Abstract
We investigated whether expression of xylosyltransferase-1 (XT-1), a key enzyme in glycosaminoglycan biosynthesis, is responsive to disk degeneration and to inhibition by the inflammatory cytokines tumor necrosis factor α and IL-1β in nucleus pulposus (NP) cells. Analysis of human NP tissues showed that XT-1 expression is unaffected by degeneration severity; XT-1 and Jun, Fos, and Sp1 mRNA were positively correlated. Cytokines failed to inhibit XT-1 promoter activity and expression. However, cytokines decreased activity of XT-1 promoters containing deletion and mutation of the –730/–723 bp AP-1 motif, prompting us to investigate the role of AP-1 and Sp1/Sp3 in the regulation of XT-1 in healthy NP cells. Overexpression and suppression of AP-1 modulated XT-1 promoter activity. Likewise, treatment with the Sp1 inhibitors WP631 and mithramycin A or cotransfection with the plasmid DN-Sp1 decreased XT-1 promoter activity. Inhibitors of AP-1 and Sp1 and stable knockdown of Sp1 and Sp3 resulted in decreased XT-1 expression in NP cells. Genomic chromatin immunoprecipitation analysis showed AP-1 binding to motifs located at –730/–723 bp and –684/–677 bp and Sp1 binding to –227/–217 bp and –124/–114 bp in XT-1 promoter. These results suggest that XT-1 expression is refractory to the disease process and to inhibition by inflammatory cytokines and that signaling through AP-1, Sp1, and Sp3 is important in the maintenance of XT-1 levels in NP cells.
Intervertebral disk degeneration is recognized as the most common cause of low-back pain,1, 2 experienced by almost 80% of the population at least once during their lifetime. The intervertebral disk is composed of gelatinous nucleus pulposus (NP) enclosed circumferentially by fibrocartilaginous annulus fibrosus (AF); the whole structure is contained superiorly and inferiorly by a layer of hyaline cartilage, the endplate. NP and AF are rich in extracellular matrix. Particularly, NP is abundant in large aggregating proteoglycans, such as aggrecan and versican, that provide the osmotic properties of the disk critical to withstanding the high compressive loads applied to the spine. Proteoglycans are composed of a core protein that is covalently attached by a variety of glycosaminoglycans (GAGs), such as chondroitin sulfate and keratan sulfate. GAGs are of critical importance in the regulation of NP cell function and organ development.3, 4, 5 GAG chain biosynthesis is initiated by the formation of a tetrasaccharide (D-GlcA-β1,3-Gal-β1,3-Gal-β1,4-Xyl-Ser) formed by the stepwise addition of individual sugar residues; the first critical step of adding xylose to serine residue of the core protein is catalyzed by xylosyltransferase-1 (XYLT1; alias XT-1) and, thus, serves as an important rate-limiting step in GAG biosynthesis.6, 7, 8
A hallmark of intervertebral disk degeneration is the marked increase in proinflammatory cytokines, mainly tumor necrosis factor α (TNF-α) and IL-1β, which suppress proteoglycan biosynthesis and promote the expression of many catabolic enzymes.9, 10, 11 The resultant decrease in proteoglycan matrix and change in matrix composition leads to gradual loss of tissue water, compromising its biomechanical function.12 Although there is some evidence to suggest that in NP cells the disease process may render the expression of GlcAT-I, one of the critical enzymes in GAG biosynthesis, less responsive to niche factors, such as transforming growth factor β, whether XT-1 expression is similarly influenced is not known. Note that altered XT-1 expression levels and, thus, changes in the biosynthesis of heparan and chondroitin sulfate have been shown to occur in diseases such as osteoarthritis, dilated cardiomyopathy, and skin fibrosis.8, 13, 14 In chondrocytes, XT-1 expression is suppressed by IL-1β, with Sp1 and Sp3 transcription factors exerting opposing effects on transcription.15, 16 However, in the case of NP cells, information on transcriptional control of XT-1 is completely lacking.
We, therefore, sought to investigate the effect of disease severity and the inflammatory cytokines TNF-α and IL-1β on XT-1 expression and to understand regulation of its expression in NP cells. These data showed that XT-1 expression in NP remained unaffected with increasing disease severity in humans; its expression positively correlated with Jun, Fos, and Sp1 levels. Unique to NP cells, XT-1 expression was refractory to the actions of TNF-α and IL-1β,15 and AP-1 and Sp1/Sp3 positively controlled its transcription.
Materials and Methods
Reagents and Plasmids
XT-1 promoter reporter luciferase constructs (−1638/+1, −797/+1, AP-1 site: −730/−723 mutation in −797/+1 and AP1Δ on −797/+1) in pGL4.10 vector were a gift from Dr. Christian Götting (University of Bochum, Bochum, Germany).17 The plasmids were gifts, as follows: A-os/DN-AP1 from Dr. Charles Vinson (NIH, Bethesda, MD) and DN-Sp1 from Dr. Gerald Thiel (University of Saarland Medical Center, Homburg, Germany). The following were obtained from Addgene (Cambridge, MA): Jun (catalog #47443) and pcDNA3-FLAG-Fos (#8966), developed by John Blenis; pN3-Sp1FL (#24543), pN3-Sp3FL (#24541), and pN3-control (#24544), by Guntram Suske; and psPAX2 (#12260) and pMD2G (#12259), by Dr. Didier Trono. Lentiviral shSp1 (clone TRCN0000020448) and shSp3 (clones TRCN0000280368 and TRCN0000280370) were purchased from Sigma-Aldrich (St. Louis, MO). The vector pRL-TK (Promega Corp., Madison, WI) containing the Renilla luciferase gene was used as an internal transfection control.17 The inhibitors WP631, mithramycin A, and tanshinone IIA were from Sigma-Aldrich. Commercially available antibodies against Sp1, Sp3, and Jun (Cell Signaling Technology Inc., Danvers, MA); XT-1 (Abcam Inc., Cambridge, MA); and glyceraldehyde-3-phosphate dehydrogenase (Novus Biologicals LLC., Littleton, CO) were used. β-Tubulin antibody was obtained from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). TNF-α and IL-1β were purchased from PeproTech (Rocky Hill, NJ). All the PCR primers were synthesized by Integrated DNA Technologies (Coralville, IA).
Human NP Tissue Collection and Grading
Twenty-five lumbar and thoracic degenerated disk tissue samples were obtained from patients undergoing elective spinal surgical procedures (Pfirrmann grade 3: n = 6, grade 4: n = 16, grade 5: n = 3). Consistent with Thomas Jefferson University's Institutional Review Board guidelines, informed consent for sample collection was obtained from each patient. The Pfirrmann grading scheme was used to assess the disease state.18
Isolation of NP Cells and Cytokine Treatments
Rat and human NP cells were isolated as described by Risbud et al.17 For isolation of human NP cells, only visually healthy, grade 1 or 2 tissues obtained from autopsy or spinal surgery were used. After isolation, cells were maintained in Dulbecco's modified Eagle's medium and 10% fetal bovine serum supplemented with ampicillin and streptomycin. To investigate the effect of cytokines and inhibitors, NP cells were cultured in serum-free medium overnight and then were treated with 50 ng/mL TNF-α, 10 ng/mL IL-1β, 50 and 100 μmol/L WP631, 100 and 1000 nmol/L mithramycin A, and 50 and 100 μmol/L tanshinone IIA for 24 hours.
