Abstract
Why the DNA-containing organelles, chloroplasts, and mitochondria, are inherited maternally is a long standing and unsolved question. However, recent years have seen a paradigm shift, in that the absoluteness of uniparental inheritance is increasingly questioned. Here, we review the field and propose a unifying model for organelle inheritance. We argue that the predominance of the maternal mode is a result of higher mutational load in the paternal gamete. Uniparental inheritance evolved from relaxed organelle inheritance patterns because it avoids the spread of selfish cytoplasmic elements. However, on evolutionary timescales, uniparentally inherited organelles are susceptible to mutational meltdown (Muller's ratchet). To prevent this, fall-back to relaxed inheritance patterns occurs, allowing low levels of sexual organelle recombination. Since sexual organelle recombination is insufficient to mitigate the effects of selfish cytoplasmic elements, various mechanisms for uniparental inheritance then evolve again independently. Organelle inheritance must therefore be seen as an evolutionary unstable trait, with a strong general bias to the uniparental, maternal, mode.
Keywords: cytoplasmic incompatibility, Muller's ratchet, organelle inheritance, organelle recombination, paternal leakage, plastome-genome incompatibility, selfish cytoplasmic elements
Introduction
The eukaryotic genome is distributed among different genetic compartments that follow contrasting modes of inheritance 1. Nuclear genes usually display Mendelian segregation. In contrast, non-Mendelian inheritance patterns are characteristic of the DNA-containing cell organelles: plastids (chloroplasts) and mitochondria. The non-Mendelian inheritance of organelles is predominantly uniparental, usually maternal. Thus, organelle inheritance can be recognized as reciprocal difference in sexual crosses (Fig. 1). Other features of organelle inheritance include somatic segregation (sorting-out) of genetically distinct organelles (Box 1; Fig. 1), and the virtual absence of recombination 1,2. Due to the different evolutionary origins and inheritance modes of the genomes of the eukaryotic cell, severe evolutionary consequences arise:
Figure 1.

Paternal leakage, biparental chloroplast inheritance, sorting-out, plastome-genome incompatibility, and gamete controlled paternal exclusion. A: Paternal leakage of plastids in tobacco seedlings detected by antibiotic selection. Green areas correspond to cells harboring spectinomycin-resistant paternal chloroplasts, whereas white sectors contain only cells with antibiotic-sensitive maternal plastids 79. Diffuse areas of green tissue indicate incomplete sorting-out of maternal and paternal plastids (Box 1). B: Biparental chloroplast inheritance in evening primroses, as evidenced by variegated progeny from the inter-specific cross Oenothera villaricae x Oe. picensis. The two species are diploid structural heterozygotes that, due to the genetic phenomenon of permanent translocation heterozygosity, inherit their haploid genomes as complete units. Oe. villaricae consists of the haploid genomes “B” and “l”, whereas Oe. picensis has the genomic composition “v” and “I”. The variegated hybrid individual shown here represents one of the possible F1 segregants and consists of the haploid genomes “l” and “v”. It is heteroplasmic for the plastids of Oe. villaricae (green sectors) and the plastids of Oe. picensis [chlorotic (virescent) sectors]. The chloroplast genome of Oe. picensis is incompatible with this hybrid nuclear background. Note that sorting-out in this particular individual is likely completed, as indicated by the sharp borders between green and chlorotic tissue sectors. C: F1 hybrid “l · v” of Oe. villaricae x Oe. picensis homoplasmic for the compatible chloroplast genome from Oe. villaricae. D: F1 hybrid “l · v” from the reciprocal cross (Oe. picensis × Oe. villaricae), homoplasmic for the incompatible chloroplast genome from Oe. picensis. Since green and variegated “l · v” individuals occur only if Oe. villaricae (“B · l”) is the mother, and the reciprocal cross with Oe. picensis (“v · I”) as maternal parent produces only incompatible homoplasmic “l · v” offspring, it can be concluded that the haploid genome “l” is unable to transmit plastids into the next generation 93. Scale bars: 0.5 mm for panel A, 5 cm for panels B-D.
Box 1
Heteroplasmy: Sorting-out and the genetic bottleneck
In contrast to the nuclear genome, organelle genomes occur at high copy numbers and are usually distributed among multiple organelles per cell. Polyploidy and free vegetative segregation of organelles and their genomes are hallmarks of cytoplasmic inheritance. Starting from a so-called “mixed cell” (a cell that is heteroplasmic for its plastid or mitochondrial genomes, due to either de novo mutation or biparental inheritance), resolution of heteroplasmy by sorting-out of the two organellar genotypes typically occurs during subsequent rounds of cell division. Since the distribution of organelles and their DNA to daughter cells is, in principle, a stochastic process, mixed cells usually disappear after a certain number of cell divisions, and homoplasmic cell lineages arise. Speed and sorting mechanisms are variable between organisms and organelles. For example, sorting-out of plastids in seed plants is a rapid process that is typically completed before flower formation (Fig. 1). In contrast, at least in some animal systems, heteroplasmy (in the germ line) can persist for several generations. Sorting-out results in intra-organismic genetic drift. The process does not change allele frequencies of neutral alleles within a population, but it does so within an organism. It further provides an opportunity for selection on particular oDNA genotypes, if a mutation is harmful or the two genome types differ in their replication speed. The phenomenon of the “genetic bottleneck” refers to an extreme intra-organismic shift in oDNA genotypes that is especially pronounced in the germline of multicellular organisms. The copy number of organelle genomes in the germline is often drastically reduced compared to the vast amount of organelle genome copies present in somatic tissues, thus resulting in rapid segregation to homoplasmy at high probability 1,12,15.
(i) Nuclear and organellar genomes differ fundamentally in their genome organization, coding capacity, mutation rate, and phylogeography 3,4. (ii) Uniparental transmission of organelles implies the existence of different mating types and sexes. However, uniparental organelle inheritance alone does not seem to represent a sufficiently strong driving force for the evolution of anisogamy and of two sexes (5,6; Box 2). (iii) Uniparental inheritance can induce genome conflicts between the nucleus and the organelles. In both plant and animal systems, an increased female fitness associated with the organellar genotype (cytotype) has been observed 7,8. This phenomenon of a sex-specific selective sieve (“mother's curse”) applies, for example, if female and male metabolic requirements are different 9. The best studied case is cytoplasmic male sterility (CMS) in plants. This typically mitochondrially encoded trait mediates sex determination in gynodioecious populations and induces a counter-selection for nuclear fertility restorer genes 8,10,11. (iv) Finally, the tight co-evolution of nuclear and organellar genomes can result in genetic incompatibilities when new genome combinations are generated through hybridization. Although the organellar genomes of related species are often very similar and typically have identical coding capacities, organelles are not freely exchangeable between species. Enforced by uniparental inheritance and lack of sexual recombination, co-evolution, and co-adaptation of the genetic compartments lead to tight genetic interdependence of the nucleus and the organelles 7,12,13. Combination of a nuclear genome with an alien mitochondrial or plastid genome thus can result in inter-specific hybrids that display so-called cytoplasmic incompatibilities (Fig. 1). Such incompatibilities can create hybridization barriers and contribute to speciation 8,13,14.
Despite being of enormous importance, the causes of the predominantly maternal inheritance mode of organelles are not fully understood (e.g. 15,16). Uniparental inheritance excludes organelles from sexual recombination. However, recombination is believed to be necessary to allow genomes to escape mutational meltdown, a process known as Muller's ratchet. Uniparental (maternal) organelle transmission should therefore be an evolutionary dead end. However, accumulating evidence for at least occasional biparental transmission (paternal leakage) provides opportunities for sporadic sexual recombination events between organellar genomes. Those could significantly slow down Muller's ratchet 16–18. The past few years have seen a paradigm shift in that the absoluteness of maternal organelle transmission is increasingly challenged 15,16,18–20. Nevertheless, there must be a selection pressure toward the evolution of uniparental transmission, for example to avoid the spreading of selfish cytoplasmic elements. Such elements can be mutant organellar genomes that replicate faster than the wild-type genome, but are maladaptive to the organism. However, whether these elements indeed represent the driving force leading to uniparental inheritance and predominance of the maternal mode has remained enigmatic. Further, the validity of the assumption that rare biparental transmission and sporadic sexual recombination of organelle DNA (oDNA) can stop the ratchet remains to be assessed.
