conflict arising from genomic imprinting, cooperation between the mother and fetus is important in regard to nutrient exchange and fetal size. If the mother fails to provide the fetus with enough resources, the fetus will have a low birth-weight, which leads to complications later in their adult life, and possibly reduced reproductive success [97,98].
Cooperation can also be observed in the maternal immune system, because it tolerates non-self-paternal antigens expressed in the placenta, which is considered a semi-allograft [18,110]. Obstetric syndromes such as pre-eclampsia have been posited to result from excessive conflict between the mother and the fetus as well as a breakdown in maternal–fetal cooperation [111]. This balance of cooperation and conflict is heightened at the human maternal–fetal interface, because the intimate haemochorial placenta has more contact with maternal blood (and by extension paternally expressed alleles), than in less intimate endotheliochorial and epitheliochorial placentas [18].
7. Conclusion
The placenta varies across eutherian species and this variation should be considered when developing animal models to study human diseases associated with placental dysfunction. While many animal models have mimicked several of the symptoms associated with placental dysfunction, it is unsure how applicable any of these results would be to the human condition [27]. One possible approach would be to examine naturally occurring conditions that in humans are associated with obstetric syndromes. For example, species with less intimate maternal–fetal interfaces (e.g. artiodactyls, lemurs) might be appropriate models for pre-eclampsia, because the syndrome is characterized by less placenta invasion [81]. Primate models, including natural examples of environmental stress in humans, are promising because factors such as hypoxia or nutritional stress modify the placenta, and therefore provide insight into complex obstetric syndromes associated with placental dysfunction.
Acknowledgements
We would like to thank Stacy Zamudio, Nick Illsley, Christopher Kuzawa and Julienne Rutherford for their contribution through useful discussion related to this topic. Lastly, we would like to thank Eric Barrington, Graham Burton and Ashley Moffett for organizing the excellent meeting entitled Human Evolution: brain, birthweight and the immune system and this issue of Philosophical Transactions of the Royal Society B.
Funding statement
We would like to acknowledge our funding provided by the National Institutes of Health through grant 1R211HD068954–01.
References
- 1.Lewis RM, Cleal JK, Hanson MA. 2012. Placenta, evolution and lifelong health. Placenta 33, S28–S32. ( 10.1016/j.placenta.2011.12.003) [DOI] [PubMed] [Google Scholar]
- 2.Lager S, Powell TL. 2012. Regulation of nutrient transport across the placenta. J. Pregnancy 2012, 179827 ( 10.1155/2012/179827) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wildman DE, Chen C, Erez O, Grossman LI, Goodman M, Romero R. 2006. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc. Natl Acad. Sci. USA 103, 3203–3208. ( 10.1073/pnas.0511344103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benirschke K, Kaufmann P. 2000. Pathology of the human placenta, 4th edn New York, NY: Springer. [Google Scholar]
- 5.Zamudio S, Palmer SK, Dahms TE, Berman JC, Young DA, Moore LG. 1995. Alterations in uteroplacental blood flow precede hypertension in preeclampsia at high altitude. J. Appl. Physiol. 79, 15–22. [DOI] [PubMed] [Google Scholar]
- 6.Lumey LH. 1992. Decreased birthweights in infants after maternal in utero exposure to the Dutch famine of 1944–1945. Paediatr. Perinat. Epidemiol. 6, 240–253. ( 10.1111/j.1365-3016.1992.tb00764.x) [DOI] [PubMed] [Google Scholar]