Quantitative Real-Time PCR Analysis
Total RNA was extracted from cells using an RNase-free DNase I set and a MinElute cleanup kit (both from Qiagen, Valencia, CA) according to the manufacturer's instructions. Two micrograms of total DNA-free RNA was used to synthesize cDNA using an EcoDry double-primed cDNA synthesis kit (Clontech Laboratories, Mountain View, CA). PCR reactions were set up in duplicate using 1 μL of cDNA with Power SYBR Green master mix (Applied Biosystems, Foster City, CA) and gene-specific forward and reverse PCR primers and were performed using a StepOnePlus real-time PCR system (Applied Biosystems). Hypoxanthine phosphoribosyltransferase 1 or glyceraldehyde 3-phosphate dehydrogenase was used to normalize the mRNA expression in rat samples. β-Actin was used to normalize mRNA expression in human samples. After normalization, expression data were presented relative to the corresponding control group.
Immunohistochemical Analysis of Disk Tissue
Paraffin-embedded sections of rat intervertebral disk tissue were deparaffinized and then rehydrated in graded ethanol. Antigen retrieval was performed by boiling in 0.01 mol/L sodium citrate buffer (pH 6.0) at approximately 100°C for 15 minutes. After blocking with 10% goat serum for 1 hour, sections were incubated with anti–XT-1 antibody at a dilution of 1:100 overnight at 4°C and then were incubated with secondary antibody (Alexa Fluor 488–conjugated anti-rabbit; Invitrogen, Carlsbad, CA) for 1 hour at room temperature. Finally, sections were mounted using DAPI-containing mounting solution (Vector Laboratories, Burlingame, CA). Cells were imaged using a fluorescence microscope (Nikon TU200; Nikon Corp, Tokyo, Japan). Sections incubated with isotype antibody or with secondary antibody alone served as negative controls.
Protein Extraction and Western Blot Analysis
After treatments, cells were placed on ice immediately and washed with ice-cold PBS containing 1× complete protease inhibitor cocktail (Roche Diagnostics Corp., Indianapolis, IN). The extraction buffer M-PER (Pierce Biotechnology, Rockford, IL) included 1× protease inhibitor cocktail, 150 mmol/L NaCl, 4 mmol/L NaF, and 20 mmol/L Na3VO4. Proteins were separated under reducing conditions on 10% to 12% polyacrylamide gels and then were transferred by electroblotting to polyvinylidene difluoride membranes (Bio-Rad Laboratories, Hercules, CA). Membranes were incubated overnight at 4°C with the antibodies at a dilution of 1:1000 (XT-1, Sp1, and Sp3) and 1:3000 (glyceraldehyde 3-phosphate dehydrogenase and β-tubulin) after blocking for 1 hour in 5% nonfat dry milk containing Tris-buffered saline and Tween 20 [50 mmol/L Tris (pH 7.6), 150 mmol/L NaCl, and 0.1% Tween 20]. Enhanced chemiluminescence reagent (GE Healthcare, Chalfont St. Giles, UK) was used to detect the immunolabeling.
Transfections and Dual-Luciferase Reporter Assay
NP cells were seeded in 48-well plates (4 × 104 cells per well) with 2% Opti-MEM medium (Invitrogen), and the following day, cells were cotransfected with 50 to 200 ng of Jun and/or Fos or DN-AP-1 or 5 to 200 ng of Sp1, Sp3, or DN-Sp1 plasmids with or without appropriate backbone vectors and 150 ng of XT-1 reporter and 150 ng of pRL-TK plasmid. For all transfection, plasmids were premixed with the transfection reagent Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, the cells were harvested, and firefly and Renilla luciferase activities were measured by a Dual-Luciferase reporter assay (Promega Corp.) using a luminometer (TD-20/20; Turner Designs Inc. Sunnyvale, CA). In addition, to determine the effect of TNF-α, IL-1β, and inhibitors, the cells were treated with 50 ng/mL TNF-α or 10 ng/mL IL-1β, 50 to 100 μmol/L WP631, 100 to 1000 nmol/L mithramycin A, and 50 to 100 μmol/L tanshinone IIA for 24 hours. All the luciferase assays were performed in triplicate, and every experiment was repeated at least three times.
Lentiviral Particle Production and Viral Transduction
HEK 293T cells were seeded in 10-cm plates (3 × 106 cells per plate) in Dulbecco's modified Eagle's medium with 10% heat-inactivated fetal bovine serum. The following day, cells were transfected with 9 μg of shRNA control or gene-specific shRNA plasmids, along with 6 μg of psPAX2 and 3 μg of pMD2.G. Sixteen hours later, the transfection medium was replaced with complete Dulbecco's modified Eagle's medium. Media containing viral particles were harvested 48 and 60 hours after transfection and were introduced into human NP cell cultures (0.5 × 106 cells per plate) along with 6 μg/mL Polybrene (Abbott Laboratories, Abbott Park, IL). The medium containing viral particles was removed after 24 hours and replaced with Dulbecco's modified Eagle's medium with 10% fetal bovine serum and antibiotics. Five days later, cells were harvested for protein or RNA extraction.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assay was performed using a ChIP-IT high-sensitivity kit (Active Motif, Carlsbad, CA). Briefly, human NP cells were cultured in 100 mm until 80% confluence. Cells were cross-linked and lyzed, and chromatin was sheared by sonication, and input DNA was generated by treating aliquots with RNase, proteinase K, and heat, followed by ethanol precipitation. DNA complexes were immunoprecipitated by incubation with rabbit polyclonal anti-Jun or anti-Sp1 antibodies or preimmune IgG overnight as negative control at 4°C followed by binding to protein G–agarose beads for 3 hours at 4°C. Cross-links were reversed by treatment with proteinase K and heat for 2.5 hours, and DNA was purified using DNA purification elution buffer (Active Motif). Real-time PCR analysis was performed using a ChIP-IT quantitative PCR analysis kit (Active Motif) and the following primer pairs (forward and reverse) for putative AP-1 and Sp1 sites in the XT-1 promoter: −730/−723 bp AP-1: 5′-TTGCCGCTATACCACTT-3′ and 5′-GTGCCAACGATGTACTAAG-3′; −684/−677 bp AP-1: 5′-CACTTAGTACATCGTTGGC-3′ and 5′-GCTTTGGGTGAGTTTCTG-3′; and −228/−224 bp and −124/−114 bp Sp1: 5′-CCTTCCTTTCCCTCCTCCTTT-3′ and 5′-CCGCCACCATCTTCGGA-3′. The negative control primers and standard curve primers used were provided with the kit. Real-time PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems). The CT values were recorded, and the data were normalized based on primer efficiency, input DNA CT values, amount of chromatin, and resuspension volume, based on the manufacturer's recommendations.