This article describes recent progress in our understanding of organelle inheritance. It discusses the current views on the driving forces and evolutionary consequences of maternal inheritance in plants, animals, algae, and fungi and highlights important unresolved problems. We suggest a unifying model of organelle inheritance, and argue that the dominance of uniparentally maternal transmission is an evolutionary unstable trait. Mutational meltdown of organelle genomes is overcome by episodes of recombination between organelle genomes. The driving force for the fall-back to strict uniparental inheritance comes from a certain type of selfish cytoplasmic elements (i.e. organellar genomes that are maladaptive, but faster replicating than the native genome). Importantly, such elements cannot be disarmed by recombination. Finally, we propose experimental strategies to test the assumptions underlying our model.
Box 2
Is organelle inheritance a by-product or the cause of two sexes? Isogamous algae can answer this question
One of the most commonly suggested models for the existence of two sexes is based on uniparental organelle inheritance. Is it assumed that two mating types exist to avoid costs of cytonuclear conflicts, for example, by competing and maladaptive cytotypes. Uniparental inheritance has first evolved in isogamous organisms and was then enforced by anisogamy to regulated uniparental inheritance via only one gamete. In this way, the organelles define the sex (23,24; see main text). However, besides the fact that various other models for the evolution of anisogamy (and two sexes) exist, the cytonuclear conflict model can be questioned. First, there are some fungi where organelle inheritance is regulated independently of gamete size or mating type. Second, it is difficult to judge if organelle inheritance is just a by-product of anisogamy. Organelle inheritance could be coupled secondarily to an already pre-existing mating type. Also, it may typically associate with the larger gamete in a quantitative manner (reviewed, e.g. in 6,25,44). If one assumes a higher mutational load of the smaller paternal gamete as driving force for the maternal predominance of organelle transmission (as we propose here), this would be a very reasonable scenario.
Most eukaryotes are unicellular and many of them are isogamous. In many isogamous species, oDNA inheritance appears to be linked to a mating type. Hence it seems reasonable to assume that the organelles indeed define the mating type. Importantly, one of the arguments standing against this view can be questioned based on our present theory of oDNA inheritance. If oDNA inheritance is phylogenetically unstable (Box 3; see main text), fungi that regulate oDNA inheritance independently of gamete size or mating type can be interpreted as derived forms. Another important point to clarify is whether uniparental organelle inheritance represents a by-product of the evolution of mating types or its cause? This question is difficult to address in organisms that carry only one organelle type (mitochondria). Disregarding biparental transmission, mitochondrial inheritance is almost always associated with the larger gamete, and so far, studies in isogamous organisms have not provided a clear answer either. However, if uniparental oDNA inheritance was a prerequisite for anisogamy, one would expect a clear linkage between mating type and organelle inheritance in those isogamous species that possess two types of organelles. This can be tested in algae that contain both plastids and mitochondria. Interestingly, the green alga Chlamydomonas reinhardtii inherits its plastid by the mt+ (“maternal”) mating type. The mitochondria, however, are inherited by the mt− (“paternal”) mating type. By contrast, Volvox, a close relative of Chlamydomonas, is oogamous (anisogamous) and displays maternal inheritance of both organelles (84; Table1). This example can be interpreted as evidence for uniparentally maternal oDNA inheritance indeed being a by-product of anisogamy. However, since oDNA inheritance is phylogenetically unstable (see main text), a much larger dataset on organelle transmission in algae should be analyzed. Unfortunately, mostly due to technical constraints, organelle transmission in (isogamous) algae is largely understudied. While data for red algae are essentially lacking, the few examples reported so far for green and brown algae argue against regular co-transmission of plastid and mitochondria in isogamous algal species 84,85.
Box 3
Modes and mechanisms of oDNA inheritance and their phylogenetic distribution
Especially in vascular plants, where chloroplast transmission has been extensively studied, a large dataset supports repeated and independent evolution of biparental plastid transmission 16,46,86. In many branches of the phylogenetic tree, plastid transmission modes vary from maternal, maternal with paternal leakage, biparental (unbiased or with maternal or paternal dominance) to paternal. Interestingly, about one third of the plant species analyzed so far display the potential for biparental plastid transmission 49,87. Relaxed uniparental maternal inheritance is also observed in ferns and algae. Moreover, mitochondria and plastids can be inherited independently of each other by different sexes. For example, plastids are maternally inherited whereas mitochondria are paternally inherited in cucumber (46,47,49,53,54,84,85; Table1). Although biparental inheritance of the mitochondria was observed in Pelargonium, compared to plastids, a higher predominance of uniparentally maternal inheritance seems to exist in plants (Table1). However, paternal or biparental mitochondrial inheritance is frequently found in fungi 22. Biparental transmission (or at least strong paternal leakage) has been reported for bees. In mussels of the genus Mytilus, a unique mechanism of so-called doubly uniparental inheritance has evolved (Table1). Thus, in addition to maternal inheritance, various other types of organelle inheritance are observed.
If inheritance is uniparental, it is often not strict. More and more evidence is accumulating that heteroplasmy and paternal leakage are quite common in natural populations of plants, animals, and fungi 15,18–20,88–91. This seems particularly frequent in inter-species crosses, where exclusion mechanisms of different species may not function properly upon hybridization 20. For a few plant species, paternal leakage frequencies of plastids could be determined experimentally. The observed leakage frequencies are rather high, and thus blur the boundary between uniparental and biparental transmission 16,79,92.
The mechanisms of how uniparental organelle transmission is achieved are also very diverse 46–49,53,54. Many different organelle exclusion mechanisms exist, and they can act either before, during or after fertilization (Fig. 2; Table1). Birky 16 lists 12 different cellular mechanisms for organelle exclusion. Often, the mechanism is not even conserved between closely related taxa. For example, in mammals such as mouse, cow or rhesus monkey, the paternal mtDNA undergoes a reduction in the sperm but is fully degraded only later during early embryogenesis (i.e. after initially biparental transmission). By contrast, in the Chinese hamster, mtDNA seems to be excluded during fertilization (reviewed in 48). In tomato, the male generative cell does not contain plastids. By contrast, in potato, the paternal plastids are eliminated at a later stage of gametogenesis. Remarkably, tomato and potato belong to the same genus (Fig. 2). These two examples indicate that uniparental (maternal) inheritance evolved repeatedly even between closely related taxa. Further, this convergent evolution frequently results in paternal gamete-controlled organelle exclusion (“killing one's own paternal cytoplasm”), which so far has been difficult to explain by modeling approaches (see main text).
The nuclear genetics of organelle exclusion also appears to be rather heterogeneous. In many plant species, the mode of chloroplast inheritance depends on the crossing direction, and varies between crosses involving different ecotypes. It can be controlled by the genetic constitution of the maternal and/or the paternal gamete (90,93–98; Fig. 1). Similar data are available for mitochondrial inheritance 15,20,99. This indicates that organelle exclusion is, in many cases, haplotype dependent.
Taken together, modes, mechanisms, phylogenetic distribution, and genetic architecture of organelle inheritance are very diverse among eukaryotes. This strongly suggests repeated and independent evolution of diverse patterns of organelle transmission.
Theoretical models for the occurrence of uniparental organelle inheritance
Although not universal, maternal inheritance is the predominant mode of organelle transmission in all eukaryotic kingdoms. This raises the question as to which evolutionary forces favor its prevalence. Currently available mathematical models typically link uniparental (maternal) organelle inheritance with the evolution of anisogamy and/or sex determination (e.g. 21–24; but also see 5,6,25; Box 2). Below, we briefly discuss the main models in the light of existing experimental evidence. We point out unsettled questions and assumptions that remain to be scrutinized. It should be emphasized that these models are not necessarily mutually exclusive.