- 7.Lumey LH. 1998. Reproductive outcomes in women prenatally exposed to undernutrition: a review of findings from the Dutch famine birth cohort. Proc. Nutr. Soc. 57, 129–135. ( 10.1079/PNS19980019) [DOI] [PubMed] [Google Scholar]
- 8.Elliot MG, Crespi BJ. 2009. Phylogenetic evidence for early hemochorial placentation in eutheria. Placenta 30, 949–967. ( 10.1016/j.placenta.2009.08.004) [DOI] [PubMed] [Google Scholar]
- 9.Mess A, Carter AM. 2006. Evolutionary transformations of fetal membrane characters in Eutheria with special reference to Afrotheria. J. Exp. Zool. B Mol. Dev. Evol. 306, 140–163. ( 10.1002/jez.b.21079) [DOI] [PubMed] [Google Scholar]
- 10.Carter AM, et al. 2008. Placentation in the Amazonian manatee (Trichechus inunguis). Reprod. Fertil. Dev. 20, 537–545. ( 10.1071/RD08009) [DOI] [PubMed] [Google Scholar]
- 11.Soma H, et al. 2013. Review: exploration of placentation from human beings to ocean-living species. Placenta 34, S17–S23. ( 10.1016/j.placenta.2012.11.021) [DOI] [PubMed] [Google Scholar]
- 12.Carter AM, Enders AC. 2004. Comparative aspects of trophoblast development and placentation. Reprod. Biol. Endocrinol. 2, 46 ( 10.1186/1477-7827-2-46) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wooding FBP, Burton G. 2008. Comparative placentation: structures, functions and evolution. Berlin, Germany: Springer. [Google Scholar]
- 14.Capellini I. 2012. The evolutionary significance of placental interdigitation in mammalian reproduction: contributions from comparative studies. Placenta 33, 763–768. ( 10.1016/j.placenta.2012.07.004) [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann P, Davidoff M. 1977. The guinea-pig placenta. Adv. Anat. Embryol. Cell Biol. 53, 5–91. [DOI] [PubMed] [Google Scholar]
- 16.Wimsatt WA, Enders AC. 1980. Structure and morphogenesis of the uterus, placenta, and paraplacental organs of the neotropical disc-winged bat Thyroptera tricolor spix (Microchiroptera, Thyropteridae). Am. J. Anat. 159, 209–243. ( 10.1002/aja.1001590208) [DOI] [PubMed] [Google Scholar]
- 17.Mess AM, et al. 2012. Placentation in the anteaters Myrmecophaga tridactyla and Tamandua tetradactyla (Eutheria, Xenarthra). Reprod. Biol. Endocrinol. 10, 102 ( 10.1186/1477-7827-10-102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sterner K, Jameson N, Wildman D. 2013. Placental development, evolution, and epigenetics of primate pregnancies. In Building babies. developments in primatology: progress and prospects, vol. 37 (eds Clancy KBH, Hinde K, Rutherford JN.), pp. 55–81. New York, NY: Springer. [Google Scholar]
- 19.Luckett WP. 1974. Comparative development and evolution of the placenta in primates. Contrib. Primatol. 3, 142–234. [PubMed] [Google Scholar]
- 20.Mess A, Carter AM. 2007. Evolution of the placenta during the early radiation of placental mammals. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 148, 769–779. ( 10.1016/j.cbpa.2007.01.029) [DOI] [PubMed] [Google Scholar]
- 21.Benirschke K. 2007. Comparative Placentation 2007 (updated 1/19/2012; cited 2007) See http://placentation.ucsd.edu/index.html.
- 22.Mossman HW. 1987. Vertebrate fetal membranes: comparative ontogeny and morphology, evolution, phylogenetic significance, basic functions, research opportunities. New Brunswick, NJ: Rutgers University Press. [Google Scholar]
- 23.Wildman DE, et al. 2007. Genomics, biogeography, and the diversification of placental mammals. Proc. Natl Acad. Sci. USA 104, 14 395–14 400. ( 10.1073/pnas.0704342104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meredith RW, et al. 2011. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 334, 521–524. ( 10.1126/science.1211028) [DOI] [PubMed] [Google Scholar]