Statistical Analysis
All the experiments were performed independently at least three times. Data are presented as means ± SEM. Differences between groups were analyzed by one-way analysis of variance and Student's t-test. RT-PCR expression data of human tissues was found to be nonparametric; therefore, the Kruskal-Wallis test was used to determine significance between 2−ΔCT values between study groups. In addition, the Spearman rank correlation was deployed to determine correlations between gene expressions for different targets. P < 0.05 was considered statistically significant.
Results
XT-1 mRNA Expression Is Unaffected by Severity of Disk Disease and Shows Correlation with Jun, Fos, and Sp1 in Human NP Tissues
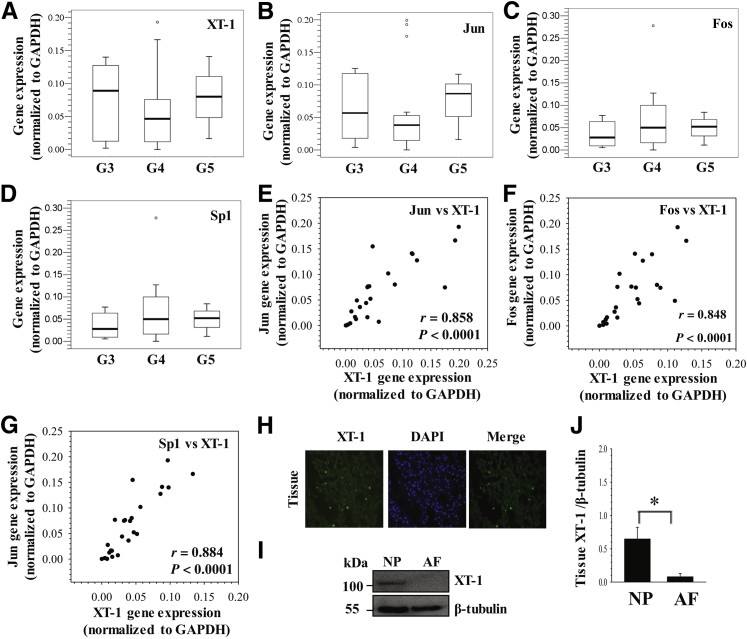
To investigate the relationship between XT-1 expression and severity of disk degeneration, we measured mRNA expression using 25 human NP tissues with increasing grades of disease. The results show that there was no significant difference in expression levels of XT-1 (Figure 1A) between NP tissues with early (grade 3) and late (grades 4 and 5) stages of degeneration. In addition, we measured mRNA expression of Jun, Fos, and Sp1, transcription factors that have been associated with XT-1 expression in other cell types.7, 13 Similar to XT-1, expression of Jun (Figure 1B), Fos (Figure 1C), and Sp1 (Figure 1D) was similar between tissues with different grades. Expression of Jun and XT-1 (Figure 1E) (r = 0.858, P < 0.0001), Fos and XT-1 (Figure 1F) (r = 0.848, P < 0.0001), and Sp1 and XT-1 (Figure 1G) (r = 0.884, P < 0.0001) significantly correlated with each other.
Figure 1.
Relationship of xylosyltransferase-1 (XT-1) mRNA expression to severity of human disk disease and Jun, Fos, and Sp1 levels. A–D: mRNA expression analysis of XT-1 (A), Jun (B), Fos (C), and Sp1 (D) from nucleus pulposus (NP) tissues with different Pfirrmann grades (G3 to G5). No significant differences were observed in any of these genes between different grade samples (N = 25; G3: n = 6, G4: n = 16, G5: n = 3). The data are represented as box and whisker plots. Open circles denote outliers that fall outside of 1.5 times the upper or lower quartile range. Each box represents the 75th–25th percentile of values, the line inside each box represents the median value separating the upper and lower quartiles, and whiskers show the maximum and minimum values excluding outliers in each set. E–G: A positive correlation in mRNA levels between Jun and XT-1 (r = 0.858, P < 0.0001) (E), Fos and XT-1 (r = 0.848, P < 0.0001) (F), and Sp1 and XT-1 (r = 0.884, P < 0.0001) (G) was observed. H: Section of the rat NP tissue treated with XT-1 antibody showed prominent cytoplasmic localization of protein in NP cells. DAPI was used to stain cell nuclei. I and J: Western blot (I) and corresponding densitometric (J) analyses of XT-1 expression in rat NP and annulus fibrosus (AF) tissues showed prominent expression in NP compared with AF tissue. Data represent means ± SEM of three independent experiments performed in triplicate. ∗P < 0.05. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To investigate the expression of XT-1 in healthy disk tissues, sagittal sections of rat disk were stained with an antibody to XT-1 to examine the expression pattern. XT-1 was prominently expressed in NP cells (Figure 1H). In addition, XT-1 expression in discal tissues of mature rat was studied using Western blot analysis. Results show that the expression level of XT-1 protein in NP tissue is significantly higher than that in the AF (Figure 1, I and J).
XT-1 Expression Is Refractory to Inhibition by TNF-α and IL-1β and Is Maintained by AP-1
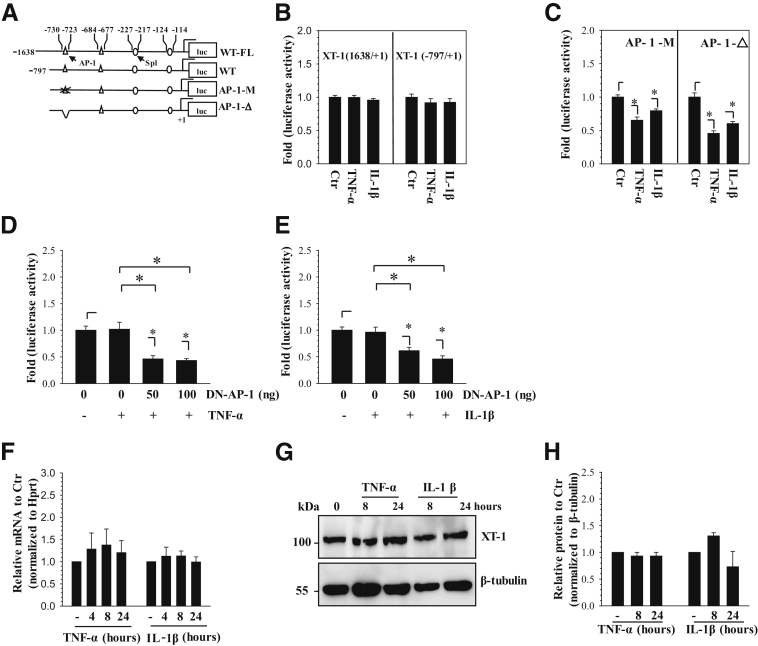
TNF-α and IL-1β are the key inflammatory cytokines that promote intervertebral disk degeneration, and IL-1β is shown to suppress XT-1 levels in chondrocytes.9, 10, 11, 15 To delineate the effect of these cytokines on XT-1 expression, we first measured the activity of two promoter fragments, a larger −1638/+1 bp and a smaller −797/+1 bp (Figure 2A) in rat NP cells after treatment with TNF-α and IL-1β. There are two conserved, functional AP-1 binding sites at −730/−723 bp and −684/−677 bp and two conserved, functional Sp1 binding sites at −227/−217 bp and −124/−114 bp within −797 bp of the human XT-1 promoter (Figure 2A).16 The activity of both the fragments was unresponsive to cytokine treatment (Figure 2B). Next, to determine whether AP-1 maintained XT-1 promoter activity in the presence of cytokines, we used reporters harboring AP-1 site mutation or deletion. Results show that unlike wild-type reporters, TNF-α and IL-1β significantly decreased the activity of these AP-1–deficient constructs (Figure 2, B and C). To confirm the contribution of AP-1 to sustain XT-1 transcription, we cotransfected rat NP cells with DN-AP-1 plasmid and measured the activity of wild-type XT-1 reporter with or without cytokines. Again, when AP-1 function was blocked, TNF-α (Figure 2D) or IL-1β (Figure 2E) significantly suppressed the promoter activity. Furthermore, in agreement with our promoter studies and in contrast to a previous report of chondrocytes,15 the cytokines did not affect XT-1 mRNA (Figure 2F). Western blot and subsequent densitometric analyses also showed a lack of suppressive effect of cytokines on XT-1 protein levels (Figure 2, G and H).