Genomic conflict models
As originally proposed by Grun 26 and based on genetic observations in evening primroses (genus Oenothera), the most frequently expressed explanation for the evolution of uniparental organelle inheritance is the avoidance of cytonuclear conflicts 21,22. The general model diverges into two types: “avoiding competition” (between organelles) and “avoiding negative interaction” (between organelle genomes and/or organelle genomes and the nuclear genome). From a modeling perspective, the two schemes cannot be fully discerned from each other 27–30.1
“Avoiding competition” models posit that two non-recombining clonal lineages (i.e. the maternal and paternal organellar genomes) will enter direct competition. Mutations in one of the genomes, for example in a locus determining the replication speed of the organelle, would allow one of the lineages to outgrow the other. Also, mutations in oDNA might arise that mar the competing organelles by their attempts to gain a competitive advantage 24,35. Metaphorically speaking, the nuclear genome does not have an “interest” in a war between organelles in the cytosol 22.
“Negative interaction” models purport that diverging cytotypes generally reduce fitness 36. As an exemplary mechanistic explanation, and somewhat overlapping with the “avoid competition” hypothesis, there could be a locus that is maladaptive to the nucleus but favors an aggressive (faster replicating) cytotype 26,30. Considering organelles alone, negative interaction could be caused by loci in the two organellar genomes that are not co-adapted to each other, but combined by sexual recombination. Alternatively, there is the possibility that different organelles harbor different alleles of one locus, and their heteroplasmic combination is maladaptive to the cell 37. Obviously, a strict uniparental inheritance of organelles largely avoids these problems. Indeed, modeling of such scenarios frequently leads to the fixation of a nuclear inheritance modifier that causes switching from an ancestral biparental to a derived uniparental mode of inheritance.
Mutation pressure and the “bottleneck model”
Another starting point toward explaining uniparental inheritance is the assumption that sexual recombination of oDNA is not the only force that counteracts Muller's ratchet (see below). Hence, strict uniparental organelle transmission may be less harmful than widely assumed. Most relevant in this context is that organelles pass a genetic bottleneck when entering the germline (Box 1). By this mechanism, organelle mutations can become purified by intra-cellular genetic drift in that genome segregation to homoplasmy occurs 38,39. Subsequently, deleterious mutations can be eliminated effectively by selection 12,40. Modeling work showed that paternal leakage (or biparental transmission) would interfere with this process 41. Interestingly, in the “bottleneck model”, absence of sexual recombination of oDNAs is, to some extent, the driving force rather than the consequence of uniparental inheritance.
Co-adaptation model
Another model that deserves consideration was postulated recently 42. The establishment of DNA-containing organelles by endosymbiosis was followed by massive gene transfer from the genome of the endosymbiont to the nuclear genome of the host cell 43. Since many of the encoded gene products are re-imported into the organelle, organellar genomes and nuclear genomes rely on tight co-evolution and co-adaptation. Mathematical modeling shows that co-adaptation is enhanced by both uniparental inheritance and the genetic bottleneck, suggesting that selection for co-adaptation was a driving force for uniparental inheritance and the evolution of two sexes. Like the other models, the co-adaptation model assumes lack of sexual oDNA recombination.
The evolutionary cause for uniparentally maternal inheritance is still unclear
In particular, the different types of genomic conflict hypotheses have been modeled extensively. From this work, several theoretical problems arose. A general argument against these hypotheses is that a mutation leading to uniparental transmission can only be advantageous if a selfish cytoplasmic element is present, but not yet fixed in the population 6,16,44. According to Hutson and Law 45, fixation of an inheritance modifier (inducing the switch from ancestral biparental inheritance to uniparental inheritance) requires a heterozygous advantage at this locus and the tight linkage to a self-incompatibility allele. Uniparental inheritance can, therefore, only evolve within rather strict boundary conditions. It seems that these problems can be solved by a recently proposed model 5. It makes the assumption that the gametes control organelle inheritance. It further takes the dynamics of the fitness costs of biparental inheritance into account in that cells do not suffer from a fixed cost of biparental inheritance, but the actual costs depend on the number of selfish or maladaptive mutations. Consequently, the model predicts that the relative advantage of uniparental inheritance declines in a mutation frequency-dependent manner within a population. This appears to be the case under very broad parameters. Hence, the model is compatible with the different inheritance patterns of oDNA, varying between (low-level) paternal leakage and regular biparental inheritance (Box 3; Table1). It can also account for genomic conflicts, mutation pressure, and nuclear-organelle co-adaptation as potential driving forces for uniparental inheritance. However, in agreement with previous modeling, it was found that an inheritance modifier that kills its own organelles cannot spread. Paternal exclusion should, therefore, be evolutionarily unstable 28. This is mainly due to the mechanistic problem that such an allele cannot be genetically linked with the fittest cytotype 5,6,44. Nevertheless, achievement of maternal inheritance by paternal exclusion of organelles (“killing one's own cytoplasm”) is frequent among plant and animal species (46–49; Fig. 2; Table1). It is, however, obviously associated with fitness costs. According to Sreedharan and Shpak 50, the trait can arise only if one assumes very high mutation rates of selfish cytoplasmic elements (5% per generation). However, in contrast to mammalian mitochondrial DNA, the nucleotide substitution frequencies in plastid and plant mitochondrial genomes are very low 51,52. Developing the idea further, the occurrence of hermaphrodites with uniparental organelle transmission (as is the case for many self-pollinating plant species) is difficult to explain. In these organisms, maternal transmission implies a costly mechanism for the organism to eliminate its own paternal cytoplasm. The second argument that can be raised against all models for uniparental inheritance is the implicit assumption that the cytotype transmitted into the hybrid (typically the maternal cytotype) is generally fitter than the excluded (paternal) cytotype (e.g. 25,46).
Table 1.
Inheritance of mitochondria and plastids in different eukaryotic taxa
| Taxon | Species | Common name | Mitochondrial inheritance | Plastid inheritance | |||||
|---|---|---|---|---|---|---|---|---|---|
| Mode | Exclusion of mt/mtDNA (sex/stage/fate) | Reference (see footnote) | Mode | Exclusion of pt/ptDNA (sex/stage/fate) | Reference (see footnote) | ||||
| Green algae | Chlamydomonas reinhardtii | U (PL) mt- | mt+/zygote/- | 1,2 | U (PL) mt+ | mt−/zygote/- | 3 | ||
| Volvox carteri | M | Male/fertilization/- | 4,5 | M | Male/fertilization/- | 4,5 | |||
| Mosses | Liverworts | Sphaerocarpos donnellii | M | male/fertilization/- | 6 | M | male/fertilization/- | 6 | |
| Ferns | Pteris vittata | Chinese brake fern | M | Male/gamete/- | 7 | M | Male/gamete/- | 7 | |
| Gymnosperms | Conifers | Pseudotsuga menziesii | Douglas-fir | M | Male/embryogenesis/exclusion | 8,9 | P | Female/embryogenesis/exclusion | 8,9 |
| Angiosperms | Monocots | Hordeum vulgare | Barley | M | Male/gamete/↓ & male/fertilization/ECB | 11,12 | M | Male/gamete/- (& male/fertilization/ECB) | 10–12 |
| Triticum aestivum | Wheat | M | Male/gamete/- | 13,14 | M | Male/gamete/- | 13,14 | ||
| Dicots | Nicotiana tabacum | Tobacco | M | Male/fertilization/ECB & male/zygote/- | 14–17 | M (PL) | Male/gamete/↓ + ECB | 14–18 | |
| Antirrhinum majus | Snapdragon | M | Male/gamete/- | 21 | M (PL) | Male/gamete/- | 19–21 | ||
| Cucumis sativus | Cucumber | P | Female/embryogenesis/sorting-out | 25,26 | M | Male/gamete/- | 24 | ||
| Medicago sativa | Alfalfa | M (PL) | Male/gamete/- | 21,22 | B (P) | Female/zygote/partial exclusion | 20–23 | ||
| Oenothera spp. | Evening primrose | M | Male/fertilization, zygote or embryogenesis/- | 27,28 | B (M) | Male/zygote/input frequency + multiplication Speed | 29,30 | ||
| Pelargonium zonale | Zonal geranium | B | No | 32,33 | B (BMP) | No | 31,32 | ||
| Amoebozoa | Slime molds | Physarum polycephalum | slime mold | U | One mating type/zygote/- | 34 | |||
| Fungi | Ascomycetes | Neurospora crassa | M | Male/fission + fusion/sorting out | 35,36 | ||||
| Saccharomyces cerevisiae | Budding yeast | B | Male + female/zygote/recombination + segregation | 37–39 | |||||
| Animals | Tunicates | Ascidia nigra | Black solitary tunicate | M | Male/fertilization/- | 40,41 | |||
| Mussels | Mytilus edulis | Blue mussel | UU | Male/embryogenesis of future female/- | 42,43 | ||||
| Arthropods | Drosophila melanogaster | Fruit fly | M (PL) | Male/gamete/↓ + “waste bag” | 44–47 | ||||
| Apis mellifera | Honey bee | B/M (PL) | Male/embryogenesis/- | 48 | |||||
| Mammals | Mus musculus | Mouse | M | Male/gamete/↓ & male/embryogenesis/- | 49,50 | ||||
| Homo sapiens | Human | M | Male/gamete/↓ & male/embryogenesis/- | 51–53 | |||||
The inheritance mode is abbreviated with U, uniparental; M, maternal; P, paternal; B, biparental; PL, paternal leakage; M, maternal predominance; P, paternal predominance; UU, doubly uniparental; BMP, biparental, maternal and paternal progeny. The mechanisms of exclusion of organelles or oDNA is denoted as “–” for complete disappearance/degradation, ↓ for decrease by digestion/reduction of copy number (by down-regulated replication or other mechanisms). ECB, enucleated cytoplasmic body; pt, plastids; mt, mitochondrion; mt+, “female” mating type; mt−, “male” mating type; “waste bag”, structure containing cytoplasmic material that is excluded from the apical end of sperm tails.
