Abstract
Single-nucleotide polymorphisms (SNPs) are the most common source of genetic variation within a species; however, few investigations demonstrate how naturally occurring SNPs may increase strain virulence. We recently used group A Streptococcus as a model pathogen to study bacteria strain genotype–patient disease phenotype relationships. Whole-genome sequencing of approximately 800 serotype M59 group A Streptococcus strains, recovered during an outbreak of severe invasive infections across North America, identified a disproportionate number of SNPs in the gene encoding multiple gene regulator of group A Streptococcus (mga). Herein, we report results of studies designed to test the hypothesis that the most commonly occurring SNP, encoding a replacement of arginine for histidine at codon 201 of Mga (H201R), significantly increases virulence. Whole transcriptome analysis revealed that the H201R replacement significantly increased expression of mga and 54 other genes, including many proven virulence factors. Compared to the wild-type strain, a H201R isogenic mutant strain caused significantly larger skin lesions in mice. Serial quantitative bacterial culture and noninvasive magnetic resonance imaging also demonstrated that the isogenic H201R strain was significantly more virulent in a nonhuman primate model of joint infection. These findings show that the H201R replacement in Mga increases the virulence of M59 group A Streptococcus and provide new insight to how a naturally occurring SNP in bacteria contributes to human disease phenotypes.
Investigating the genetic basis for altered virulence phenotypes of bacteria is crucial to our ability to understand human infectious diseases. These molecular pathogenesis data are needed to generate new clinical tools, such as diagnostics and vaccines,1–3 and they may guide public health maneuvers during outbreaks.4,5 Within this context, single-nucleotide polymorphisms (SNPs) are the most common source of genomic variation within a particular species or serotype of bacteria.6–8 However, little data exist bearing on the effect of naturally occurring SNPs on host-pathogen interactions and increased strain virulence.9–12 Most investigations of strain genotype–disease phenotype relationships have focused on large regions of genetic variation, such as pathogenicity islands, recombination events, and mobile genetic elements (ie, plasmids), that introduce new gene content or multiple polymorphisms to a strain.13–16 In comparison, the effect of SNPs on strain virulence has been less intensely studied, with most investigations relying on random mutagenesis or animal passage experiments to discover mutations that alter strain phenotypes in vitro. Herein, we demonstrate that a naturally occurring SNP in a key regulatory gene in an important human pathogen significantly increases the expression of multiple virulence factors, and as a result, significantly increases strain virulence.
Group A Streptococcus (GAS; Streptococcus pyogenes), a human-specific pathogen, is a major cause of morbidity and mortality worldwide.17 GAS infections range in severity from mild pharyngitis (strep throat) to life-threatening necrotizing fasciitis (flesh-eating disease), toxic shock syndrome, and synovitis.18 GAS strains are taxonomically categorized by serologic- or sequence-based typing of the highly variable emm gene that encodes the M-protein virulence factor.19,20 To date, >200 M-types have been described.21 Although some GAS serotypes, such as M1 and M3, are globally disseminated,16 M59 strains are an uncommon cause of human disease, representing <1% of GAS recovered from large population-based studies of pharyngitis or invasive infections.20,21 However, a hypervirulent serotype M59 GAS clone recently emerged, and its progeny rapidly spread across Canada and some parts of the United States to cause several hundred severe invasive infections.22–25 To investigate the population genetic structure of strains comprising the epidemic and understand the host-pathogen interactions underlying strain virulence, our laboratory sequenced the genome of approximately 800 serotype M59 GAS strains.22–24 An unexpected discovery was that the most highly polymorphic gene was the multiple gene regulator of group A Streptococcus (mga), a key transcriptional regulator that influences approximately 10% to 20% of the GAS transcriptome, depending on the strain or serotype studied.26–28 More important, every SNP in mga encoded a missense (amino acid changing) or nonsense (premature protein termination) codon, suggesting selection for altered Mga sequences. Furthermore, one particular SNP encoding an arginine to histidine replacement in amino acid 201 of Mga (H201R) arose independently at least five times and was identified in 34 strains overall.22 These genomic data led us to hypothesize that the Mga H201R amino acid replacement significantly alters the virulence phenotype of M59 GAS. We used genome-wide transcript studies, a mouse model of skin and soft tissue infection, and a newly developed nonhuman primate model of joint infection to compare Mga wild-type and H201R strains. Results demonstrated that the Mga H201R amino acid replacement significantly increases M59 GAS virulence.
Materials and Methods
Bacterial Strains and Growth Conditions
GAS strains were grown routinely on trypticase soy agar containing 5% sheep blood or in Todd-Hewitt broth containing 0.2% yeast extract (THY; Becton Dickinson and Company, Franklin Lakes, NJ) at 37°C with 5% CO2. When appropriate, chloramphenicol was added to a final concentration of 8 μg/mL.
Genome-Wide Transcript Analysis
Serotype M59 GAS strains MGAS15252 (reference) and MGAS18055 (naturally occurring Mga H201R sequence) were recovered from patients in Canada with invasive infections.23,25 Strain MGAS15252 was selected as the reference because its genome has been sequenced to closure, it expresses mga and Mga-regulated genes at levels similar to other M59 GAS strains (Kachroo P), and it has been used in numerous mouse and nonhuman primate virulence studies.23,24 Genome-wide transcript analysis was performed as previously described.16 Briefly, triplicate cultures were grown in THY overnight, diluted 1:50 in fresh THY media, and harvested at late-logarithmic (OD600 = 1) phase of growth. RNA was stabilized (RNAprotect Bacteria Reagent; Qiagen, Valencia, CA), cells were homogenized using ballistic disintegration (Lysing Matrix B and FastPrep96 Automated Homogenizer; MP Biomedicals, Santa Ana, CA), and nucleic acids were extracted using standard methods (RNeasy96; Qiagen). RNA quality (model 2100 Bioanalyzer; Agilent Technologies, Santa Clara, CA) and quantity (Qubit 2.0 fluorometer; Life Technologies, Carlsbad, CA) were assessed. RNA sequencing libraries were prepared according to manufacturer's instructions (ScriptSeq version 2; EpiCentre Biotechnologies, Madison, WI), and sequenced using a MiSeq Instrument (MiSeq reagent kit; Illumina Inc., San Diego, CA). RNA transcripts were quantified as previously described (CLC Bio, Cambridge, MA)29 using reference strain MGAS15252 (http://www.ncbi.nlm.nih.gov/nuccore; Accession number CP003116). Differences >1.7-fold and P < 0.05, using Baggerly's test and Bonferroni's correction for multiple comparisons, were considered statistically significant (Supplemental Table S1).