Figure 2.
Xylosyltransferase-1 (XT-1) expression in nucleus pulposus (NP) cells is refractory to inhibition by tumor necrosis factor α (TNF-α) and IL-1β and is maintained through AP-1 signaling. A: Schematic of −1638/+1 bp and −797/+1 bp human XT-1 promoter constructs showing AP-1 and Sp1 binding sites used for the studies. B and C: Rat NP cells transfected with both wild-type XT-1 promoter constructs showed no change in promoter activity in response to TNF-α and IL-1β treatment (B). NP cells transfected with the −797/+1 bp construct harboring either mutation (B) or deletion (C) of the −730/−723 bp AP-1 site showed a decrease in activity in response to cytokines. D and E: NP cells were cotransfected with DN-AP-1 and the −797/+1 bp XT-1 promoter construct and were treated with either TNF-α (D) or IL-1β (E). The XT-1 promoter showed no change in activity in response to cytokines alone; however, activity declined after cytokine treatment in the presence of DN-AP-1. F–H: Quantitative PCR (F) and Western blot (G) and corresponding densitometric (H) analyses of NP cells treated with TNF-α or IL-1β until 24 hours. Cytokine treatment showed no significant change in XT-1 mRNA (F) and protein (G and H) levels. Data represent means ± SEM of three independent experiments performed in triplicate. ∗P < 0.05. Ctr, control; luc, luciferase; WT, wild type; WT-FL, wild type-full length.
AP-1 Signaling Pathway Controls Basal XT-1 Transcription in NP Cells
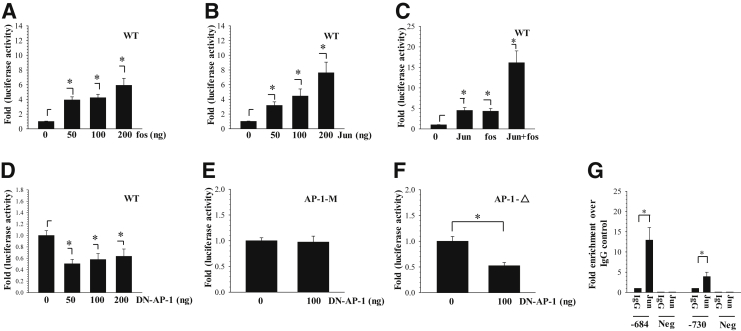
To determine whether AP-1 controls XT-1 promoter activity, we performed classical gain- and loss-of-function experiments by cotransfecting −797/+1 bp XT-1 promoter construct with Jun and/or Fos and DN-AP1 plasmids. Results show that increasing concentrations of Fos (Figure 3A) as well as Jun (Figure 3B) robustly enhanced promoter activity in a dose-dependent manner. Moreover, co-expression of Jun and Fos produced much stronger induction of promoter activity (Figure 3C). On the other hand, cotransfection with DN-AP-1 resulted in strong suppression of promoter activity; even at a dose of 50 ng, a suppressive effect was evident (Figure 3D). However, DN-AP-1 evidenced no effect on XT-1 promoter harboring mutation in the AP-1 motif located at −730/−723 bp (Figure 3E). DN-AP-1 suppressed the activity of XT-1 promoter with deletion of the AP1 binding site at −730/−723 bp (Figure 3F). These results indicated that the AP-1 (Jun/Fos) controls human XT-1 promoter function. To delineate whether AP-1 binds to the XT-1 gene promoter, we assessed its binding to cognate AP-1 motifs by ChIP analysis. There was approximately 13-fold enrichment in binding of Jun to a site located at −684/−677 bp compared with the IgG control, whereas fourfold enrichment of Jun binding was detected at a motif located at −730/−723 bp of the human XT-1 promoter (Figure 3G). As expected, negative control primers showed almost undetectable amplification.
Figure 3.
AP-1 signaling controls basal XT-1 promoter activity in nucleus pulposus (NP) cells. A: Effect of Fos overexpression on XT-1-wild-type (WT) promoter activity. Note the dose-dependent increase in XT-1 reporter activity with increasing concentrations of Fos. B: Jun overexpression induced XT-1 promoter activity in a dose-dependent manner. C: Cotransfection of Jun and Fos significantly increased the XT-1 promoter activity compared with their individual addition. D: DN-AP-1 diminished the activity of XT-1 promoter in a dose-dependent manner. E: DN-AP-1 has no effect on XT-1 reporter activity harboring a mutation in −730/−723 bp AP-1 motif. F: DN-AP-1 suppresses activity of the XT-1 promoter with deletion of −730/−723 bp AP-1 motif. G: Genomic chromatin immunoprecipitation analysis to detect AP-1 binding to XT-1 promoter in human NP cells. Pull down using Jun antibody clearly showed significant enrichment of binding to AP-1 motifs at −730/−723 bp and −684/−677 bp over IgG alone. Negative primers (Neg) showed little or no amplification for experimental and IgG control samples. Values shown are means ± SEM of three independent experiments. ∗P < 0.05.