- 25.Bininda-Emonds OR, et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512. ( 10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 26.Enders AC. 1965. Formation of syncytium from cytotrophoblast in the human placenta. Obstet. Gynecol. 25, 378–386. [PubMed] [Google Scholar]
- 27.Moffett A, Loke C. 2006. Immunology of placentation in eutherian mammals. Nat. Rev. Immunol. 6, 584–594. ( 10.1038/nri1897) [DOI] [PubMed] [Google Scholar]
- 28.Warning JC, et al. 2011. A balancing act: mechanisms by which the fetus avoids rejection by the maternal immune system. Reproduction 141, 715–724. ( 10.1530/REP-10-0360) [DOI] [PubMed] [Google Scholar]
- 29.Harris A, Seckl J. 2011. Glucocorticoids, prenatal stress and the programming of disease. Horm. Behav. 59, 279–289. ( 10.1016/j.yhbeh.2010.06.007) [DOI] [PubMed] [Google Scholar]
- 30.Khashan AS, McNamee R, Abel KM, Pedersen MG, Webb RT, Kenny LC, Mortensen PB, Baker PN. 2008. Reduced infant birthweight consequent upon maternal exposure to severe life events. Psychosom. Med. 70, 688–694. ( 10.1097/PSY.0b013e318177940d) [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Miller DC, Harman R, Antczak DF, Clark AG. 2013. Paternally expressed genes predominate in the placenta. Proc. Natl Acad. Sci. USA 110, 10 705–10 710. ( 10.1073/pnas.1308998110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Miller DC, Clark AG, Antczak DF. 2012. Random X inactivation in the mule and horse placenta. Genome Res. 22, 1855–1863. ( 10.1101/gr.138487.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Jonathan N, Mershon JL, Allen DL, Steinmetz RW. 1996. Extrapituitary prolactin: distribution, regulation, functions, and clinical aspects. Endocr. Rev. 17, 639–669. [DOI] [PubMed] [Google Scholar]
- 34.Emera D, Casola C, Lynch VJ, Wildman DE, Agnew D, Wagner GP. 2012. Convergent evolution of endometrial prolactin expression in primates, mice, and elephants through the independent recruitment of transposable elements. Mol. Biol. Evol. 29, 239–247. ( 10.1093/molbev/msr189) [DOI] [PubMed] [Google Scholar]
- 35.Rawn SM, Cross JC. 2008. The evolution, regulation, and function of placenta-specific genes. Annu. Rev. Cell Dev. Biol. 24, 159–181. ( 10.1146/annurev.cellbio.24.110707.175418) [DOI] [PubMed] [Google Scholar]
- 36.Simpson ER, et al. 1994. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr. Rev. 15, 342–355. [DOI] [PubMed] [Google Scholar]
- 37.Furbass R, Selimyan R, Vanselow J. 2008. DNA methylation and chromatin accessibility of the proximal Cyp 19 promoter region 1.5/2 correlate with expression levels in sheep placentomes. Mol. Reprod. Dev. 75, 1–7. ( 10.1002/mrd.20756) [DOI] [PubMed] [Google Scholar]
- 38.Kamat A, Mendelson CR. 2001. Identification of the regulatory regions of the human aromatase P450 (CYP19) gene involved in placenta-specific expression. J. Steroid Biochem. Mol. Biol. 79, 173–180. ( 10.1016/S0960-0760(01)00156-X) [DOI] [PubMed] [Google Scholar]
- 39.Vanselow J, Zsolnai A, Fesus L, Furbass R, Schwerin M. 1999. Placenta-specific transcripts of the aromatase encoding gene include different untranslated first exons in sheep and cattle. Eur. J. Biochem. 265, 318–324. ( 10.1046/j.1432-1327.1999.00734.x) [DOI] [PubMed] [Google Scholar]
- 40.Fournet-Dulguerov N, MacLusky NJ, Leranth CZ, Todd R, Mendelson CR, Simpson ER, Naftolin F. 1987. Immunohistochemical localization of aromatase cytochrome P-450 and estradiol dehydrogenase in the syncytiotrophoblast of the human placenta. J. Clin. Endocrinol. Metab. 65, 757–764. ( 10.1210/jcem-65-4-757) [DOI] [PubMed] [Google Scholar]