Reference list: 1. Boynton et al. 1987, Proc Natl Acad Sci USA 84: 2391; 2. Aoyama et al. 2006, Protoplasma 228: 231; 3. Kuroiwa et al. 1982, Nature 298: 481; 4. Adams et al. 1990 Curr Genet 18: 141; 5. Kuroiwa 2010, J Plant Res 123: 207; 6. Diers 1967a, Planta 72: 119; 7. Kuroiwa et al. 1988, Protoplasma 146: 89; 8. Owens and Morris 1990, Am J Bot 77: 433; 9. Owens and Morris 1991, Am J Bot 78: 1515; 10. Mogensen and Rusche 1985, Protoplasma 128: 1; 11. Mogensen 1988, Proc Natl Acad Sci USA 85: 2594; 12. Sodmergen et al. 2002, Planta 216: 235; 13. Hagemann and Schröder 1989, Protoplasma 152: 57; 14. Miyamura et al. 1987, Protoplasma 141: 149; 15. Yu et al. 1994, Sex Plant Reprod 7: 312; 16. Yu and Russell 1994a, Sex Plant Reprod 7: 324; 17. Yu and Russell 1994b, Planta 193: 115; 18. Medgyesy et al. 1986, Proc Natl Acad Sci USA 82: 6960; 19. Diers 1967b, Mol Gen Genet 100: 56; 20. Corriveau et al. 1990, Curr Genet 17: 439; 21. Nagata et al. 1999, Planta 209: 53; 22. Forsthoefel et al. 1992, J Hered 83: 342; 23. Mogensen 1996, Am J Bot 83: 383; 24. Corriveau and Coleman 1988, Am J Bot 75: 1443; 25. Matsuura 1995, Rep Cucurbit Genet Coop 18: 31; 26. Havey 1997, J Hered 88: 232; 27. Brennicke and Schwemmle 1984, Z Naturforsch 39c: 191; 28. Sodmergen et al. 1997, Protoplasma 198: 66; 29. Meyer and Stubbe 1974, Ber Deutsch Bot Ges 87: 29; 30. Chiu and Sears 1988, Curr Genet 13: 181; 31. Metzlaff et al. 1981, Theor Appl Genet 60: 37; 32. Sodmergen et al. 1992, Protoplasma 186: 73; 33. Weihe et al. 2009, Mol Genet Genom 282: 587; 34. Moriyama and Kawano 2003, Genetics 164: 963; 35. Mannella et al. 1979, J Bacteriol 137: 1449; 36. Reich and Luck 1966, Proc Natl Acad Sci USA 55: 1600; 37. Birky et al. 1982, in Mitochondrial Genes: 333; 38. Birky 2001, Annu Rev Genet 35: 125; 39. Solieri 2010, Trends Microbiol 18: 521; 40. Ursprung and Schabtach 1965, J Exp Zool 159: 379; 41. Schabtach and Ursprung 1965, J Exp Zool 159: 357; 42. Breton et al. 2007, Trends Genet 23: 465; 43. Zouros et al. 1994 Proc Natl Acad Sci USA 91: 7463; 44. Reilly and Thomas 1980, Plasmid 3: 109; 45. DeLuca and O‘Farrell 2012, Dev Cell 22: 660; 46. Politi et al. 2014, Dev Cell 29: 305; 47. Kondo et al. 1990, Genetics 126: 657; 48. Meusel and Moritz 1993, Curr Genet 24: 539; 49. Kaneda et al. 1995, Proc Natl Acad Sci USA 92: 4542; 50. Cummins et al. 1997, Zygote 5: 301; 51. Giles et al. 1980, Proc Natl Acad Sci USA 77: 6715; 52. Larsson et al. 1997, Hum Mol Genet 6: 185; 53. Cummins 1998, Rev Reprod 3: 172.
Figure 2.
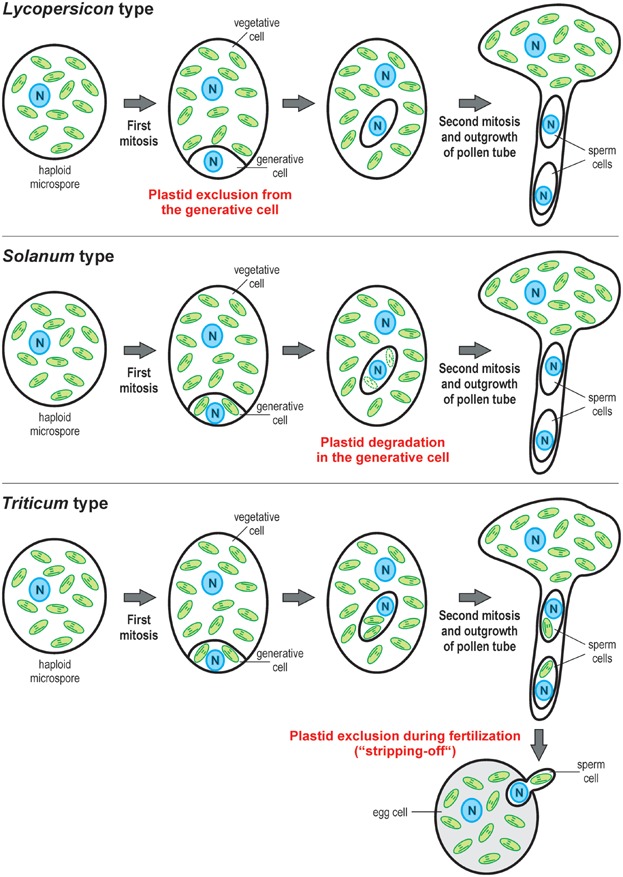
Different cytological mechanisms can result in maternal inheritance of plastids in angiosperms 120. Species belonging to the Lycopersicon type (tomato type), exclude plastids in pollen mitosis I. As the result of an unequal cell division, the resulting large vegetative cell receives all plastids, whereas the generative cell is devoid of plastids. Species of the Solanum type (potato type) exclude plastids after pollen mitosis I. Their generative cell contains a few plastids which, however, are selectively degraded (by an unknown mechanism) prior to division of the generative cell into the two sperm cells in pollen mitosis II. Both mechanisms must be under genetic control of the paternal gamete. Species of the Triticum type (wheat type) produce sperm cells that still contain plastids. However, the plastids are stripped off upon fertilization and thus do not enter the cytoplasm of the egg cell. Alternative mechanisms are possible in which the paternal plastids enter the egg cell, but do not contribute to the embryo. The close phylogenetic relatedness of tomato and potato, which belong to the same family (Solanaceae; nightshade family) and, according to the most recent taxonomy, even to the same genus (tomato, formerly called Lycopersicon esculentum, was renamed Solanum lycopersicum), suggests significant evolutionary flexibility and repeated independent evolution of the mechanisms leading to (paternally controlled) maternal plastid inheritance.