Sp1/Sp3 Transcription Factor Controls XT-1 Promoter Activity in NP Cells
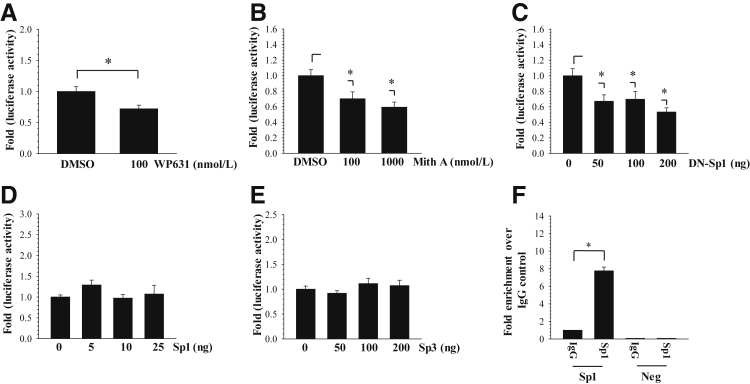
To further examine whether the Sp1 and Sp3 transcription factors control XT-1 promoter activity, we measured the activity of −797/+1 XT-1 promoter first in the presence of the Sp1 inhibitor WP631 or mithramycin A, which through binding to GC-rich DNA motifs displaces bound Sp1 from promoters and serves as its transcriptional inhibitor. The results showed that WP631 (Figure 4A) and mithramycin A (Figure 4B) significantly suppressed the activity of XT-1 promoter in rat NP cells. In parallel, we cotransfected NP cells with DN-Sp1 to assess the effect of lowered Sp1 activity on XT-1 promoter. It was evident that the addition of DN-Sp1 significantly reduced the basal activity of XT-1 promoter (Figure 4C). To complement these loss-of-function experiments, we performed gain-of-function assays by cotransfecting XT-1 promoter along with full-length Sp1 and Sp3. The results showed that exogenously overexpressed Sp1 (Figure 4D) or Sp3 (Figure 4E) did not increase the activity of XT-1 promoter in NP cells. To determine whether Sp1 binds to its cognate sites in XT-1 promoter, we performed ChIP analysis using human NP cells. When Sp1 antibody was used for the chromatin pull down, approximately eightfold enrichment in amplicon encompassing both −227/−217 bp and −124/−114 bp binding sites was detected over the IgG control (Figure 4F).
Figure 4.
Sp1 controls XT-1 promoter activity in nucleus pulposus (NP) cells. A and B: Effect of Sp1 inhibitors WP631 (A) and mithramycin A (Mith A) (B) on XT-1 promoter activity in rat NP cells. Both inhibitors significantly suppressed basal XT-1 promoter activity. C: Transfection of rat NP cells with increasing doses of DN-Sp1 suppressed XT-1 promoter. D and E: Overexpression of Sp1 (D) or Sp3 (E) did not influence the activity of XT-1 promoter in NP cells. F: Genomic chromatin immunoprecipitation analysis to detect Sp1 binding to XT-1 promoter in human NP cells. Pull down using Sp1 antibody showed significant enrichment of binding to Sp1 motifs at −227/−217 bp and −124/−114 bp over IgG alone. Negative primers (Neg) showed little or no amplification for experimental and IgG control samples. Values shown are means ± SEM of three independent experiments. ∗P < 0.05. DMSO, dimethyl sulfoxide.
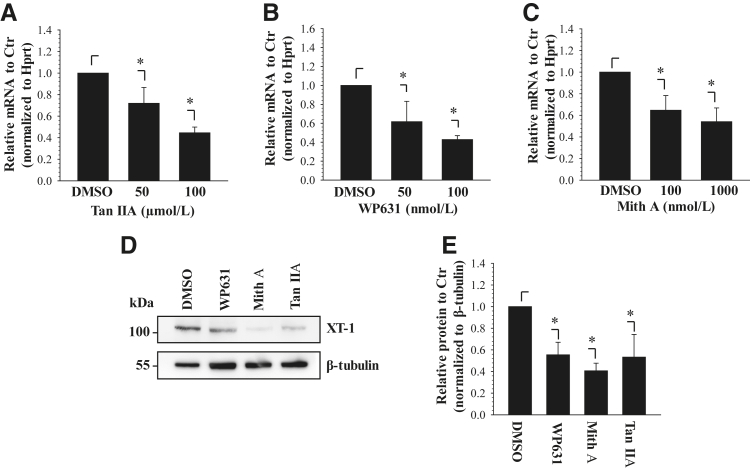
Treatment with AP-1 and Sp1 Inhibitors Suppresses XT-1 Expression in NP Cells
After the promoter studies, to investigate the effects of AP-1 inhibition on XT-1 expression we treated rat NP cells with tanshinone IIA, an AP-1 inhibitor, for 24 hours and measured mRNA levels. Treatment with 50 and 100 μmol/L tanshinone IIA decreased XT-1 mRNA expression to 72% and 45%, respectively, of untreated control (Figure 5A). To further test the inhibitory role of Sp1 inhibition on the XT-1 mRNA expression, NP cells were treated with WP631 or mithramycin A for 24 hours. The results show that treatment with WP631 resulted in a dose-dependent decrease in XT-1 mRNA expression (Figure 5B). Similarly, with increasing concentrations of mithramycin A, XT-1 mRNA levels were decreased to 65% and 54%, respectively (Figure 5C). To determine whether the inhibition of AP-1 and Sp1 affects XT-1 protein levels, we performed Western blot analysis after the treatment of NP cells with 100 μmol/L WP631, 1 μmol/L mithramycin A, or 100 μmol/L tanshinone IIA for 24 hours. The results show that compared with nontreated cells, the inhibitor treatment resulted in a significant decrease in XT-1 protein levels (Figure 5, D and E).
Figure 5.
AP-1 and Sp1 inhibitors suppress xylosyltransferase-1 (XT-1) expression in nucleus pulposus (NP) cells. A–C: Effect of AP-1 inhibitor tanshinone IIA (Tan IIA) (A) and Sp1 inhibitors WP631 (B) and mithramycin A (Mith A) (C) on XT-1 mRNA expression in rat NP cells. Real-time RT-PCR analysis showed that the inhibitors significantly suppress XT-1 mRNA in a dose-dependent manner. D: Western blot analysis of rat NP cells treated with the inhibitors WP631, Mith A, and Tan IIA. XT-1 protein levels decreased with inhibitor treatment. E: Multiple blots shown in experiment D were quantified by densitometric analysis. β-Tubulin was used as a loading control and to calculate relative expression levels to untreated control (Ctr). Values shown are means ± SEM of three independent experiments. ∗P < 0.05. DMSO, dimethyl sulfoxide.