Construction of Isogenic Mutants
Isogenic mutant strains were generated using MGAS15249 because its genome has been sequenced, it is genomically representative of the epidemic serotype M59 GAS clone, and it has the most common allele (wild-type sequence) for all major regulatory genes.23 An isogenic mutant strain lacking the gene encoding mga was generated by in-frame insertional inactivation with a spectinomycin-resistance cassette, as previously described (Supplemental Figure S1).10 The mga-deleted strain was then complemented in trans using the pDC123 low-copy plasmid30 containing either no insert (designated Δmga) or the coding sequence of the full-length mga gene with its native promoter (designated wild type). To generate an isogenic mutant strain carrying the Mga H201R sequence (designated H201R), the plasmid containing the wild-type allele was used as a template for site-directed mutagenesis, according to the manufacturer's instructions (QuikChange II; Stratagene, Life Technologies). The sequence of mga in all isogenic strains was verified by Sanger dideoxynucleotide sequencing (Big Dye Terminator and ABI 3730 DNA Analyzer; Life Technologies). No differences in growth were observed among the isogenic strains (Supplemental Figure S2A). A real-time PCR assay using total genomic DNA extracted from the isogenic strains confirmed that no differences in mga gene copy number were introduced by the in trans complementation strategy (Supplemental Figure S2B). The sequence of all oligonucleotides used in this research is listed in Table 1.
Table 1.
Probes and Primers Used for This Research
| Primer | Sequence | Application |
|---|---|---|
| delMga A | 5′-ATAGCGAGTGTTGCCATGTTAG-3′ | Upstream fragment of mga for isogenic deletion |
| delMga B | 5′-GTTATAGTTATTATAACATGTATTATGCATTAACTTCATGTCCTTATC-3′ | Upstream fragment of mga for isogenic deletion |
| delMga C | 5′-CTATTTAAATAACAGATTAAAAAAATTATAAACATCATCATAGGATTTCAGACGT-3′ | Downstream fragment of mga for isogenic deletion |
| delMga D | 5′-AATTTCCTCAGTCTTAGAGGCATCT-3′ | Downstream fragment of mga for isogenic deletion |
| delMga E | 5′-CATTTTCAAGAGCTAATGTTGGT-3′ | Sequencing and PCR confirmation of deletion mga |
| delMga F | 5′-TATACTTGTCGCTAGATTCTCT-3′ | Sequencing and PCR confirmation of deletion mga |
| delMga G | 5′-AGCTGCCTGCCTGTTGACCAATC-3′ | Sequencing and PCR confirmation of deletion mga |
| delMga H | 5′-ATCTAGCTTAGCTTGCAGATCAGTC-3′ | Sequencing and PCR confirmation of deletion mga |
| Mga-spc forward | 5′-GATAAGGACATGAAGTTAATGCATAATACATGTTATAATAACTATAAC-3′ | Amplification of spec cassette |
| Mga-spc reverse | 5′-ACGTCTGAAATCCTATGATGATGTTTATAATTTTTTTAATCTGTTATTTAAATAG-3′ | Amplification of spec cassette |
| pDC-Mga top | 5′-GGAAGATCTTAGAGTAATAGGTCAAATAATC-3′ | Pmga-mga cloning into pDC |
| pDC-Mga bottom | 5′-GGGAATCCATATGCTATGATGATGTTGCTTGC-3′ | Pmga-mga cloning into pDC |
| pDC-Mga Top2 | 5′-ATGGTTCATACGGACTTG-3′ | Sequencing of mga |
| pDC-Mga Top3 | 5′-TGCTATTAGTATCGTGACAAG-3′ | Sequencing of mga |
| pDC-Mga Bottom2 | 5′-GATCAATCAGCTCACTT-3′ | Sequencing of mga |
| H201R top | 5′-GTAGATGTCAAAGTTCGTTTTACACTATTTCAG-3′ | Site-directed mutagenesis to introduce H201R |
| H201R bottom | 5′-CATCTACAGTTTCAAGCAAAATGTGATAAAGTC-3′ | Site-directed mutagenesis to introduce H201R |
| mga qRT forward | 5′-TCAATCAAGACCCGACATCA-3′ | RT-qPCR analysis of mga |
| mga qRT reverse | 5′-GGTCACGGCAACTTCGTATT-3′ | RT-qPCR analysis of mga |
| mga Probe | 5′-GCTCAATCTCAGCATCACCA-3′ | RT-qPCR analysis of mga |
| tufA qRT forward | 5′-CAACTCGTCACTATGCGCACAT-3′ | RT-qPCR analysis of tufA |
| tufA qRT reverse | 5′-GAGCGGCACCAGTGATCAT-3′ | RT-qPCR analysis of tufA |
| tuf A probe | 5′-CTCCAGGACACGCGGACTACGTTAAAAA-3′ | RT-qPCR analysis of tufA |
qPCR, real-time quantitative PCR.
GAS–Human Epithelial Cell Adherence Assay
Epithelial cell adherence of the isogenic M59 GAS strains (Δmga, wild-type, and H201R) was measured as previously described.31 Briefly, the human primary keratinocyte cell line HaCaT (Life Technologies, Grand Island, NY) was cultured in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 2 mmol/L glutamine and 10% calf serum. HaCaT cells were resuspended at a concentration of 1 × 106/mL, and 900 μL was seeded into each well of a 12-well tissue culture plate and incubated for 24 hours at 37°C with 5% CO2. GAS strains were prepared by growing each strain to OD600 = 0.5 (early logarithmic phase of growth) and resuspended in an equal volume of phosphate-buffered saline, and 100 μL was added to each well containing the lawn of HaCaT cells. After incubating for 2 minutes, the supernatant was removed and the wells were washed five times to remove nonadherent GAS. Then, 1 mL of phosphate-buffered saline containing 1% saponin was added to each well, and the plate was incubated for 30 minutes at 37°C to lyse the HaCaT cells. GAS strains (adherent to the HaCaT cells) recovered from each well were enumerated by serial dilution and plating. Mean GAS recovered from eight biological replicates of each strain was compared using the Mann-Whitney test (Prism version 6; GraphPad Software, La Jolla, CA), with P < 0.05 considered to be statistically significant.
Virulence in a Mouse Model of Skin and Soft Tissue Infection
Virulence of the isogenic M59 GAS strains (Δmga, wild-type, and H201R) was tested using a mouse model of skin and soft tissue infection, as previously described.23 Briefly, 4- to 5-week-old, 18- to 20-g, immunocompetent SKH1-hrBR hairless female mice (Charles River BRF, Houston, TX) were inoculated in the s.c. tissue overlying the neck with 1 × 108 colony-forming units (CFUs; n = 12 mice per strain). Lesions were measured daily using a digital caliper for 10 days, and then on days 12 and 14 after inoculation. Mean abscess area caused by each strain was compared using two-way analysis of variance (Prism version 6), with P < 0.05 considered to be statistically significant. For histopathological evaluation, skin lesions were excised en bloc and processed using standard methods. Slides were examined independently by two pathologists (M.S., R.J.O.) blinded to the strain treatment groups, as previously described.23 Representative micrographs were obtained using a BX5 microscope fitted with a DB70 digital camera (Olympus, Tokyo, Japan). The study protocol was approved by the Houston Methodist Research Institute (Houston, TX) Animal Care and Use Committee.