In summary, the available models of organelle inheritance fail to explain why the uniparentally paternal mode of organelle inheritance is rare 46 and why “killing one's own (paternal) cytoplasm” occurs. Hence, the current theoretical problem connected with organelle inheritance is not its sex linkage per se, but rather the dominance of the maternal over the paternal mode and in many cases its control by the paternal gamete. Arguing that gamete size simply determines organelle inheritance in a largely quantitative manner (in that female gametes are larger and, therefore, harbor more organelles), is not satisfactory either. Especially in plants, many examples exist for (i) contrasting modes of plastid DNA (ptDNA) and mitochondrial DNA (mtDNA) inheritance, and (ii) biparental or predominantly paternal transmission, implying a high organelle load in the paternal gamete (46,49,53,54; Table1; Box 3).
Alternative explanations for uniparental organelle DNA inheritance do not apply to the whole eukaryotic domain
In view of the problems outlined above, some authors assume that the current models do not provide a fully satisfactory explanation for the prevalence of uniparental transmission of plastids and mitochondria in the entire eukaryotic domain 16,42,44,47. Organelle genomes of plants and animals as well as those of unicellular and multicellular eukaryotes differ greatly in genome organization, coding capacity, copy number per cell and mutation rate, as do cell and gamete sizes and ecological niches. In theory, modes of organelle transmission could even be explained as an evolutionary by-product of selection forces shaping organellar genomes in a lineage-specific manner 16.
On the other hand, the predominance of maternal organelle transmission, along with the virtual absence of sexual recombination between organelles in most lineages of eukaryotic evolution, is striking. It thus appears likely that there is a general explanation for the observed pattern (but also see 55). The exclusion of organelles from the germline is an active process and should be costly 46–49. Also, it has likely evolved repeatedly (16,46,49; Fig. 2; Box 3; see below). Hence, there must be a strong, general selection pressure maintaining this trait.
By arguing from a physiological point of view, a possible explanation was offered by Allen 56. It posits that only the maternal organelle DNA is maintained because it is protected from oxidative damage (as caused by the electron transfer reactions in photosynthesis and respiration). Since the sessile egg cell has a lower energy demand than the mobile sperm, the paternal oDNA may suffer from higher oxidative damage and, therefore, is excluded from inheritance. By contrast, the maternal germline cells are protected in specialized tissues, where organelles would display low metabolic rates. This assumption seems to be true for a wide range of animal systems 57, and likely also for proplastids in plant meristems. However, since the meristem confers plant growth and cellular differentiation, it has a high energy demand. Therefore, its mitochondria should not be protected from reactive oxygen species. Also, the hypothesis cannot apply to unicellular organisms. Thus, like most of the genetic models described above, the theory falls short of explaining organelle inheritance patterns for all eukaryotes. Even though it provides an elegant explanation for why paternal exclusion of cytoplasms could be frequent, the theory cannot explain the widespread occurrence of biparental transmission of chloroplast, paternal leakage, and cases of paternal oDNA inheritance (46,49,53; Table1).
A unifying model for organelle inheritance
Taking a number of theoretical considerations into account, we propose here a unifying model for organelle inheritance (Fig. 3). We argue that uniparental inheritance arises to avoid the spread of selfish (faster replicating) organelle genomes that are maladaptive and/or incompatible with the host nucleus. However, uniparental inheritance is evolutionarily unstable, because organelles are subject to Muller's ratchet. This drives a relaxation of strict maternal inheritance by paternal leakage or regular biparental transmission. Biparental inheritance is again susceptible to the evolution of selfish genomes and, therefore, is repeatedly lost and restored over evolutionary timeframes. In other words, the mutational meltdown by Muller's ratchet is escaped from by episodes (or longer periods) of sexual recombination between organelle genomes. Importantly, sexual oDNA recombination is not sufficient to stop the spread of selfish cytoplasmic elements. The observed maternal predominance in uniparental transmission is due to a higher mutational load of paternal cytotypes, which in turn is caused by oxidative damage and/or genetic drift and is most pronounced if oDNA copy numbers in the sperm cell are small. The load is high enough to favor the evolution of paternal gamete-controlled organelle exclusion mechanisms (“killing one's own cytoplasm”). What is the actual evidence for these assumptions?
Figure 3.
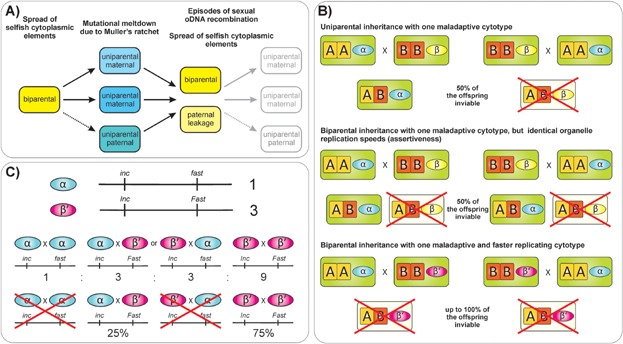
Repeated origin and loss of uniparental organelle inheritance in evolution and selection pressures for uniparental and biparental organelle transmission. A: Biparental organelle inheritance likely represented the ancestral stage. It is selected against to avoid the spread of selfish cytoplasmic elements (left panel). This drives evolution for uniparental inheritance. It is typically maternal and, due to its lineage-dependent evolution, realized by various cellular mechanisms (indicated by different colors). Uniparental paternal inheritance (dashed arrow) can evolve, if the mutational load for paternally inherited organelles is low and/or comparable to that of organelles in the egg cell. Strict uniparental inheritance leads to organelle genome susceptibility to mutational meltdown (middle panel). This, in turn, provides a driving force for a fall-back to relaxed organelle inheritance patterns to allow (low levels of) sexual oDNA recombination. Repeated evolution of uniparental inheritance is necessary, since biparental transmission allows the spread of selfish cytoplasmic elements, even if organelle genomes undergo sexual recombination (right panel). B: Selection pressure for uniparental organelle inheritance as caused by an aggressive and maladaptive cytoplasm. Organelle genomes α and β are both compatible with their nuclear host genomes AA and BB, respectively. Consider that cytotype β is incompatible with the hybrid nuclear genome AB, whereas cytotype α is compatible. Upon uniparental inheritance of the two organelles, reciprocal crosses will give 50% viable offspring (top panel). Identical offspring viability is achieved if both organelles are inherited biparentally and have identical multiplication speeds (i.e. assertiveness rates in the zygote and the F1 generation; middle panel). The situation changes dramatically, if in the cytotype that is incompatible to the hybrid a mutation arises (β') that can overgrow the compatible cytotype α in the offspring. If transmitted biparentally, it will effectively eliminate the compatible cytotype α. This situation would provide a strong selection pressure for the evolution of uniparental inheritance (lower panel). C: Spread of maladaptive and aggressive cytoplasmic genotypes cannot be prevented by sexual oDNA recombination. Assume that the compatible cytotype α carries two genetically unlinked loci (cf. Box 4) that confer compatibility with the hybrid nucleus (inc) and normal replication speed (fast). The incompatible and aggressive cytotype β' harbors the alleles Inc and Fast, conferring incompatibility in the hybrid nuclear background and faster replication. Further assume that the allele Fast shifts the input ratio of the two cytoplasms α and β' into the zygote from 1:1 (upon biparental inheritance with no maternal or paternal bias) to 1:3. Since in an organelle cross, input frequencies reflect output frequencies and homologous recombination can occur between genomes (Box 4), the allele combinations inc/fast, inc/Fast, Inc/fast and Inc/Fast will occur in a 1:3:3:9 frequency. [The α and β' genomes can recombine with themselves, resulting in 1 × 1 α (inc/fast), and 3 × 3 β' (Inc/Fast) genotypes. Recombination between α and β' results 1 × 3 in the allele combinations inc/Fast and Inc/fast, respectively.] If all oDNA genomes carrying the allele fast are overgrown by Fast genotypes during ontogenesis, the only two remaining genotypes will be inc/Fast (25%) and Inc/Fast (75%). The latter is incompatible with the host nuclear genome, but substantially overrepresented in the hybrid population, thus conferring a strong selective disadvantage.