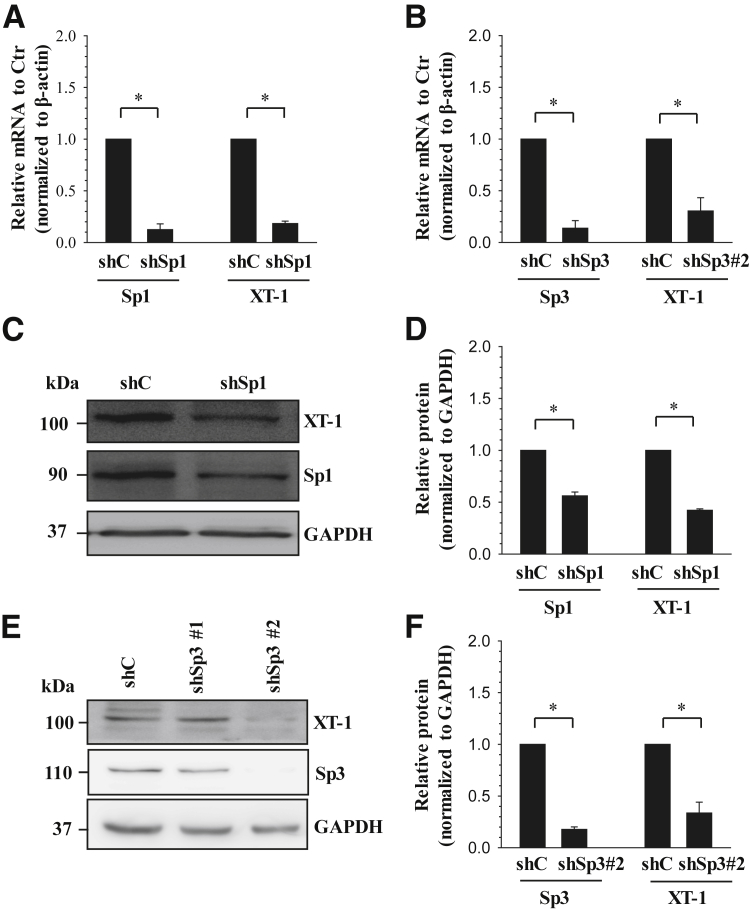
Stable Silencing of Sp1 and Sp3 Decreases XT-1 Expression
To further delineate the novel complementary role of Sp1 and Sp3 in controlling XT-1 expression in NP cells, we performed Sp1 and Sp3 loss-of-function studies using lentiviral delivery of gene-targeting shRNAs. As expected, in human NP cells transduced with shSp1 or shSp3#2, the mRNA expression of Sp1 or Sp3 significantly decreased by approximately 88% and 86%, respectively, compared with cells transduced with control shRNA. One of two shRNAs targeting Sp3 (shSp3#1) did not result in the suppression of Sp3 levels. In Sp1- and Sp3-supressed cells, XT-1 mRNA expression is markedly reduced by approximately 82% (Figure 6A) and 70% (Figure 6B), respectively, of the control cells. Furthermore, we investigated the expression of XT-1 protein in the Sp1- and Sp3-silenced cells using Western blot analysis. Compared with the NP cells transduced with control shRNA, shSp1 or shSp3 resulted in a significant decrease in Sp1 (Figure 6, C and D) or Sp3 (Figure 6, E and F) protein. Consistent with the changes in mRNA, XT-1 protein expression in Sp1- (Figure 6C) or Sp3- (Figure 6E) silenced cells was decreased by approximately 70% (Figure 6D) and 66% (Figure 6F), respectively, of the control cells.
Figure 6.
Stable silencing of Sp1 or Sp3 using lentivirally delivered shRNAs decreases xylosyltransferase-1 (XT-1) expression in human nucleus pulposus (NP) cells. A and B: Real-time PCR analysis of human NP cells transfected with LV-sh-control (LV-shC) or LV-shSp1 (A) or LV-shSp3#2 (B). XT-1 mRNA expression was significantly suppressed by LV-shSp1 and LV-shSp3#2 compared with cells transduced with control lentivirus (LV-shC). Strong suppression in Sp1 and Sp3 mRNA expression was observed with corresponding gene-targeting shRNAs. C–F: Western blot (C and E) and corresponding densitometric (D and F) analyses of NP cells transduced with LV-shC or LV-shSp1 (C and D) or LV-shSp3 (E and F). XT-1 expression levels were suppressed by LV-shSp1 and LV-shSp3#2 compared with cells transduced with control shRNA. Targeting sequence shSp3#1 did not result in the suppression of Sp3 levels. As expected, transduction of cells with shSp1 or shSp3#2 showed a corresponding decrease in Sp1 or Sp3 protein levels. Data represent means ± SEM of three independent experiments performed in triplicate. ∗P < 0.05.
Discussion
The experiments described in this investigation demonstrate for the first time that the expression of XT-1, a key enzyme that catalyzes the initial step in GAG biosynthesis, is refractory to actions of the inflammatory cytokines TNF-α and IL-1β and that XT-1 expression is similar between the early and late stages of the degenerative process. A second major observation was that Jun/AP-1 and complementary actions of members of the Sp transcription factor family, Sp1 and Sp3, are essential for the maintenance of XT-1 expression in NP cells through interaction with respective cognate binding sites in XT-1 promoter. Jun, Fos, and Sp1 expression positively correlated with XT-1 expression in human NP tissues. Taken together, these findings, for the first time, present novel insights into the expression and regulation of XT-1 in the intervertebral disk and highlight clear differences compared with other cartilaginous tissues.
Biomechanical function of the intervertebral disk is directly related to its ability to bind and retain water in the aggrecan-rich matrix of the NP; therefore, understanding the mechanisms that control GAG synthesis in NP cells is of paramount importance. Note that increased proteoglycan catabolism and reduced overall aggrecan and GAG content are hallmarks of intervertebral disk degeneration.19 Because XT-1 represents the first and most important rate-limiting step in GAG biosynthesis, we investigated whether the expression was responsive to the disease process. To our surprise, and in contrast to osteoarthritis, the lack of difference in XT-1 levels in NP tissues with early and late grades of degeneration suggested that expression was refractory to disease progression.13 Because AP1 and Sp1/Sp3 have been shown to control XT-1 expression in other cell types,8, 15 we also measured their expression levels in human NP tissues. AP-1 components Fos and Jun are reported to be expressed in extrusions, sequesters, and protrusions of human intervertebral disks.20 In agreement, the present studies confirmed that Jun, Fos, and Sp1 are expressed in degenerate human NP tissues. Although no apparent difference in expression of these transcription factors was observed, a strong positive correlation between XT-1 and Jun or Fos as well as Sp1 suggested that they are likely involved in maintaining a steady state of XT-1 expression in NP cells during disease. Moreover, robust expression of XT-1 in healthy NP tissue compared with AF supported the hypothesis that GAG biosynthesis is likely to be more active in mature native NP tissue than in AF.
Tissue analysis raised an important question: How do inflammatory cytokines affect XT-1 expression in NP cells? TNF-α and IL-1β are the key inflammatory cytokines associated with degenerative disk disease; our recent results also showed that they promote matrix degradation through the expression and activation of many catabolic enzymes.9, 11, 21 Moreover, IL-1β is shown to affect GAG synthesis by down-regulating the expression of GlcAT-I and XT-1 in chondrocytes,15, 22 wherein XT-1 suppression is mediated by displacement of Sp1 and enhanced recruitment of Sp3 to the gene promoter.15 We, therefore, investigated whether such repression of XT-1 by cytokines is observed in NP cells. These results revealed that in NP cells, XT-1 promoter activity and its mRNA and protein levels were unaffected by TNF-α or IL-1β. This may be explained by a previous observation that Sp1 DNA binding is unaffected by TNF-α in NP cells.23 Moreover, decreased activity of promoter with AP-1 –730/–723 bp binding site mutation or deletion by TNF-α or IL-1β suggested that AP-1 was responsible for sustaining transcription in the presence of cytokines.