- 41.Mangeney M, et al. 2007. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl Acad. Sci. USA 104, 20 534–20 539. ( 10.1073/pnas.0707873105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee X, et al. 2001. Downregulation of placental syncytin expression and abnormal protein localization in pre-eclampsia. Placenta 22, 808–812. ( 10.1053/plac.2001.0722) [DOI] [PubMed] [Google Scholar]
- 43.Dupressoir A, Marceau G, Vernochet C, Benit L, Kanellopoulos C, Sapin V, Heidmann T. 2005. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl Acad. Sci. USA 102, 725–730. ( 10.1073/pnas.0406509102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang X, et al. 2014. Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nat. Commun. 5, 3966 ( 10.1038/ncomms4966) [DOI] [PubMed] [Google Scholar]
- 45.Cheng YH, Handwerger S. 2005. A placenta-specific enhancer of the human syncytin gene. Biol. Reprod. 73, 500–509. ( 10.1095/biolreprod.105.039941) [DOI] [PubMed] [Google Scholar]
- 46.Huang Q, Chen H, Li J, Oliver M, Ma X, Byck D, Gao Y, Jiang S-W. 2014. Epigenetic and non-epigenetic regulation of syncytin-1 expression in human placenta and cancer tissues. Cell. Signal. 26, 648–656. ( 10.1016/j.cellsig.2013.11.002) [DOI] [PubMed] [Google Scholar]
- 47.Yu C, et al. 2002. GCMa regulates the syncytin-mediated trophoblastic fusion. J. Biol. Chem. 277, 50 062–50 068. ( 10.1074/jbc.M209316200) [DOI] [PubMed] [Google Scholar]
- 48.Sun Y, Duckworth ML. 1999. Identification of a placental-specific enhancer in the rat placental lactogen II gene that contains binding sites for members of the Ets and AP-1 (activator protein 1) families of transcription factors. Mol. Endocrinol. 13, 385–399. ( 10.1210/mend.13.3.0243) [DOI] [PubMed] [Google Scholar]
- 49.Steger DJ, Buscher M, Hecht JH, Mellon PL. 1993. Coordinate control of the alpha- and beta-subunit genes of human chorionic gonadotropin by trophoblast-specific element-binding protein. Mol. Endocrinol. 7, 1579–1588. [DOI] [PubMed] [Google Scholar]
- 50.Cecati M, Giannubilo SR, Emanuelli M, Tranquilli AL, Saccucci F. 2011. HLA-G and pregnancy adverse outcomes. Med. Hypotheses 76, 782–784. ( 10.1016/j.mehy.2011.02.017) [DOI] [PubMed] [Google Scholar]
- 51.Flicek P, et al. 2014. Ensembl 2014. Nucleic Acids Res. 42(Database issue), D749–D755. ( 10.1093/nar/gkt1196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fant M, Barerra-Saldana H, Dubinsky W, Poindexter B, Bick R. 2007. The PLAC1 protein localizes to membranous compartments in the apical region of the syncytiotrophoblast. Mol. Reprod. Dev. 74, 922–929. ( 10.1002/mrd.20673) [DOI] [PubMed] [Google Scholar]
- 53.Cocchia M, Huber R, Pantano S, Chen EY, Ma P, Forabosco A, Ko MSH, Schlessinger D. 2000. PLAC1, an Xq26 gene with placenta-specific expression. Genomics 68, 305–312. ( 10.1006/geno.2000.6302) [DOI] [PubMed] [Google Scholar]
- 54.Massabbal E, Parveen S, Weisoly DL, Nelson DM, Smith SD, Fant M. 2005. PLAC1 expression increases during trophoblast differentiation: evidence for regulatory interactions with the fibroblast growth factor-7 (FGF-7) axis. Mol. Reprod. Dev. 71, 299–304. ( 10.1002/mrd.20272) [DOI] [PubMed] [Google Scholar]
- 55.Than NG, Romero R, Kim CJ, McGowen MR, Papp Z, Wildman DE. 2012. Galectins: guardians of eutherian pregnancy at the maternal–fetal interface. Trends Endocrinol. Metab. 23, 23–31. ( 10.1016/j.tem.2011.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Than NG, et al. 2009. A primate subfamily of galectins expressed at the maternal–fetal interface that promote immune cell death. Proc. Natl Acad. Sci. USA 106, 9731–9736. ( 10.1073/pnas.0903568106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Than NG, et al. 2014. Evolutionary origins of the placental expression of chromosome 19 cluster galectins and their complex dysregulation in preeclampsia. Placenta 35, 855–865. ( 10.1016/j.placenta.2014.07.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie L, Mouillet JF, Chu T, Parks WT, Sadovsky E, Knöfler M, Sadovsky Y. 2014. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology 155, 4975–4985. ( 10.1210/en.2014-1501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sunderland N, Hennessy A, Makris A. 2011. Animal models of pre-eclampsia. Am. J. Reprod. Immunol. 65, 533–541. ( 10.1111/j.1600-0897.2010.00929.x) [DOI] [PubMed] [Google Scholar]