Virulence in a Nonhuman Primate Model of Joint Infection
Virulence of the isogenic M59 GAS strains (wild-type and H201R) was also assessed using a new nonhuman primate joint infection model. Cynomolgus macaques (Macaca fasicularis; Charles River BRF) were inoculated in the intra-articular space with 1 × 108 CFUs of the wild-type strain in the right elbow and the isogenic H201R strain in the left elbow (n = 3). By using this strategy, each animal serves as its own control. The animals were observed continuously for 7 days. On days 1, 2, 4, and 7 (one animal was examined on day 8 rather than day 7 to accommodate vivarium scheduling) after inoculation, each animal was sedated, a thorough physical examination was conducted, blood and synovial fluid were collected, and radiological examination was performed using magnetic resonance (MR) imaging. For quantitative culture, synovial fluid aspirated from each elbow was serially diluted, inoculated in duplicate on trypticase soy agar, and grown as described above. CFUs recovered per volume (mL) synovial fluid were calculated, with P < 0.05 considered statistically significant using repeated-measures analysis of variance (XLSTAT; Addinsoft, New York, NY). Blood cultures performed on each sampling day confirmed that the animals were not bacteremic. For radiological examination, each arm was immobilized using a soft cast, placed in a human wrist coil, and imaged using a 3.0-T MR imaging instrument (Ingenia; Philips Healthcare, Amsterdam, the Netherlands). To compare virulence of the isogenic wild-type (right elbow) and H201R (left elbow) strains, the volume fraction of inflamed tissue of each limb at each time point was calculated from a series of five coronal images centered on the elbow joint. The T2 maps were scaled to make optimal use of the available dynamic range, normalized and pseudocolored. The scaling factor was extracted from the image headers. A lower threshold (threshold 1) of 20 was established to capture all pixels across the entire limb. An upper threshold (threshold 2) of 104 was established to capture all inflammation above the baseline condition. Then, the volume fraction of inflamed tissue was calculated as the sum total of pixels greater than threshold 2/the sum total of pixels greater than threshold 1. This formula yielded a volume fraction of inflamed tissue <0.05 for all joints at the baseline MR imaging studies acquired before GAS inoculation. A disease model for each strain was then calculated using nonlinear least squares regression (R Foundation for Statistical Computing, Vienna, Austria). All parameters in the disease progression model were significantly different from 0 (P < 0.001), indicating that this model effectively captures the progress of inflammation. The study protocol was approved by the Houston Methodist Research Institute Animal Care and Use Committee.
Results
The Mga H201R Amino Acid Replacement Significantly Increases Expression of mga and Mga-Regulated Genes
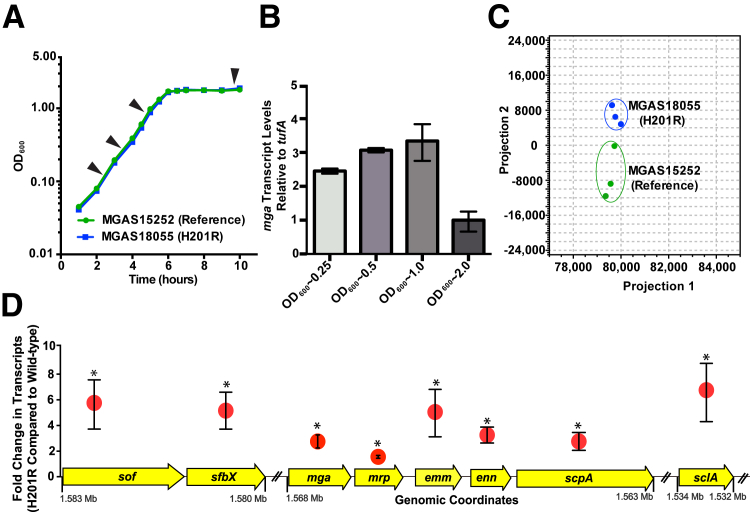
One possible mechanism to markedly alter strain virulence with a single amino acid replacement is to target a major transcriptional regulator and, thus, alter the expression of all genes under its control.32 In other GAS serotype strains, Mga has been implicated in the regulation of 10% to 20% of the transcriptome.26–28 To begin testing the hypothesis that the Mga H201R amino acid replacement significantly alters the M59 GAS virulence phenotype, genome-wide transcript studies were performed using a reference and Mga H201R strain. GAS strains were grown to late-logarithmic phase of growth, the point in the GAS growth curve when mga is maximally expressed (Figure 1, A and B). Analysis of RNA sequence data revealed a highly similar genome-wide expression profile that is compatible with the clonal background of epidemic M59 GAS (Figure 1C). Compared to the reference strain, 55 transcripts demonstrated significantly altered expression in the H201R strain, including 48 genes with increased expression and 7 genes with decreased expression (Supplemental Table S1). Transcripts with significantly increased expression by the H201R strain include mga, which is known to autoregulate its expression, and seven proven virulence factors that are also regulated by Mga in other GAS serotype strains (Figure 1D).26–28 These gene expression data are consistent with the idea that the Mga H201R amino acid replacement may increase M59 GAS virulence by increasing the expression of mga and Mga-regulated virulence factors.
Figure 1.
Genome-wide transcript analysis of serotype emm59 group A Streptococcus (GAS) strains MGAS15252 (reference) and MGAS18055 (Mga H201R amino acid replacement). A: Growth curve of strains MGAS15252 and MGAS18055. Arrowheads indicate the 4 time points that were tested for gene transcript studies. B:mga expression is highest at late-logarithmic phase (OD600 = 1.0). C: Principal component analysis is shown for genome-wide transcript profiles of strains grown to late-logarithmic phase. D: The expression of mga and seven Mga-regulated genes was significantly increased in strain MGAS18055 (H201R) compared to strain MGAS15252. Mean X-fold change in transcripts and genomic coordinates are shown for each gene. ∗P < 0.05 compared to MGAS15252 (Baggerly's test after applying Bonferroni's correction for multiple comparisons).
The Mga H201R Amino Acid Replacement Significantly Increases GAS Adherence to Human Epithelial Cells
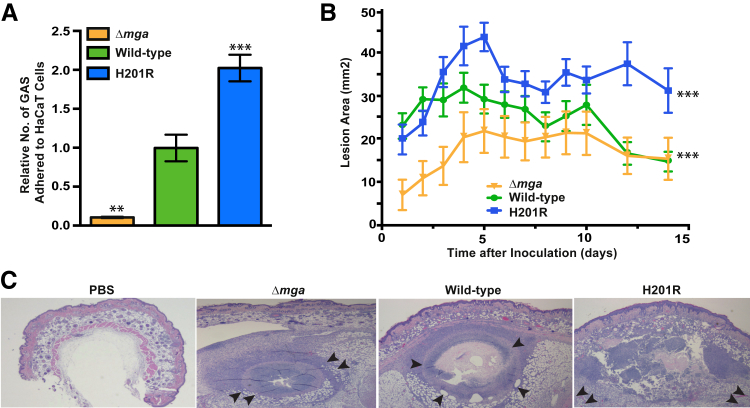
Adhesion to human tissues is a key virulence activity underlying pathogen transmission, colonization, and invasion. Our genome-wide transcript studies identified several genes with significantly increased expression by the Mga H201R strain, including emm, mrp, and enn, which have been implicated in GAS adhesion to host cells.33–35 To test the hypothesis that the Mga H201R amino acid replacement alters GAS adhesion to human tissues, GAS–epithelial cell adhesion assays were performed using isogenic Mga-deficient (Δmga), wild-type, and H201R strains. Consistent with our hypothesis, significantly more of the isogenic Mga H201R GAS organisms adhered to the human cells in vitro (Figure 2A). Results also demonstrated that deletion of Mga significantly decreases GAS adhesion compared to either comparator strain (Figure 2A). Taken together, these data are consistent with the hypothesis that the Mga H201R amino acid replacement results in increased adhesion to human tissues and, thus, may increase the virulence of M59 GAS.