AP-1 transcription factor has been shown to control the expression of GAG biosynthetic enzymes and matricellular proteins important for intervertebral disk function.24, 25 In XT-1 promoter, several binding sites of AP-1 that are active in transcription have been reported.1, 15, 16 We, therefore, hypothesized that transcription factor AP-1 may also be involved in regulating basal XT-1 expression even in healthy NP cells. Gain- and loss-of-function studies using XT-1 promoter clearly indicated that Jun and Fos activated the transcription. Site-directed mutagenesis suggested that AP-1 binding to motif at −730/−723 bp is the preferential site for maintenance of the promoter activity. However, unlike the mutant, deletion of the −730/−723 bp AP-1 site resulted in promoter gaining responsiveness to suppression by DN-AP-1, indicating that Jun recruited at −684/−677 bp could control transcription perhaps owing to better accessibility to the transcriptional machinery. Results of genomic ChIP analysis that showed enrichment of Jun binding to both AP-1 motifs in NP cells provided further support to the functional studies and the hypothesis that AP-1 recruitment to XT-1 promoter controls its basal transcription. Decreased levels of mRNA and protein in the presence of AP-1 inhibitor tanshinone IIA confirmed that AP-1 maintains basal expression of XT-1 in NP cells.
Sp1 and Sp3, members of the Sp protein family, have a similar structure and bind to the same cognate Sp1 binding sites. However, their DNA-binding properties and regulatory functions are different.26 In many cell types, Sp1 and Sp3 exert opposing effects on gene transcription, and the ratio of these two proteins determines the expression level of the target gene, as demonstrated for collagen II transcription in chondrocytes.27, 28, 29 In the intervertebral disk, recent results have shown that transcription factors AP-1 and Sp1 are involved in controlling the basal expression of GlcAT-I, an enzyme that controls GAG synthesis.25 Relevant to this discussion, in SW1353 cells, Sp1 knockdown had no detectable effect on the XT-1 mRNA levels, whereas knockdown of Sp3 led to a highly significant reduction.16 Mutation analysis of Sp1 binding sites of XT-1 promoter in primary chondrocytes showed that disruption of the Sp1 –227/–217 bp core sequence resulted in 80% loss of XT-1 promoter activity, whereas mutation of Sp1 –124/–114 bp stimulated XT-1 promoter activity by 66%, suggesting that Sp1 –124/–114 bp may be essential for repression of the XT-1 gene expression.15 To understand the role of Sp1 and Sp3 in the regulation of XT-1 expression in NP cells, we performed loss- and gain-of-function studies. The studies conclusively showed that suppression of Sp1 transcriptional activity using either a dominant-negative approach or inhibitors resulted in decreased XT-1 promoter activity. In contrast to AP-1, apparent unresponsiveness of XT-1 promoter activity to overexpression of Sp1 and Sp3 may suggest saturating levels of Sp1/Sp3 present in NP cells. These functional studies were further supported by a genomic ChIP analysis that demonstrated enriched binding of Sp1 to the XT-1 promoter fragment that contained the −227/−217 bp and −124/−114 bp binding sites. Unlike the previously suggested differential role of Sp1 and Sp3 in XT-1 transcription and other matrix genes,15, 27, 28 a robust decrease in XT-1 mRNA and protein after inhibitor treatment and stable silencing of Sp1 or Sp3 supported the positive contribution of these factors in XT-1 expression in NP cells. Taken together, the results of this study clearly show that AP-1, Sp1, and Sp3 are critical for the maintenance of XT-1 expression under physiologic and pathophysiologic conditions in NP cells.
Footnotes
Supported by NIH grants AR055655, AR064733, and AR050087 (M.V.R.) and partly by grant 81101385 from the National Natural Science Foundation of China (W.Y. and J.Z.).
W.Y. and J.Z. contributed equally to this work.
Disclosures: None declared.
References
- 1.Maniadakis N., Gray A. The economic burden of back pain in the UK. Pain. 2000;84:95–103. doi: 10.1016/S0304-3959(99)00187-6. [DOI] [PubMed] [Google Scholar]
- 2.Luoma K., Riihimäki H., Luukkonen R., Raininko R., Viikari-Juntura E., Lamminen A. Low back pain in relation to lumbar disc degeneration. Spine (Phila Pa 1976) 2000;25:487–492. doi: 10.1097/00007632-200002150-00016. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi S., Meir A., Urban J. Effect of cell density on the rate of glycosaminoglycan accumulation by disc and cartilage cells in vitro. J Orthop Res. 2008;26:493–503. doi: 10.1002/jor.20507. [DOI] [PubMed] [Google Scholar]
- 4.Hiyama A., Gajghate S., Sakai D., Mochida J., Shapiro I.M., Risbud M.V. Activation of TonEBP by calcium controls β1,3-glucuronosyltransferase-I expression, a key regulator of glycosaminoglycan synthesis in cells of the intervertebral disc. J Biol Chem. 2009;284:9824–9834. doi: 10.1074/jbc.M807081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel K.P., Sandy J.D., Akeda K., Miyamoto K., Chujo T., An H.S., Masuda K. Aggrecanases and aggrecanase-generated fragments in the human intervertebral disc at early and advanced stages of disc degeneration. Spine (Phila Pa 1976) 2007;32:2596–2603. doi: 10.1097/BRS.0b013e318158cb85. [DOI] [PubMed] [Google Scholar]
- 6.Kuhn J., Götting C., Schnölzer M., Kempf T., Brinkmann T., Kleesiek K. First isolation of human UDP-D-xylose: proteoglycan core protein beta-D-xylosyltransferase secreted from cultured JAR choriocarcinoma cells. J Biol Chem. 2001;276:4940–4947. doi: 10.1074/jbc.M005111200. [DOI] [PubMed] [Google Scholar]
- 7.Venkatesan N., Barré L., Magdalou J., Mainard D., Netter P., Fournel-Gigleux S., Ouzzine M. Modulation of xylosyltransferase I expression provides a mechanism regulating glycosaminoglycan chain synthesis during cartilage destruction and repair. FASEB J. 2009;23:813–822. doi: 10.1096/fj.08-118166. [DOI] [PubMed] [Google Scholar]
- 8.Prante C., Milting H., Kassner A., Farr M., Ambrosius M., Schön S., Seidler D.G., Banayosy A.E., Körfer R., Kuhn J., Kleesiek K., Götting C. Transforming growth factor beta1-regulated xylosyltransferase I activity in human cardiac fibroblasts and its impact for myocardial remodeling. J Biol Chem. 2007;282:26441–26449. doi: 10.1074/jbc.M702299200. [DOI] [PubMed] [Google Scholar]