Patterns of organelle inheritance are phylogenetically unstable
If uniparental inheritance is evolutionarily unstable, three major patterns in organelle inheritance should be observable. First, biparental transmission should evolve repeatedly and independently. Second, paternal leakage should be relatively frequent. Third, the switch back to sex-specific organelle exclusion (uniparental inheritance) should occur by diverse mechanisms that can differ between closely related species or even between haplotypes. Strikingly, these patterns are indeed observed, throughout the eukaryotic domain (Box 3; Fig. 1; Table1).
In addition, paternal gamete-controlled organelle exclusion certainly plays an important role in organelle exclusion. As for the observed predominance of maternal organelle transmission (see above), any general theory of organelle inheritance must therefore give a reasonable explanation for “killing one's own (paternal) cytoplasm”.
The predominance of maternal oDNA transmission might be due to higher mutational load in the male gamete
In all sexually reproducing eukaryotes, the zygote develops through fusion of an egg cell with a (usually motile) sperm cell. It subsequently undergoes rapid divisions that incur a high energy demand. Hence, in agreement with the “oxidative damage model”, there should be an immediate selection for the fittest organelle genotype. This should be the case for both mtDNA and ptDNA, because in many seed plant taxa, embryos are green (at least in the early stages of seed development the embryo is exposed to light) and perform photosynthesis 58. If selection in the zygote is the driving force for paternal exclusion, one must, however, assume higher mutation rates for paternally inherited organelle genotypes. This can be tested in paternally inherited cytotypes as found in gymnosperms. Strikingly, oDNA mutation rates are indeed higher in these taxa, suggesting that, compared to the egg cell, organelles in the pollen carry a higher mutational load 52,59,60. The “oxidative damage” assumption 56 could be relevant to this observation. However, since oxidative damage fails to explain relaxed maternal organelle inheritance patterns (see above), it cannot be the sole and universal driving force for the observed patterns of oDNA inheritance. However, paternal oDNA copy numbers in the sperm cell are typically substantially smaller than maternal copy numbers in the larger egg cell. Hence, genetic drift of oDNAs due to stronger genetic bottlenecking at the level of the gamete might represent an additional relevant factor. Consequently, paternal or biparental oDNA inheritance should be associated with (i) high organelle numbers and oDNA copy numbers in the pollen, as it is the case, for example, in alfalfa, melon or Pelargonium 49,61, and/or (ii) a much larger population size of the pollen compared to the egg, as it is the case in gymnosperms which are wind-pollinated. This view is in line with theoretical considerations, arguing that the higher mutational load of organelle genomes in general is not due to asexuality per se, but is the result of the small effective population size of organellar genomes 62.
Taken together, maternal dominance in organelle inheritance could be due to a lower mutational load, since in most organisms, more oDNA copies are inherited by the mother. However, in organisms where bottlenecking is less severe for the male gamete, paternal oDNA inheritance can evolve, thus potentially explaining why contrasting modes of organelle inheritance exist. This especially applies to isogamous organisms, carrying two organelles such as green algae (Box 2). Moreover, the higher mutational load in the paternal oDNA might be the selection force for the evolution of mechanisms that “kill one's own (paternal) cytoplasm”. Finally, uniparental (maternal) inheritance must be seen as a consequence of, rather than the underlying reason for, anisogamy (Box 2).
Possible selection pressures for uniparental organelle inheritance
Maternal dominance of uniparental inheritance could be explained by a higher mutational load of the paternal gamete. However, why does uniparental inheritance exist at all, and what are the selection forces, leading to uniparental (maternal) inheritance?
Deleterious interactions between co-existing organelle genomes
A commonly suggested putative selection force for uniparental inheritance is deleterious epistatic interaction between co-existing organelle genomes. In the case of mitochondria, a possible mechanistic scenario could, for example, involve the unscheduled onset of apoptosis. That can be triggered by production of reactive oxygen species if improperly co-adapted subunits of the mitochondrial respiratory chain are combined with each other 63. However, to what extent deleterious epistatic interactions between co-existing organelles occur in nature, is currently unclear. At least for plastids of seed plants, such interactions are difficult to image, since plastids usually do not undergo fusion 16,46,54. There is no molecular or cell biological evidence for negative interactions between co-existing plastids, even though some classic genetic evidence could be interpreted in this direction (pages 154–155 of 26, 64). Negative interactions between mitochondria in plants and animals seem to be possible, and were reported in some cell fusions 65,66. Recently, it was demonstrated that heteroplasmic mice display reduced respiratory activity and behavioral phenotypes, whereas mice homoplasmic for either of the two mitochondrial genotypes had no phenotype 36. Taken together, inter-organellar epistasis seems to exist, although its mechanisms are largely enigmatic. However, analyses on sexual oDNA recombination in yeast and Chlamydomonas somewhat argue against the widespread occurrence of deleterious epistasis between oDNA alleles, since the expected segregation distortion is not normally observed (Box 4). Given the few documented examples, the general significance of deleterious epistatic interactions between organelles is currently questionable.
Box 4
Sexual recombination of oDNA: experimentally determined frequencies and occurrence in natural populations
Recombination between biparentally inherited organelle genomes has been reported for some taxa, but is controversial in others (15,16,18,20 for references). It is important to note that, upon strict uniparental inheritance (and lack of sexual recombination), selective sweeps should be frequent in organellar genomes, but are barely observed 12. Detailed genetic linkage analyses based on sexual oDNA recombination were conducted in yeast (e.g. 100), and genetic linkage maps were also established for ptDNA and mtDNA in the green alga Chlamydomonas (e.g. 101–103). Both organisms display biparental inheritance or paternal leakage of their organelles (Table1). Linkage analyses in yeast and Chlamydomonas uncovered remarkable differences of oDNA recombination compared to recombination mapping in the nuclear genome. Similar to “phage crosses”, where two phages are mixed and allowed to recombine in bacteria upon double infection, the maximum recombination frequency in an inter-organelle cross is 25% (rather than 50%), because half of the recombination events occur between identical genotypes. This value is supported by experimental data in that the maximal recombination frequencies observed are indeed in the range of 20–25%. However, in contrast to a phage cross where titers and double infection rates can be easily determined, models for oDNA recombination usually assume that oDNA contribution from both parents is equal and that there is no intra-cellular selection for or against particular recombinants. Furthermore, random pairing of oDNA molecules, multiple rounds of paring and recombination, and random segregation of oDNA copies is assumed 104. In spite of these uncertainties, genetic distances obtained from segregation analyses usually correlate well with the physical distances of the genetic markers 102,105,106. Although generally high, recombination frequencies of oDNAs vary between species (on average, 15–20% recombinant clones are observed within a given sequence interval of 1 kb in brewer's yeast, 3.2% in Chlamydomonas mtDNA, 1.6% in fission yeast, and 1% in Chlamydomonas ptDNA; 102). Since the lowest observed recombination frequency of 1% within 1 kb reflects the existence of a linkage group only for a 25 kb distance (because 25% recombination is the maximum possible frequency), it appears that, if sexual recombination within oDNA occurs, large portions of the genome can be genetically unlinked. Also, recombination hotspots can exist in oDNAs 107. Another important finding from genetic analyses was that oDNA recombination events are mostly non-reciprocal at the level of the individual, but reciprocal at the level of the population 101,108. This is likely due to gene conversion, but other factors may be involved as well 109.
The general features of sexual oDNA recombination as worked out for unicellular organisms may be transferable to many multicellular eukaryotes. However, there are some limitations concerning the frequency of organelle mixing and fusion. For example, plastids of seed plants do not seem to regularly undergo recombination in crosses, not even in organisms with biparental plastid inheritance 110. However, especially in cell fusion experiments ptDNA recombination was occasionally seen (e.g. 111,112, but see also 3,64). It appears likely that recombination between plastomes in sexual crosses of seed plants is largely prevented by the absence of plastid fusion in the zygote 46,47,54. This is in contrast to Chlamydomonas, where plastid fusion occurs after syngamy.