Figure 2.
The Mga H201R amino acid replacement significantly increases strain virulence in vitro and in a mouse model of skin and soft tissue infection. A: The isogenic Mga-deficient (Δmga), wild-type, and H201R strains were incubated with human epithelial keratinocytes (HaCaT cells), and adhesion was calculated by quantitative culture. B: The H201R amino acid replacement significantly increases lesion size in a mouse model of skin and soft tissue infection. C: Microscopic examination of skin lesions collected at day 7 after inoculation. The abscesses caused by strains wild-type and Δmga have confined borders (inward-facing arrowheads). In comparison, the abscess caused by strain H201R destroys more tissue and extends beyond the lateral and deep margins of the microscopic field (outward-facing arrowheads). Hematoxylin and eosin staining was used. ∗∗P < 0.01, ∗∗∗P < 0.001 versus wild-type using the Mann-Whitney test (A); ∗∗∗P < 0.001 versus wild-type using two-way analysis of variance (B). Original magnification, ×4 (C).
The Mga H201R Amino Acid Replacement Significantly Increases GAS Virulence in a Mouse Model of Invasive Skin and Soft Tissue Infection
Animal infection models are important for testing hypotheses that bear on genetic alterations and strain virulence. To test the hypothesis that the Mga H201R amino acid replacement increases M59 GAS virulence, the isogenic Mga-deficient (Δmga), wild-type, and H201R strains were compared using a mouse model of invasive skin and soft tissue infection. Results demonstrated that all strains caused necrotic lesions centered at the inoculation site (Figure 2, B and C). Consistent with our hypothesis, the isogenic Mga H201R strain caused lesions that were significantly larger than either comparator strain (Figure 2B). We next hypothesized that the increased virulence of the Mga H201R strain would manifest as significantly altered histopathological characteristics of the resulting abscesses. Consistent with this hypothesis, microscopic examination of the skin lesions collected on day 7 after inoculation (the time point when differences in lesion character were most evident by visual examination) revealed that the isogenic Mga H201R strain caused more tissue destruction and dissemination than the comparator strains (Figure 2C). The wild-type strain had a virulence phenotype intermediate of the Mga-deficient and H201R strain (Figure 2, B and C).
The Mga H201R Amino Acid Replacement Significantly Increases GAS Virulence in a Monkey Model of Joint Infection
GAS is a host-specific pathogen with exquisite specificity for human molecules. Several proven and putative GAS virulence factors have modest to no activity against homologous targets in mice.36,37 Therefore, mouse and other lower vertebrate animal models, although widely used in molecular pathogenesis research, may provide an incomplete understanding of the complex host-pathogen interactions that underlie human-GAS disease. To this end, the cynomolgus macaque has proved to be an excellent phenocopy of GAS pharyngitis, pneumonia, and necrotizing fasciitis in humans, and it has been successfully used in numerous GAS molecular pathogenesis research studies.10,16,22,38–40 That is, the cynomolgus macaque is the most human relevant model possible to study GAS virulence.
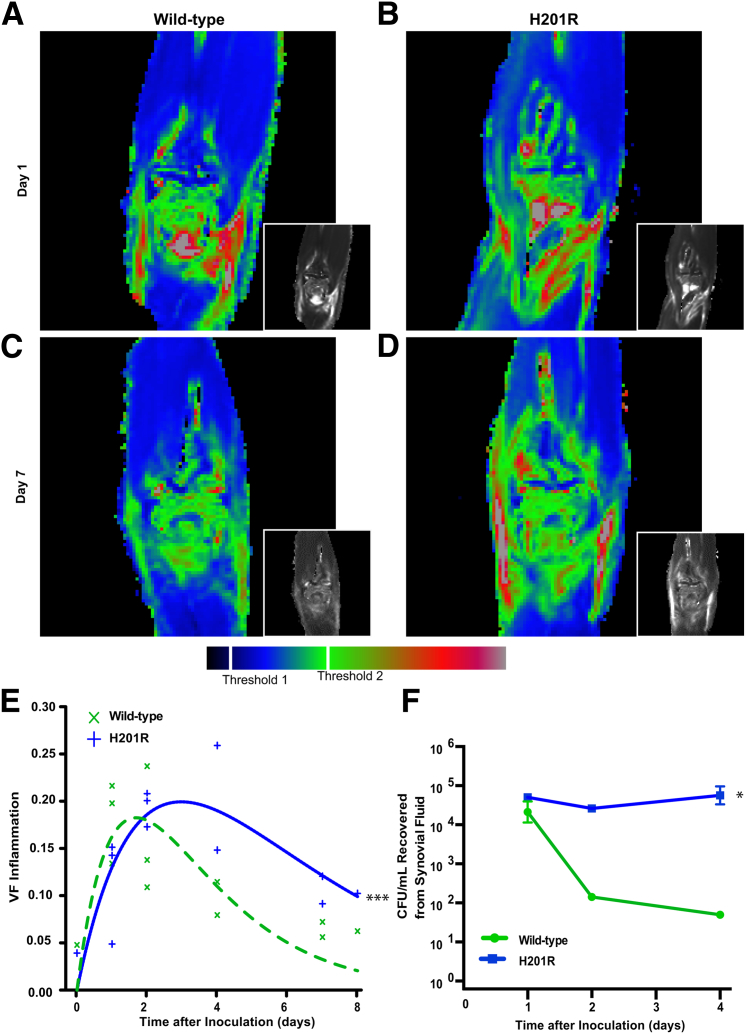
Most invasive disease caused by M59 GAS manifests as bacteremia, skin and soft tissue infection, and synovitis.22,25 To further test the hypothesis that the Mga H201R amino acid replacement increases GAS virulence, we generated a new cynomolgus macaque model of joint infection. For this experiment, each monkey served as its own control, receiving the isogenic Mga wild-type strain in one elbow and the H201R strain in the contralateral elbow. Physical examination results revealed signs of joint infection in all animals, including swollen and erythematous joints bilaterally, altered posturing, and reluctance to use the infected limbs. Noninvasive serial MR imaging confirmed that the isogenic Mga wild-type and H201R strains both caused marked intra-articular and peri-articular inflammation and edema at 1 and 2 days after inoculation (Figure 3, A and B). However, the synovial disease rapidly dissipated in elbows infected with the Mga wild-type strain (Figure 3C). In comparison, the isogenic H201R strain caused inflammation and edema that persisted for 7 days (Figure 3D). Consistent with our hypothesis, the Mga H201R strain caused significantly more tissue damage, inflammation, and edema overall (Figure 3E). Similarly, significantly more CFUs were recovered from synovial fluid collected from joints infected with the isogenic Mga H201R strain compared to those receiving the wild-type strain (Figure 3F).
Figure 3.
The Mga H201R amino acid replacement significantly increases strain virulence in a nonhuman primate model of joint infection. Cynomolgus macaques were inoculated in the right and left elbow with the isogenic wild-type and H201R strains, respectively, and observed for 7 days. A–D: Magnetic resonance imaging was used for noninvasive serial measurement of inflammation and edema. Representative T2 pseudocolored coronal maps collected on days 1 and 7 after inoculation are shown. E: The volume fraction of inflammation (VF) caused by each strain was calculated using thresholds 1 and 2, and disease was modeled using nonlinear least squares. F: Synovial fluid [colony-forming unit (CFU)/mL] was quantified. ∗P < 0.05 compared to wild-type using repeated-measures analysis of variance; ∗∗∗P < 0.001 versus wild-type using analysis of variance.