- 60.Carter AM. 2007. Animal models of human placentation—a review. Placenta 28(Suppl. A), S41–S47. ( 10.1016/j.placenta.2006.11.002) [DOI] [PubMed] [Google Scholar]
- 61.Ahmed A, Singh J, Khan Y, Seshan SV, Girardi G. 2010. A new mouse model to explore therapies for preeclampsia. PLoS ONE 5, e13663 ( 10.1371/journal.pone.0013663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clark DA. 2008. Immunological factors in pregnancy wastage: fact or fiction. Am. J. Reprod. Immunol. 59, 277–300. ( 10.1111/j.1600-0897.2008.00580.x) [DOI] [PubMed] [Google Scholar]
- 63.Clark DA, Arck PC, Chaouat G. 1999. Why did your mother reject you? Immunogenetic determinants of the response to environmental selective pressure expressed at the uterine level. Am. J. Reprod. Immunol. 41, 5–22. ( 10.1111/j.1600-0897.1999.tb00071.x) [DOI] [PubMed] [Google Scholar]
- 64.Clark DA. 2014. The use and misuse of animal analog models of human pregnancy disorders. J. Reprod. Immunol. 103, 1–8. ( 10.1016/j.jri.2014.02.006) [DOI] [PubMed] [Google Scholar]
- 65.Rossant J, Cross JC. 2001. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2, 538–548. ( 10.1038/35080570) [DOI] [PubMed] [Google Scholar]
- 66.Morrison JL. 2008. Sheep models of intrauterine growth restriction: fetal adaptations and consequences. Clin. Exp. Pharmacol. Physiol. 35, 730–743. ( 10.1111/j.1440-1681.2008.04975.x) [DOI] [PubMed] [Google Scholar]
- 67.Nanaev A, Chwalisz K, Frank HG, Kohnen G, Hegele-Hartung C, Kaufmann P. 1995. Physiological dilation of uteroplacental arteries in the guinea pig depends on nitric oxide synthase activity of extravillous trophoblast. Cell Tissue Res. 282, 407–421. ( 10.1007/BF00318873) [DOI] [PubMed] [Google Scholar]
- 68.Carter AM. 1993. Current topic: restriction of placental and fetal growth in the guinea-pig. Placenta. 14, 125–135. ( 10.1016/S0143-4004(05)80255-3) [DOI] [PubMed] [Google Scholar]
- 69.Hill PM, Young M. 1973. Net placental transfer of free amino acids against varying concentrations. J. Physiol. 235, 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saito S, Nakashima A. 2014. A review of the mechanism for poor placentation in early-onset preeclampsia: the role of autophagy in trophoblast invasion and vascular remodeling. J. Reprod. Immunol. 101–102, 80–88. ( 10.1016/j.jri.2013.06.002) [DOI] [PubMed] [Google Scholar]
- 71.Khong TY, De Wolf F, Robertson WB, Brosens I. 1986. Inadequate maternal vascular response to placentation in pregnancies complicated by pre-eclampsia and by small-for-gestational age infants. Br. J. Obstet. Gynaecol. 93, 1049–1059. ( 10.1111/j.1471-0528.1986.tb07830.x) [DOI] [PubMed] [Google Scholar]
- 72.Roberts JM, Gammill HS. 2005. Preeclampsia: recent insights. Hypertension 46, 1243–1249. ( 10.1161/01.HYP.0000188408.49896.c5) [DOI] [PubMed] [Google Scholar]
- 73.Redman CW, Sargent IL. 2005. Latest advances in understanding preeclampsia. Science 308, 1592–1594. ( 10.1126/science.1111726) [DOI] [PubMed] [Google Scholar]
- 74.Lyall F, Robson SC, Bulmer JN. 2013. Spiral artery remodeling and trophoblast invasion in preeclampsia and fetal growth restriction: relationship to clinical outcome. Hypertension 62, 1046–1054. ( 10.1161/HYPERTENSIONAHA.113.01892) [DOI] [PubMed] [Google Scholar]