- 9.Tian Y., Yuan W., Fujita N., Wang J., Wang H., Shapiro I.M., Risbud M.V. Inflammatory cytokines associated with degenerative disc disease control aggrecanase-1 (ADAMTS-4) expression in nucleus pulposus cells through MAPK and NF-κB. Am J Pathol. 2013;182:2310–2321. doi: 10.1016/j.ajpath.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H., Tian Y., Wang J., Phillips K.L., Binch A.L., Dunn S., Cross A., Chiverton N., Zheng Z., Shapiro I.M., Le Maitre C.L., Risbud M.V. Inflammatory cytokines induce NOTCH signaling in nucleus pulposus cells: implications in intervertebral disc degeneration. J Biol Chem. 2013;288:16761–16774. doi: 10.1074/jbc.M112.446633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Wang H., Yang H., Li J., Cai Q., Shapiro I.M., Risbud M.V. TNF-α and IL-1β dependent MMP-3 expression in nucleus pulposus cells requires cooperative signaling via syndecan 4 and MAPK-NF-κB-axis: implications in inflammatory disc disease. Am J Pathol. 2014;184:2560–2572. doi: 10.1016/j.ajpath.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts S., Evans H., Trivedi J., Menage J. Histology and pathology of the human intervertebral disc. J Bone Joint Surg Am. 2006;88S:10–14. doi: 10.2106/JBJS.F.00019. [DOI] [PubMed] [Google Scholar]
- 13.Venkatesan N., Barré L., Bourhim M., Magdalou J., Mainard D., Netter P., Fournel-Gigleux S., Ouzzine M. Xylosyltransferase-I regulates glycosaminoglycan synthesis during the pathogenic process of human osteoarthritis. PLoS One. 2012;7:e34020. doi: 10.1371/journal.pone.0034020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faust I., Roch C., Kuhn J., Prante C., Knabbe C., Hendig D. Human xylosyltransferase-I - a new marker for myofibroblast differentiation in skin fibrosis. Biochem Biophys Res Commun. 2013;436:449–454. doi: 10.1016/j.bbrc.2013.05.125. [DOI] [PubMed] [Google Scholar]
- 15.Khair M., Bourhim M., Barré L., Li D., Netter P., Magdalou J., Fournel-Gigleux S., Ouzzine M. Regulation of xylosyltransferase I gene expression by interleukin 1β in human primary chondrocyte cells: mechanism and impact on proteoglycan synthesis. J Biol Chem. 2013;288:1774–1784. doi: 10.1074/jbc.M112.419887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller B., Prante C., Kleesiek K., Götting C. Identification and characterization of the human xylosyltransferase I gene promoter region. J Biol Chem. 2009;284:30775–30782. doi: 10.1074/jbc.M109.016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Risbud M.V., Guttapalli A., Stokes D.G., Hawkins D., Danielson K.G., Schaer T.P., Albert T.J., Shapiro I.M. Nucleus pulposus cells express HIF-1 alpha under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J Cell Biochem. 2006;98:152–159. doi: 10.1002/jcb.20765. [DOI] [PubMed] [Google Scholar]
- 18.Pfirrmann C.W., Metzdorf A., Zanetti M., Hodler J., Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 2001;26:1873–1878. doi: 10.1097/00007632-200109010-00011. [DOI] [PubMed] [Google Scholar]
- 19.Hoogendoorn R., Doulabi B.Z., Huang C.L., Wuisman P.I., Bank R.A., Helder M.N. Molecular changes in the degenerated goat intervertebral disc. Spine (Phila Pa 1976) 2008;33:1714–1721. doi: 10.1097/BRS.0b013e31817d2468. [DOI] [PubMed] [Google Scholar]
- 20.Tolonen J., Grönblad M., Virri J., Seitsalo S., Rytömaa T., Karaharju E. Oncoprotein c-Fos and c-Jun immunopositive cells and cell clusters in herniated intervertebral disc tissue. Eur Spine J. 2002;11:452–458. doi: 10.1007/s00586-001-0383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Markova D., Anderson D.G., Zheng Z., Shapiro I.M., Risbud M.V. TNF-α and IL-1β promote a disintegrin-like and metalloprotease with thrombospondin type I motif-5-mediated aggrecan degradation through syndecan-4 in intervertebral disc. J Biol Chem. 2011;286:39738–39749. doi: 10.1074/jbc.M111.264549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouze J.N., Bordji K., Gulberti S., Terlain B., Netter P., Magdalou J., Fournel-Gigleux S., Ouzzine M. Interleukin-1beta down-regulates the expression of glucuronosyltransferase I, a key enzyme priming glycosaminoglycan biosynthesis: influence of glucosamine on interleukin-1beta-mediated effects in rat chondrocytes. Arthritis Rheum. 2001;44:351–360. doi: 10.1002/1529-0131(200102)44:2<351::AID-ANR53>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 23.Séguin C.A., Pilliar R.M., Madri J.A., Kandel R.A. TNF-alpha induces MMP2 gelatinase activity and MT1-MMP expression in an in vitro model of nucleus pulposus tissue degeneration. Spine (Phila Pa 1976) 2008;33:356–365. doi: 10.1097/BRS.0b013e3181642a5e. [DOI] [PubMed] [Google Scholar]
- 24.Tran C.M., Markova D., Smith H.E., Susarla B., Ponnappan R.K., Anderson D.G., Symes A., Shapiro I.M., Risbud M.V. Regulation of CCN2/connective tissue growth factor expression in the nucleus pulposus of the intervertebral disc: role of Smad and activator protein 1 signaling. Arthritis Rheum. 2010;62:1983–1992. doi: 10.1002/art.27445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiyama A., Gogate S.S., Gajghate S., Mochida J., Shapiro I.M., Risbud M.V. BMP-2 and TGF-beta stimulate expression of beta1,3-glucuronosyl transferase 1 (GlcAT-1) in nucleus pulposus cells through AP1, TonEBP, and Sp1: role of MAPKs. J Bone Miner Res. 2010;25:1179–1190. doi: 10.1359/jbmr.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu B., Datta P.K., Bagchi S. Stability of the Sp3-DNA complex is promoter-specific: Sp3 efficiently competes with Sp1 for binding to promoters containing multiple Sp-sites. Nucleic Acids Res. 2003;31:5368–5376. doi: 10.1093/nar/gkg706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chadjichristos C., Ghayor C., Kypriotou M., Martin G., Renard E., Ala-Kokko L., Suske G., de Crombrugghe B., Pujol J.P., Galéra P. Sp1 and Sp3 transcription factors mediate interleukin-1 beta down-regulation of human type II collagen gene expression in articular chondrocytes. J Biol Chem. 2003;278:39762–39772. doi: 10.1074/jbc.M303541200. [DOI] [PubMed] [Google Scholar]
- 28.Ghayor C., Chadjichristos C., Herrouin J.F., Ala-Kokko L., Suske G., Pujol J.P., Galera P. Sp3 represses the Sp1-mediated transactivation of the human COL2A1 gene in primary and de-differentiated chondrocytes. J Biol Chem. 2001;276:36881–36895. doi: 10.1074/jbc.M105083200. [DOI] [PubMed] [Google Scholar]
- 29.Chadjichristos C., Ghayor C., Kypriotou M., Martin G., Renard E., Ala-Kokko L., Suske G., de Crombrugghe B., Pujol J.P., Galéra P. Down-regulation of human type II collagen gene expression by transforming growth factor-beta 1 (TGF-β1) in articular chondrocytes involves SP3/SP1 ratio. J Biol Chem. 2002;277:43903–43917. doi: 10.1074/jbc.M206111200. [DOI] [PubMed] [Google Scholar]