Discussion
Our laboratory has recently used whole-genome sequencing to investigate the population genetic structure of GAS and identify strain genotype–disease phenotype relationships.16,23,41 An emerging theme is that GAS organisms can dramatically alter their virulence phenotype via SNPs in major regulatory genes. That is, a single-nucleotide change in a transcriptional regulator can exert wide-ranging effects by altering the expression of multiple virulence factors under its control. For example, naturally occurring polymorphisms in the gene-encoding regulator of protease B (ropB) alter 10% to 25% of the GAS transcriptome.32,42,43 These ropB polymorphisms significantly decrease expression of the potent secreted protease virulence factor SpeB that is crucial for tissue destruction and systemic dissemination, and decrease GAS virulence in mice and invasive infections in humans.32,42,43 Similarly, Olsen et al10 demonstrated that a naturally occurring SNP in the gene encoding the metal transporter of Streptococcus regulator (mtsR) significantly decreases the ability of GAS to cause necrotizing fasciitis in nonhuman primates and humans. In comparison, naturally occurring amino acid replacements in the control of virulence regulator/sensor (covR/S), an extensively studied two-component transcriptional regulator in GAS, are well known to increase strain virulence.44 Herein, we demonstrated that the Mga H201R amino acid replacement also significantly increases GAS virulence. These findings have important implications to vaccine design. Many virulence factors regulated by regulator of protease B (ropB), metal transporter of Streptococcus regulator (mtsR), control of virulence regulator/sensor, and Mga have been proposed for inclusion in vaccines.45–47 This strategy needs to be carefully considered. If naturally occurring mutations in transcription factors frequently decrease the expression of some virulence factors in vivo, then those molecules may not be ideal targets for intervention. Rather, vaccines that include virulence factors that are highly expressed in wild-type and mutant strains may be more effective.
Our whole-genome sequencing studies demonstrated mga to be the most highly polymorphic gene among epidemic M59 GAS strains. In total, 48 different SNPs in mga have been identified.22,23 Each is predicted to change the encoded amino acid or result in premature termination, suggesting selection for altered Mga sequences. Although less common among the epidemic M59 GAS strains, SNPs have also been identified in the covR, covS, ropB, and rofA transcriptional regulators.23 Similar to the Mga H201R amino acid substitution that arose independently at least five times, a single-nucleotide deletion that introduces a premature stop codon in covS is also present in multiple subclonal lineages.22 However, the biological effect of these mutant alleles on strain virulence is unknown.
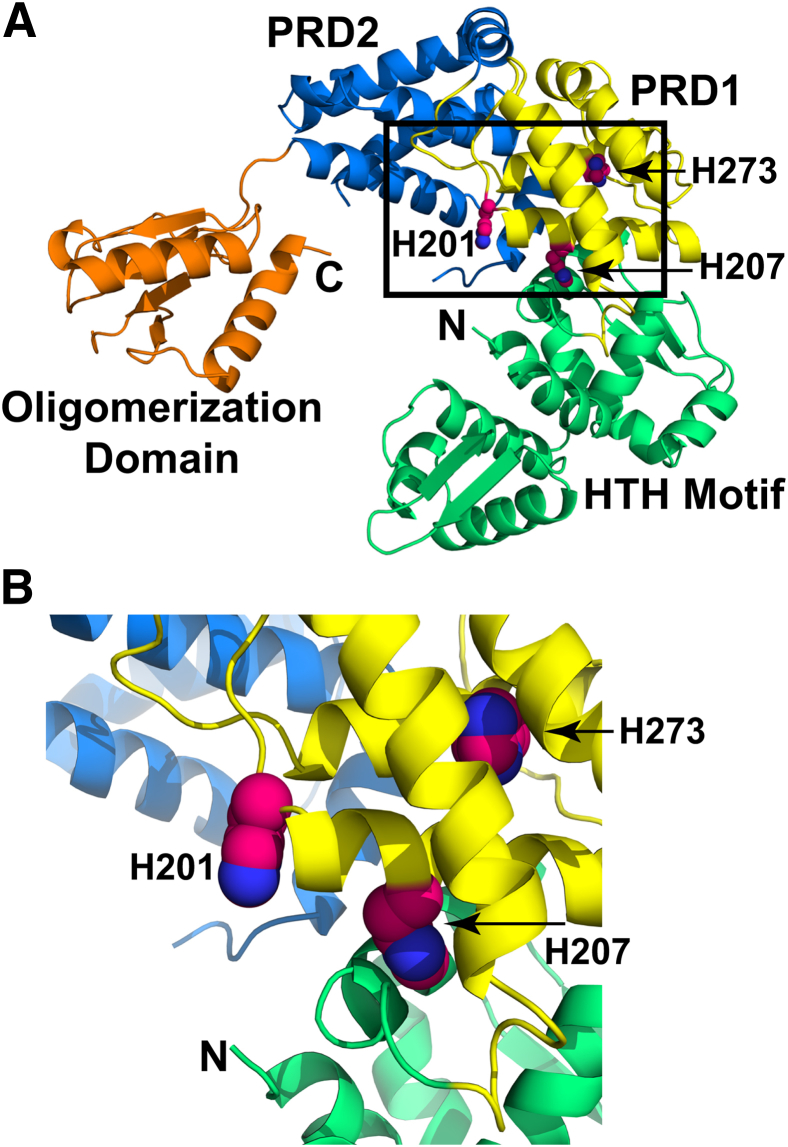
One possible explanation for the H201R amino acid replacement leading to increased strain virulence is disruption of the normal pathways that regulate transcriptional activity by Mga. Results of the genome-wide transcript studies (Figure 1) and animal infection models (Figures 2 and 3) unambiguously demonstrated that the Mga H201R amino acid replacement significantly increases expression of mga and Mga-regulated genes and GAS virulence. To determine the possible molecular mechanism underlying the increased virulence phenotype of strains carrying the Mga H201R sequence, molecular modeling simulations were performed using the I-TASSER structure prediction server (http://zhanglab.ccmb.med.umich.edu/I-TASSER, last accessed October 3, 2014).48 The model, using the M-protein transacting positive regulator HTH domain protein from Enterococcus faecalis as a template, is predicted to have a reliable template modeling score of 0.65 (Figure 4). Mga consists of four domains, including an amino-terminal DNA-binding domain with helix-turn-helix motifs, a central domain with two phosphotransferase system regulatory domains (PRD-1 and PRD-2), and a C-terminal oligomerization domain.49 Hondorp et al49 recently demonstrated that phosphorylation of two histidines in the PRD-1 of Mga, H207 and H273, serve as a regulatory switch for decreasing transcription activation. That is, transcription activation by Mga is greatest when amino acids H207 and H273 are not phosphorylated. The model structure predicts that amino acid 201 lies on the same α helix as the phosphorylatable H207 residue and in close proximity to the phosphorylatable H273 residue (Figure 4). Because the solubility of Mga with a histidine or arginine at codon 201 is similar (data not shown), it is unlikely that the amino acid replacement causes any structural defect. Rather, the replacement of arginine, which has a longer and positively charged side chain compared to histidine, may prevent phosphorylation of one or both PRD-1 histidines. Alternatively, the H201R replacement may block phosphorylation-dependent signal transduction from the PRD-1 domain to the amino-terminal DNA-binding domain. As a result, M59 GAS strains with the Mga H201R sequence are significantly more virulent. Compared to wild-type M59 GAS strains, Mga H201R strains have an increased ability to adhere to host tissue, grow in situ, destroy tissue, and cause inflammation and edema.