- 75.Varki NM, Strobert E, Dick EJ, Jr, Benirschke K, Varki A. 2011. Biomedical differences between human and nonhuman hominids: potential roles for uniquely human aspects of sialic acid biology. Annu. Rev. Pathol. 6, 365–393. ( 10.1146/annurev-pathol-011110-130315) [DOI] [PubMed] [Google Scholar]
- 76.Roberts VH, Rasanen JP, Novy MJ, Frias A, Louey S, Morgan TK, Thornburg KL, Spindel ER, Grigsby PL. 2012. Restriction of placental vasculature in a non-human primate: a unique model to study placental plasticity. Placenta 33, 73–76. ( 10.1016/j.placenta.2011.10.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robillard PY, Dekker GA, Hulsey TC. 2002. Evolutionary adaptations to pre-eclampsia/eclampsia in humans: low fecundability rate, loss of oestrus, prohibitions of incest and systematic polyandry. Am. J. Reprod. Immunol. 47, 104–111. ( 10.1034/j.1600-0897.2002.1o043.x) [DOI] [PubMed] [Google Scholar]
- 78.Carter AM, Pijnenborg R. 2011. Evolution of invasive placentation with special reference to non-human primates. Best Pract. Res. Clin. Obstet. Gynaecol. 25, 249–257. ( 10.1016/j.bpobgyn.2010.10.010) [DOI] [PubMed] [Google Scholar]
- 79.Pijnenborg R, Vercruysse L, Carter AM. 2011. Deep trophoblast invasion and spiral artery remodelling in the placental bed of the lowland gorilla. Placenta 32, 586–591. ( 10.1016/j.placenta.2011.05.007) [DOI] [PubMed] [Google Scholar]
- 80.Pijnenborg R, Vercruysse L, Carter AM. 2011. Deep trophoblast invasion and spiral artery remodelling in the placental bed of the chimpanzee. Placenta 32, 400–408. ( 10.1016/j.placenta.2011.02.009) [DOI] [PubMed] [Google Scholar]
- 81.Crosley EJ, Elliot MG, Christians JK, Crespi BJ. 2013. Placental invasion, preeclampsia risk and adaptive molecular evolution at the origin of the great apes: evidence from genome-wide analyses. Placenta 34, 127–132. ( 10.1016/j.placenta.2012.12.001) [DOI] [PubMed] [Google Scholar]
- 82.Moore LG, Shriver M, Bemis L, Hickler B, Wilson M, Brutsaert T, Parra E, Vargas E. 2004. Maternal adaptation to high-altitude pregnancy: an experiment of nature--a review. Placenta 25(Suppl. A), S60–S71. ( 10.1016/j.placenta.2004.01.008) [DOI] [PubMed] [Google Scholar]
- 83.Zamudio S. 2003. The placenta at high altitude. High Alt. Med. Biol. 4, 171–191. ( 10.1089/152702903322022785) [DOI] [PubMed] [Google Scholar]
- 84.Moore LG, Armaza F, Villena M, Vargas E. 2000. Comparative aspects of high-altitude adaptation in human populations. Adv. Exp. Med. Biol. 475, 45–62. ( 10.1007/0-306-46825-5_6) [DOI] [PubMed] [Google Scholar]
- 85.Beall CM. 2007. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc. Natl Acad. Sci. USA 104(Suppl. 1), 8655–8660. ( 10.1073/pnas.0701985104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beall CM. 2007. Detecting natural selection in high-altitude human populations. Respir. Physiol. Neurobiol. 158, 161–171. ( 10.1016/j.resp.2007.05.013) [DOI] [PubMed] [Google Scholar]
- 87.Mathieu-Costello O. 2001. Muscle adaptation to altitude: tissue capillarity and capacity for aerobic metabolism. High Alt. Med. Biol. 2, 413–425. ( 10.1089/15270290152608598) [DOI] [PubMed] [Google Scholar]
- 88.Cheviron ZA, Brumfield RT. 2012. Genomic insights into adaptation to high-altitude environments. Heredity 108, 354–361. ( 10.1038/hdy.2011.85) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bigham A, et al. 2010. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 6, e1001116 ( 10.1371/journal.pgen.1001116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. 2003. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 22, 4082–4090. ( 10.1093/emboj/cdg392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Julian CG, Hageman JL, Wilson MJ, Vargas E, Moore LG. 2011. Lowland origin women raised at high altitude are not protected against lower uteroplacental O2 delivery during pregnancy or reduced birth weight. Am. J. Hum. Biol. 23, 509–516. ( 10.1002/ajhb.21167) [DOI] [PubMed] [Google Scholar]