Organelle recombination of plant mitochondrial genomes was repeatedly demonstrated in protoplast fusion 3,65,113 and preliminary evidence for recombination in sexual crosses has also been obtained 114. If biparental transmission occurs, sexual recombination of plant mtDNA is expected, because plant mitochondria regularly undergo fusion (and fission), and homologous recombination events seem to occur frequently in mitochondrial genomes 115,116. In contrast to plants and fungi (reviewed in 20), occurrence and evolutionary relevance of mitochondrial genome recombination in animals are still controversial. Mixed evidence is available in that recombination was detected in some animal species, but not in others 2,15,117,118.
The general presence of sexual oDNA recombination has gained some support from investigations of natural populations. Circumstantial phylogenetic evidence points to sexual recombination in both plant and animal systems, but the currently available data are still sparse and a bit controversial 1,3,15,18,89. While genetic studies in natural populations of campion (genus Silene) suggest presence of recombination 19, somewhat contradicting evidence has been obtained for fruit flies and fungi 91,119. More rigorous and systematic investigations of oDNA recombination in natural populations and hybrid zones are needed that, for example, also take into account the possibility of selection against recombinant genotypes.
In summary, it seems possible that sexual recombination of oDNA is widespread and perhaps even a general phenomenon. As paternal leakage of plastids occurs at least occasionally in many, if not all, species, sexual recombination of plastids in seed plants may be limited by the rarity of plastid fusion events. In contrast, the limiting factor in sexual recombination of mtDNA may be paternal leakage and reduced recombination ability, at least in some animal taxa, most notably in mammals (cf. 2,3,15,18–20). It is noteworthy in this respect that mammalian mitochondrial genomes have considerably higher nucleotide substitution rates than plastid genomes and plant mitochondrial genomes. Interestingly, plant mitochondria, which are likely subject to paternal leakage and regularly undergo fusion and mtDNA recombination, display one of the lowest nucleotide substitution rates known in nature 51,52. However, whether or not oDNA recombination frequencies in all organisms, and especially in mammalian mitochondria and seed plant plastids, are high enough to overcome Muller's ratchet, remains to be determined (see main text).
Selfish cytoplasmic elements, nuclear-cytoplasmic co-adaptation, and their interplay
Another common posit is that uniparental inheritance has evolved to avoid the spread of selfish cytoplasmic elements. Some solid datasets are available for competition between organelle genomes in both plant and animal systems. Examples have come from cell fusion events, oDNA mutants and sexual crosses 6,12,18,67,68. For example, in evening primroses, plastids display different multiplication speeds in sexual crosses depending on the plastid genotype 64. If the avoidance of competition between organelles was the major driving force for the evolution of uniparental inheritance, a replication race between oDNAs must be harmful to the nucleus. Although some human diseases are associated with altered mtDNA copy numbers 69, in most eukaryotic systems studied so far, the amounts of oDNA versus nuclear DNA remain constant within a rather narrow range and are likely under nuclear control 1,12,69–71. Hence, it appears unlikely that solely differences in oDNA replication speeds provide sufficient driving force for the evolution of uniparental inheritance.
A likely much stronger selection force for uniparental inheritance will arise if an organelle with a higher replication speed carries a genotype that is incompatible with or maladaptive to the host nucleus. Prime examples are some of the petite mutants of yeast, which lack the capability for respiration due to large deletions in the mitochondrial genome. Although yeast can grow anaerobically, growth rates achieved by fermentation are substantially lower. However, due to the presence of more replication origins and/or their smaller genome size, these petite mutant mitochondria are able to overgrow the wild-type mitochondria 72. In evening primroses, the competitive advantage of specific plastid genotypes is largely independent of the nuclear background and is also observed when the more competitive plastid genotype is deleterious 73,74, exemplifying a naturally occurring aggressive and maladaptive cytotype 26.
Strikingly, cytotypes that are maladaptive to the nucleus are well known in plants, fungi, and animals. They lead to cytoplasmic incompatibilities, which are the result of diverging evolution between the organellar and nuclear genomes involved (7,8,13,14; Fig. 1). Together with potentially ubiquitously present differences in organelle replication speeds, this can lead to a hitchhiking of cytoplasmic incompatibilities. Potentially, this provides a strong selection force for uniparental organelle inheritance (26,30; Fig. 3B) which, however, cannot be disarmed by sexual recombination of oDNAs (Fig. 3C).
How much sexual oDNA recombination is needed to overcome Muller's ratchet?
As summarized by Birky 16, the assumption that sexual recombination in oDNA is required to counteract Muller's ratchet has been challenged. Hence, the virtual absence of recombination may be less harmful than widely assumed. A major argument is that organelles generally undergo a genetic bottleneck when entering the germline. Thus, organelle genomes become purified by intra-organismic genetic drift, rapidly segregate to homoplasmy, and therefore malfunctioning genotypes can be eliminated effectively by selection (12,38,39; Box 1). Also, organelles may have very efficient DNA repair mechanisms that might have evolved to cope with the constant exposure to high levels of reactive oxygen species that are generated as unavoidable by-products of respiratory and photosynthetic electron transfer reactions. In plants, nucleotide substitution rates in ptDNA and mtDNA are much lower than in the nucleus, thus defying Muller's ratchet 51,52. High genome copy numbers, together with active gene conversion, seem to be effective mechanisms for slowing down the ratchet, at least in plant oDNAs 75. Nonetheless, the ratchet should still be clicking, raising the question how much recombination is needed to stop it. According to Charlesworth et al. 76, assuming a population size of no more than 100 diploid individuals and a chromosome with 1,000 loci, 10−5 cross-over events per locus and generation are sufficient (for review see 62). If applied to a sexual oDNA recombination frequency of 3.2% within 1 kb (Chamydomonas mtDNA, with each base pair representing a locus; Box 4), and the paternal leakage frequency of tobacco mitochondria (10−4 to 10−5; 77), this would result in 3.2 × 10−6 to 3.2 × 10−7 recombination events per locus and generation. Although this estimate may be an over-simplification 78, the calculated frequency comes close to the value expected to suffice. Based on this value, it also seems clear that, for the chloroplast genome (upon leakage frequencies between 10−4 and 10−5; 77,79), due to rarity of plastid fusion (Box 4 and see above), paternal leakage might be insufficient to stop the ratchet from clicking. This could explain why uniparental inheritance of plastids is evolutionarily particularly unstable, and biparental transmission is more frequently observed for plastids than for mitochondria.
Killing one's own cytoplasm
As mentioned above, a major problem with the current theoretical modeling of the occurrence of uniparental inheritance lies in the frequent occurrence of paternal gamete-controlled exclusion of organelles. However, the present models might be too simple to reflect the true pattern of organelle inheritance. For example, the probably best theoretical approximation to the naturally observed organelle inheritance patterns 5 assumes a unicellular organism, a simple single-locus genetics of nuclear control of organelle inheritance, and the absence of sexual recombination of oDNA. Furthermore, no sex-specific mutational load is assumed. However, organelle inheritance can be controlled by multiple nuclear loci 80,81, the mutational load in paternal oDNA may be elevated, and sexual recombination of oDNA is known to occur in many systems (Box 4). In addition, improved models should take into account more complex patterns of sorting-out, as they occur in multicellular eukaryotes, and the underlying population genetics. Although this unavoidably complicates the modeling, a higher paternal mutational load and the possibility for sexual recombination of oDNA might explain why “killing one's own cytoplasm” is frequent in nature. In the presence of occasional sexual oDNA recombination, the fittest alleles of the paternal cytotype might be able to escape a uniparental inheritance modifier. The key question then will be whether the theoretical values that can be deduced from refined modeling approaches are in agreement with observed paternal leakage frequencies, oDNA recombination rates, and the strength of the selection pressures for uniparental inheritance.