Figure 4.
Model of group A Streptococcus (GAS) Mga. A: Individual domains of Mga are color coded: N-terminal DNA-binding domain with the helix-turn-helix motifs in green, PRD-1 in yellow, PRD-2 in blue, and C-terminal oligomerization domain in orange. The N- and C-terminal ends are labeled. The side chains of H201, H207, and H273 are shown as spheres. B: Magnified view of amino acid H201 and its relative proximity to the phosphorylatable H207 and H273 residues.
We generated a new nonhuman primate model of joint infection to test hypotheses bearing on the virulence of a human-specific pathogen. The use of nonhuman primates, the most human-relevant animal model possible, combined with MR imaging in a dedicated translational imaging suite was key to the success of this investigation. Although imaging technologies have been extensively used to study cancer and inflammatory disorders, they have yet to become widely applied to infectious disease research.50–52 MR imaging is the modality of choice for imaging soft tissue infection, particularly synovitis, because it can depict inflammation and edema.53 Still, the current literature for MR imaging of invasive infections is mostly limited to small animal models, selected case reports, or case series with a few cases.54–56 Herein, we used MR imaging to monitor disease progression and compare the virulence of isogenic mutant strains, with results directly bearing on the overarching hypothesis. The ability to perform serial real-time imaging allowed each animal to serve as its own control, thereby reducing intrahost variation, increasing statistical power, and decreasing costs. Performing the study on a clinical MR human 3.0 T scanner housed in a dedicated translational imaging facility also enables us to design custom MR acquisition protocols that can be readily applied to human imaging and facilitates the refinement of our model for future investigations. Serial noninvasive imaging data make practical the interpretation with a mathematical model of disease progression over time in a given host. Translating our mathematical approach to research and diagnostic applications in patients is thereafter straightforward.
In summary, we used whole-genome sequencing,22–24 genome-wide transcript analysis, and mouse and nonhuman primate models of infection to demonstrate that a naturally occurring single amino acid replacement significantly increases the virulence of M59 GAS. This discovery provides a genetic basis, in part, for understanding differences in virulence phenotypes among closely related strains. The investigative strategy should be used with other medically important microbial pathogens, such as Staphylococcus aureus and Klebsiella pneumoniae. Taken together, these data will improve our understanding of strain genotype–disease phenotype relationships and can be used to generate new clinical tools, such as diagnostics and vaccines.
Acknowledgments
We thank Nam Yu for helpful discussions; Jamie Champagne, Leslie Jenkins, and Annessa Raiford for veterinary assistance; and Kathryn Stockbauer for editorial assistance. The magnetic resonance imaging was performed at the Houston Methodist Research Institute MRI Core.
M.S., M.K., N.F., J.M.M., and R.J.O. designed the study. M.S., B.E.O., P.K., J.R.A., A.R.F., C.V., C.C.C., N.M., C.K, and R.J.O. performed the research. M.S., J.M.M., and R.J.O. wrote the first complete draft of the report. All authors provided contributions and suggestions and read and approved the final version.
Footnotes
Supported by the Fondren Foundation, Department of Pathology and Genomic Medicine, Houston Methodist Hospital, in part by School of Medicine and Health Sciences, Monterrey Institute of Technology grant 421460/263885 (M.S.), and in part by NIH, National Institute of Allergy and Infectious Diseases, grant R21 AI103708 (M.K.).
Disclosures: None declared.
Supplemental Data
Cloning scheme to generate isogenic Mga-deficient (Δmga), wild-type, and H201R strains. A: Molecular genetic strategy for inactivating mga in strain MGAS15249. The mga was replaced with a gene conferring spectinomycin resistance. B: pDC123 with no insert was used to generate strain designated as Δmga. C: The mga was cloned into pdDC123 and used in trans to generate strain designated as wild-type. D: Quick-change site-directed mutagenesis was used to introduce the H201R amino acid replacement to generate strain H201R. E. coli, Escherichia coli.
A: Growth curves of isogenic group A Streptococcus (GAS) strains Δmga, wild-type, and H201R. B: Relative mga copies per GAS cell were measured using quantitative real-time PCR assay.
References
- 1.Croucher N.J., Harris S.R., Fraser C., Quail M.A., Burton J., van der Linden M., McGee L., von Gottberg A., Song J.H., Ko K.S., Pichon B., Baker S., Parry C.M., Lambertsen L.M., Shahinas D., Pillai D.R., Mitchell T.J., Dougan G., Tomasz A., Klugman K.P., Parkhill J., Hanage W.P., Bentley S.D. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eppinger M., Mammel M.K., Leclerc J.E., Ravel J., Cebula T.A. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A. 2011;108:20142–20147. doi: 10.1073/pnas.1107176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koser C.U., Holden M.T., Ellington M.J., Cartwright E.J., Brown N.M., Ogilvy-Stuart A.L., Hsu L.Y., Chewapreecha C., Croucher N.J., Harris S.R., Sanders M., Enright M.C., Dougan G., Bentley S.D., Parkhill J., Fraser L.J., Betley J.R., Schulz-Trieglaff O.B., Smith G.P., Peacock S.J. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med. 2012;366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasko D.A., Webster D.R., Sahl J.W., Bashir A., Boisen N., Scheutz F., Paxinos E.E., Sebra R., Chin C.S., Iliopoulos D., Klammer A., Peluso P., Lee L., Kislyuk A.O., Bullard J., Kasarskis A., Wang S., Eid J., Rank D., Redman J.C., Steyert S.R., Frimodt-Moller J., Struve C., Petersen A.M., Krogfelt K.A., Nataro J.P., Schadt E.E., Waldor M.K. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilmour M.W., Graham M., Van Domselaar G., Tyler S., Kent H., Trout-Yakel K.M., Larios O., Allen V., Lee B., Nadon C. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics. 2010;11:120. doi: 10.1186/1471-2164-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beres S.B., Musser J.M. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One. 2007;2:e800. doi: 10.1371/journal.pone.0000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakobsson M., Scholz S.W., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., Bras J.M., Schymick J.C., Hernandez D.G., Traynor B.J., Simon-Sanchez J., Matarin M., Britton A., van de Leemput J., Rafferty I., Bucan M., Cann H.M., Hardy J.A., Rosenberg N.A., Singleton A.B. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 8.Deleo F.R., Chen L., Porcella S.F., Martens C.A., Kobayashi S.D., Porter A.R., Chavda K.D., Jacobs M.R., Mathema B., Olsen R.J., Bonomo R.A., Musser J.M., Kreiswirth B.N. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A. 2014;111:4988–4993. doi: 10.1073/pnas.1321364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao W., Chua K., Davies J.K., Newton H.J., Seemann T., Harrison P.F., Holmes N.E., Rhee H.W., Hong J.I., Hartland E.L., Stinear T.P., Howden B.P. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 2010;6:e1000944. doi: 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen R.J., Sitkiewicz I., Ayeras A.A., Gonulal V.E., Cantu C., Beres S.B., Green N.M., Lei B., Humbird T., Greaver J., Chang E., Ragasa W.P., Montgomery C.A., Cartwright J., Jr., McGeer A., Low D.E., Whitney A.R., Cagle P.T., Blasdel T.L., DeLeo F.R., Musser J.M. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc Natl Acad Sci U S A. 2010;107:888–893. doi: 10.1073/pnas.0911811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otsuki H., Kaneko O., Thongkukiatkul A., Tachibana M., Iriko H., Takeo S., Tsuboi T., Torii M. Single amino acid substitution in Plasmodium yoelii erythrocyte ligand determines its localization and controls parasite virulence. Proc Natl Acad Sci U S A. 2009;106:7167–7172. doi: 10.1073/pnas.0811313106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores A.R., Jewell B.E., Olsen R.J., Shelburne S.A., 3rd, Fittipaldi N., Beres S.B., Musser J.M. Asymptomatic carriage of group A streptococcus is associated with elimination of capsule production. Infect Immun. 2014;82:3958–3967. doi: 10.1128/IAI.01788-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S., Green N.M., Sitkiewicz I., Lefebvre R.B., Musser J.M. Identification and characterization of an antigen I/II family protein produced by group A Streptococcus. Infect Immun. 2006;74:4200–4213. doi: 10.1128/IAI.00493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt H., Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev. 2004;17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan R., Reeves P.R. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 2000;8:396–401. doi: 10.1016/s0966-842x(00)01791-1. [DOI] [PubMed] [Google Scholar]
- 16.Nasser W., Beres S.B., Olsen R.J., Dean M.A., Rice K.A., Long S.W., Kristinsson K.G., Gottfredsson M., Vuopio J., Raisanen K., Caugant D.A., Steinbakk M., Low D.E., McGeer A., Darenberg J., Henriques-Normark B., Van Beneden C.A., Hoffmann S., Musser J.M. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A. 2014;111:E1768–E1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ralph A.P., Carapetis J.R. Group a streptococcal diseases and their global burden. Curr Top Microbiol Immunol. 2013;368:1–27. doi: 10.1007/82_2012_280. [DOI] [PubMed] [Google Scholar]
- 18.Olsen R.J., Musser J.M. Molecular pathogenesis of necrotizing fasciitis. Annu Rev Pathol. 2010;5:1–31. doi: 10.1146/annurev-pathol-121808-102135. [DOI] [PubMed] [Google Scholar]
- 19.Lancefield R.C. A serological differentiation of human and other groups of hemolytic streptococci. J Exp Med. 1933;57:571–595. doi: 10.1084/jem.57.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shea P.R., Ewbank A.L., Gonzalez-Lugo J.H., Martagon-Rosado A.J., Martinez-Gutierrez J.C., Rehman H.A., Serrano-Gonzalez M., Fittipaldi N., Beres S.B., Flores A.R., Low D.E., Willey B.M., Musser J.M., Group A. Streptococcus emm gene types in pharyngeal isolates, Ontario, Canada, 2002-2010. Emerg Infect Dis. 2011;17:2010–2017. doi: 10.3201/eid1711.110159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steer A.C., Law I., Matatolu L., Beall B.W., Carapetis J.R. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9:611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 22.Fittipaldi N., Tyrrell G., Low D., Martin E., Lin D., Kumar L.H., Musser J.M. Integrated whole-genome sequencing and temporostapial analysis of a continuing Group A Streptococcus epidemic. Emerg Microb Infect. 2013;2:e13. doi: 10.1038/emi.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fittipaldi N., Beres S.B., Olsen R.J., Kapur V., Shea P.R., Watkins M.E., Cantu C.C., Laucirica D.R., Jenkins L., Flores A.R., Lovgren M., Ardanuy C., Linares J., Low D.E., Tyrrell G.J., Musser J.M. Full-genome dissection of an epidemic of severe invasive disease caused by a hypervirulent, recently emerged clone of group A Streptococcus. Am J Pathol. 2012;180:1522–1534. doi: 10.1016/j.ajpath.2011.12.037. [DOI] [PubMed] [Google Scholar]
- 24.Fittipaldi N., Olsen R.J., Beres S.B., Van Beneden C., Musser J.M. Genomic analysis of emm59 group A Streptococcus invasive strains, United States. Emerg Infect Dis. 2012;18:650–652. doi: 10.3201/eid1804.111803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyrrell G.J., Lovgren M., St Jean T., Hoang L., Patrick D.M., Horsman G., Van Caeseele P., Sieswerda L.E., McGeer A., Laurence R.A., Bourgault A.M., Low D.E. Epidemic of group A Streptococcus M/emm59 causing invasive disease in Canada. Clin Infect Dis. 2010;51:1290–1297. doi: 10.1086/657068. [DOI] [PubMed] [Google Scholar]
- 26.Hondorp E.R., McIver K.S. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol. 2007;66:1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- 27.Luo F., Lizano S., Banik S., Zhang H., Bessen D.E. Role of Mga in group A streptococcal infection at the skin epithelium. Microb Pathog. 2008;45:217–224. doi: 10.1016/j.micpath.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIver K.S., Heath A.S., Green B.D., Scott J.R. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J Bacteriol. 1995;177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 30.Chaffin D.O., Rubens C.E. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene. 1998;219:91–99. doi: 10.1016/s0378-1119(98)00396-5. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z., Trevino J., Ramirez-Pena E., Sumby P. The small regulatory RNA FasX controls pilus expression and adherence in the human bacterial pathogen group A Streptococcus. Mol Microbiol. 2012;86:140–154. doi: 10.1111/j.1365-2958.2012.08178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll R.K., Shelburne S.A., 3rd, Olsen R.J., Suber B., Sahasrabhojane P., Kumaraswami M., Beres S.B., Shea P.R., Flores A.R., Musser J.M. Naturally occurring single amino acid replacements in a regulatory protein alter streptococcal gene expression and virulence in mice. J Clin Invest. 2011;121:1956–1968. doi: 10.1172/JCI45169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellen R.P., Gibbons R.J. M protein-associated adherence of Streptococcus pyogenes to epithelial surfaces: prerequisite for virulence. Infect Immun. 1972;5:826–830. doi: 10.1128/iai.5.5.826-830.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollingshead S.K., Arnold J., Readdy T.L., Bessen D.E. Molecular evolution of a multigene family in group A streptococci. Mol Biol Evol. 1994;11:208–219. doi: 10.1093/oxfordjournals.molbev.a040103. [DOI] [PubMed] [Google Scholar]