- 92.Stein Z, Susser M. 1975. The Dutch famine, 1944–1945, and the reproductive process. I. Effects on six indices at birth. Pediatr. Res. 9, 70–76. [DOI] [PubMed] [Google Scholar]
- 93.Waterland RA, et al. 2010. Season of conception in rural Gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet. 6, e1001252 ( 10.1371/journal.pgen.1001252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tobi EW, et al. 2011. DNA methylation of IGF2, GNASAS, INSIGF and LEP and being born small for gestational age. Epigenetics 6, 171–176. ( 10.4161/epi.6.2.13516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. 2008. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl Acad. Sci. USA 105, 17 046–17 049. ( 10.1073/pnas.0806560105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smith FM, Garfield AS, Ward A. 2006. Regulation of growth and metabolism by imprinted genes. Cytogenet. Genome Res. 113, 279–291. ( 10.1159/000090843) [DOI] [PubMed] [Google Scholar]
- 97.Barker DJ. 1998. In utero programming of chronic disease. Clin. Sci. 95, 115–128. ( 10.1042/CS19980019) [DOI] [PubMed] [Google Scholar]
- 98.Hoy WE, Rees M, Kile E, Mathews JD, Wang Z. 1999. A new dimension to the Barker hypothesis: low birthweight and susceptibility to renal disease. Kidney Int. 56, 1072–1077. ( 10.1046/j.1523-1755.1999.00633.x) [DOI] [PubMed] [Google Scholar]
- 99.Gluckman PD, Hanson MA, Buklijas T. 2010. A conceptual framework for the developmental origins of health and disease. J. Dev. Origins Health Dis. 1, 6–18. ( 10.1017/S2040174409990171) [DOI] [PubMed] [Google Scholar]
- 100.Trivers RL. 1972. Mother–offspring conflict. Am. Zool. 12, 648. [Google Scholar]
- 101.Trivers RL. 1974. Parent–offspring conflict. Am. Zool. 14, 249–264. [Google Scholar]
- 102.Haig D. 2002. Genomic imprinting and the evolution of human childhood. Am. J. Hum. Biol. 14, 113. [DOI] [PubMed] [Google Scholar]
- 103.Moore T, Haig D. 1991. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 7, 45–49. ( 10.1016/0168-9525(91)90230-N) [DOI] [PubMed] [Google Scholar]
- 104.Pijnenborg R, Vercruysse L, Hanssens M. 2008. Fetal–maternal conflict, trophoblast invasion, preeclampsia, and the Red Queen. Hypertens. Pregnancy 27, 183–196. ( 10.1080/10641950701826711) [DOI] [PubMed] [Google Scholar]
- 105.Haig D. 1993. Genetic conflicts in human pregnancy. Q. Rev. Biol. 68, 495–532. ( 10.1086/418300) [DOI] [PubMed] [Google Scholar]
- 106.Sha K. 2008. A mechanistic view of genomic imprinting. Annu. Rev. Genomics Hum. Genet. 9, 197–216. ( 10.1146/annurev.genom.122007.110031) [DOI] [PubMed] [Google Scholar]
- 107.Ishida M, Moore GE. 2013. The role of imprinted genes in humans. Mol. Aspects Med. 34, 826–840. ( 10.1016/j.mam.2012.06.009) [DOI] [PubMed] [Google Scholar]
- 108.Haig D. 1997. Parental antagonism, relatedness asymmetries, and genomic imprinting. Proc. R. Soc. Lond. B 264, 1657–1662. ( 10.1098/rspb.1997.0230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Del Giudice M. 2012. Fetal programming by maternal stress: insights from a conflict perspective. Psychoneuroendocrinology 37, 1614–1629. ( 10.1016/j.psyneuen.2012.05.014) [DOI] [PubMed] [Google Scholar]
- 110.Medawar PB. (eds). 1953. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. Symp. Soc. Exp. Biol. 7, 320–338. [Google Scholar]
- 111.Romero R. 2009. Prenatal medicine: the child is the father of the man (Reprinted from Prenatal and Neonatal Medicine, vol. 1, pg 8–11, 1996). J. Matern. Fetal Neo M. 22, 636–639. ( 10.1080/14767050902784171) [DOI] [PubMed] [Google Scholar]