- 35.Courtney H.S., Ofek I., Penfound T., Nizet V., Pence M.A., Kreikemeyer B., Podbielski A., Hasty D.L., Dale J.B. Relationship between expression of the family of M proteins and lipoteichoic acid to hydrophobicity and biofilm formation in Streptococcus pyogenes. PLoS One. 2009;4:e4166. doi: 10.1371/journal.pone.0004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agniswamy J., Lei B., Musser J.M., Sun P.D. Insight of host immune evasion mediated by two variants of group a Streptococcus Mac protein. J Biol Chem. 2004;279:52789–52796. doi: 10.1074/jbc.M410698200. [DOI] [PubMed] [Google Scholar]
- 37.Sumby P., Zhang S., Whitney A.R., Falugi F., Grandi G., Graviss E.A., Deleo F.R., Musser J.M. A chemokine-degrading extracellular protease made by group A Streptococcus alters pathogenesis by enhancing evasion of the innate immune response. Infect Immun. 2008;76:978–985. doi: 10.1128/IAI.01354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen R.J., Kobayashi S.D., Ayeras A.A., Ashraf M., Graves S.F., Ragasa W., Humbird T., Greaver J.L., Cantu C., Swain J.L., Jenkins L., Blasdel T., Cagle P.T., Gardner D.J., DeLeo F.R., Musser J.M. Lack of a major role of Staphylococcus aureus Panton-Valentine leukocidin in lower respiratory tract infection in nonhuman primates. Am J Pathol. 2010;176:1346–1354. doi: 10.2353/ajpath.2010.090960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Virtaneva K., Graham M.R., Porcella S.F., Hoe N.P., Su H., Graviss E.A., Gardner T.J., Allison J.E., Lemon W.J., Bailey J.R., Parnell M.J., Musser J.M. Group A Streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect Immun. 2003;71:2199–2207. doi: 10.1128/IAI.71.4.2199-2207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sumby P., Tart A.H., Musser J.M. A non-human primate model of acute group a Streptococcus pharyngitis. Methods Mol Biol. 2008;431:255–267. doi: 10.1007/978-1-60327-032-8_20. [DOI] [PubMed] [Google Scholar]
- 41.Beres S.B., Carroll R.K., Shea P.R., Sitkiewicz I., Martinez-Gutierrez J.C., Low D.E., McGeer A., Willey B.M., Green K., Tyrrell G.J., Goldman T.D., Feldgarden M., Birren B.W., Fofanov Y., Boos J., Wheaton W.D., Honisch C., Musser J.M. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci U S A. 2010;107:4371–4376. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen R.J., Laucirica D.R., Watkins M.E., Feske M.L., Garcia-Bustillos J.R., Vu C., Cantu C., Shelburne S.A., 3rd, Fittipaldi N., Kumaraswami M., Shea P.R., Flores A.R., Beres S.B., Lovgren M., Tyrrell G.J., Efstratiou A., Low D.E., Van Beneden C.A., Musser J.M. Polymorphisms in regulator of protease B (RopB) alter disease phenotype and strain virulence of serotype M3 group A Streptococcus. J Infect Dis. 2012;205:1719–1729. doi: 10.1093/infdis/jir825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hollands A., Aziz R.K., Kansal R., Kotb M., Nizet V., Walker M.J. A naturally occurring mutation in ropB suppresses SpeB expression and reduces M1T1 group A streptococcal systemic virulence. PLoS One. 2008;3:e4102. doi: 10.1371/journal.pone.0004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horstmann N., Sahasrabhojane P., Suber B., Kumaraswami M., Olsen R.J., Flores A., Musser J.M., Brennan R.G., Shelburne S.A., 3rd Distinct single amino acid replacements in the control of virulence regulator protein differentially impact streptococcal pathogenesis. PLoS Pathog. 2011;7:e1002311. doi: 10.1371/journal.ppat.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lei B., Liu M., Chesney G.L., Musser J.M. Identification of new candidate vaccine antigens made by Streptococcus pyogenes: purification and characterization of 16 putative extracellular lipoproteins. J Infect Dis. 2004;189:79–89. doi: 10.1086/380491. [DOI] [PubMed] [Google Scholar]
- 46.Musser J.M., Shelburne S.A., 3rd A decade of molecular pathogenomic analysis of group A Streptococcus. J Clin Invest. 2009;119:2455–2463. doi: 10.1172/JCI38095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanderson-Smith M., De Oliveira D.M., Guglielmini J., McMillan D.J., Vu T., Holien J.K., Henningham A., Steer A.C., Bessen D.E., Dale J.B., Curtis N., Beall B.W., Walker M.J., Parker M.W., Carapetis J.R., Van Melderen L., Sriprakash K.S., Smeesters P.R., M Protein Study Group A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis. 2014;210:1325–1338. doi: 10.1093/infdis/jiu260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hondorp E.R., Hou S.C., Hause L.L., Gera K., Lee C.E., McIver K.S. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A streptococcus. Mol Microbiol. 2013;88:1176–1193. doi: 10.1111/mmi.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guermazi A., Roemer F.W., Haugen I.K., Crema M.D., Hayashi D. MRI-based semiquantitative scoring of joint pathology in osteoarthritis. Nat Rev Rheumatol. 2013;9:236–251. doi: 10.1038/nrrheum.2012.223. [DOI] [PubMed] [Google Scholar]
- 51.Kauppinen R.A., Peet A.C. Using magnetic resonance imaging and spectroscopy in cancer diagnostics and monitoring: preclinical and clinical approaches. Cancer Biol Ther. 2011;12:665–679. doi: 10.4161/cbt.12.8.18137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung C.B., Boucher R., Resnick D. MR imaging of synovial disorders of the knee. Semin Musculoskelet Radiol. 2009;13:303–325. doi: 10.1055/s-0029-1242186. [DOI] [PubMed] [Google Scholar]
- 53.Turecki M.B., Taljanovic M.S., Stubbs A.Y., Graham A.R., Holden D.A., Hunter T.B., Rogers L.F. Imaging of musculoskeletal soft tissue infections. Skeletal Radiol. 2010;39:957–971. doi: 10.1007/s00256-009-0780-0. [DOI] [PubMed] [Google Scholar]
- 54.Nikolaou M., Zampakis P., Vervita V., Almaloglou K., Adonakis G., Marangos M., Decavalas G. Necrotizing fasciitis complicating pregnancy: a case report and literature review. Case Rep Obstet Gynecol. 2014;2014:505410. doi: 10.1155/2014/505410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saiag P., Le Breton C., Pavlovic M., Fouchard N., Delzant G., Bigot J.M. Magnetic resonance imaging in adults presenting with severe acute infectious cellulitis. Arch Dermatol. 1994;130:1150–1158. [PubMed] [Google Scholar]
- 56.Rahmouni A., Chosidow O., Mathieu D., Gueorguieva E., Jazaerli N., Radier C., Faivre J.M., Roujeau J.C., Vasile N. MR imaging in acute infectious cellulitis. Radiology. 1994;192:493–496. doi: 10.1148/radiology.192.2.8029421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cloning scheme to generate isogenic Mga-deficient (Δmga), wild-type, and H201R strains. A: Molecular genetic strategy for inactivating mga in strain MGAS15249. The mga was replaced with a gene conferring spectinomycin resistance. B: pDC123 with no insert was used to generate strain designated as Δmga. C: The mga was cloned into pdDC123 and used in trans to generate strain designated as wild-type. D: Quick-change site-directed mutagenesis was used to introduce the H201R amino acid replacement to generate strain H201R. E. coli, Escherichia coli.
A: Growth curves of isogenic group A Streptococcus (GAS) strains Δmga, wild-type, and H201R. B: Relative mga copies per GAS cell were measured using quantitative real-time PCR assay.