Experimental model systems
As suggested above, the avoidance of spreading of an incompatible but aggressive cytoplasm with a faster replicating genotype might be a major driving force for uniparental organelle inheritance. However, for a full understanding of oDNA inheritance patterns, one needs to assess the fitness effects of all potential driving forces of uniparental inheritance. It further will be necessary to identify the nuclear factors responsible for organelle exclusion as well as the organellar loci controlling replication speed and the loci conferring deleterious epistatic interactions between co-existing organelles or between organelles and the nucleus. The interplay of these genetic factors in natural populations must be studied, taking sexual oDNA recombination into account. Currently, excellent experimental models are available to study sexual oDNA recombination, especially yeast and Chlamydomonas. Also campions (genus Silene) and fruit flies have proved to be valuable systems for studying paternal oDNA leakage and oDNA recombination in natural populations. High-throughput application of next-generation sequencing technologies will certainly increase our understanding of organelle inheritance. Paternal leakage, sorting-out of genome types, selection against deleterious variants as well as oDNA stability and recombination dynamics are now accessible at much finer scale (Boxes 1 and 4) and at all levels: in cells, tissues, individuals, and entire populations. Nevertheless, there is currently a shortage of suitable models that would allow the investigation of fitness effects and evolutionary consequences of relaxed oDNA inheritance in natural populations. For plants, evening primroses provide such a model. Major principles of chloroplast genetics were initially worked out in evening primroses. Moreover, early theoretical considerations on the selection forces of uniparental inheritance were formulated based on data from evening primroses by Grun 26. Evening primroses also represent a uniquely suited system to test possible selection pressures for uniparental transmission at the population level for several reasons. First, the genus offers an extremely well characterized formal genetics at the population level. Second, biparental transmission of chloroplasts is the rule in evening primroses. Third, plastome-genome incompatibility occurs frequently in inter-specific hybrids. These incompatibilities are associated with genetically distinguishable plastome types (I–V), which are already known to differ in their multiplication speeds. Of particular interest is the common evening primrose (Oenothera biennis), a hybrid species that is naturally distributed in the eastern half of North America. It harbors the basic nuclear genomes A and B (in a stable heterozygous state) associated with either plastome type II or III in overlapping subpopulations 13. Due to hybridization within the population, the incompatible combinations AA-III and BB-II are sometimes observed, building asymmetric hybridization barriers of different strengths (Fig. 1). Other observed combinations, such as AA-II, BB-III, AB-II, or AB-III, are compatible. Since plastids with plastome III are multiplying faster than those with plastome II, they have the potential to outcompete plastome II plastids in this population. Moreover, this constellation provides a hitchhiking opportunity for a maladaptive trait in inter-specific hybridization events 8,13,82,83. In view of all these attractive features, the Oenothera system is clearly one of the most suitable models for testing some of the key predictions of our current hypotheses on organelle inheritance.
Conclusions and outlook
Here, we propose a unifying, potentially universal and testable model, to explain the evolution of organelle inheritance. We argue that uniparentally maternal organelle inheritance is an evolutionarily unstable trait. In anisogamous organisms, the maternal predominance seems to be due to a higher mutational load of the paternal gamete. The major driving force for uniparental inheritance could come from selfish cytoplasmic genomes that are maladaptive to the host nucleus but replicate faster than the native cytoplasmic genome. The model is in line with the various inheritance patterns observed in nature. To test the underlying assumptions, the factors involved in and/or leading to uniparental (maternal) organelle inheritance need to be identified and quantified. We must understand nature and function of selfish mutations and determine their strength in selection. Also, it will be necessary to measure sexual oDNA recombination rates in natural populations, to identify the genes involved in organelle exclusion and to investigate their population genetics. Further, age and extinction rates of lineages displaying uniparental or biparental inheritance need to be determined (cf. 16). All these parameters should then be used in advanced modeling approaches, to solve one of the most fundamental and puzzling questions in genetics and evolutionary biology.
Acknowledgments
We thank Dr. Barbara B. Sears (Michigan State University) for critical reading and fruitful discussion and Dr. Stephanie Ruf (MPI-MP) for images illustrating paternal leakage and help with artwork. Research on organelle inheritance and organelle-nuclear interactions by the authors is supported by the Max Planck Society and the Deutsche Forschungsgemeinschaft (DFG). We apologize to all colleagues whose work could not be discussed due to space constraints.
Glossary
- mtDNA
mitochondrial DNA
- oDNA
organelle DNA
- ptDNA
plastid DNA
Footnotes
Integrated in some models is the idea that uniparental inheritance might also help with reducing the negative impact of cytoplasmic parasites 31,32. However, with few exceptions (e.g. Wolbachia and related infectious bacteria in arthropods and nematodes), the frequent presence and vertical transmission of cytoplasmic parasites is not documented for many eukaryotes. In addition, the assumption that mixing of such parasites generally reduces host fitness is doubtable 33. Moreover, uniparental transmission may exclude organelles from vertical transmission, but not necessarily parasites at the same time. For example, paternal transmission of a virus was observed in barley 34, a species that inherits its organelles maternally (Table1).
References
- Birky CW. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu Rev Genet. 2001;35:125–48. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- Hagström E, Freyer C, Battersby BJ, Stewart JB. No recombination of mtDNA after heteroplasmy for 50 generations in the mouse maternal germline. Nucleic Acids Res. 2014;42:1111–6. doi: 10.1093/nar/gkt969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Vendramin GG. Plant phylogeography based on organelle genes: an introduction. In: Weiss S, Ferrand N, editors. Phylogeography of Southern European Refugia. Dordrecht: Springer; 2007. pp. 23–97. [Google Scholar]
- Bazin E, Glémin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–2. [Google Scholar]
- Hadjivasiliou Z, Lane N, Seymour RM, Pomiankowski A. Dynamics of mitochondrial inheritance in the evolution of binary mating types and two sexes. Proc R Soc Lond B. 2013;280:20131920. doi: 10.1098/rspb.2013.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra RF. Nucleo-cytoplasmic conflict and the evolution of gamete dimorphism. In: Togashi T, Cox PA, editors. The evolution of anisogamy. Cambridge: Cambridge University Press; 2011. pp. 111–30. [Google Scholar]
- Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 2004;19:645–53. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Greiner S, Rauwolf U, Meurer J, Herrmann RG. The role of plastids in plant speciation. Mol Ecol. 2011;20:671–91. doi: 10.1111/j.1365-294X.2010.04984.x. [DOI] [PubMed] [Google Scholar]
- Wolff JN, Gemmell NJ. Mitochondria, maternal inheritance, and asymmetric fitness: why males die younger. BioEssays. 2013;35:93–9. doi: 10.1002/bies.201200141. [DOI] [PubMed] [Google Scholar]
- Budar F, Touzet P, De Paepe R. The nucleo-mitochondrial conflict in cytoplasmic male sterilities revisited. Genetica. 2003;117:3–16. doi: 10.1023/a:1022381016145. [DOI] [PubMed] [Google Scholar]
- Chase CD. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 2007;23:81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Rand DM. The units of selection of mitochondrial DNA. Annu Rev Ecol Syst. 2001;32:415–48. [Google Scholar]
- Greiner S, Bock R. Tuning a ménage à trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. BioEssays. 2013;35:354–65. doi: 10.1002/bies.201200137. [DOI] [PubMed] [Google Scholar]
- Burton RS, Pereira RJ, Barreto FS. Cytonuclear genomic interactions and hybrid breakdown. Annu Rev Ecol Evol Syst. 2013;44:281–302. [Google Scholar]
- White DJ, Wolff JN, Pierson M, Gemmell NJ. Revealing the hidden complexities of mtDNA inheritance. Mol Ecol. 2008;17:4925–42. doi: 10.1111/j.1365-294X.2008.03982.x. [DOI] [PubMed] [Google Scholar]
- Birky CW. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci USA. 1995;92:11331–8. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra RF. Evolutionary origin and consequences of uniparental mitochondrial inheritance. Hum Reprod. 2000;15:102–11. doi: 10.1093/humrep/15.suppl_2.102. [DOI] [PubMed] [Google Scholar]
- Barr CM, Neiman M, Taylor DR. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005;168:39–50. doi: 10.1111/j.1469-8137.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- McCauley DE. Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytol. 2013;200:966–77. doi: 10.1111/nph.12431. [DOI] [PubMed] [Google Scholar]
- Xu J. The inheritance of organelle genes and genomes: patterns and mechanisms. Genome. 2005;48:951–8. doi: 10.1139/g05-082. [DOI] [PubMed] [Google Scholar]
- Cosmides LM, Tooby J. Cytoplasmic inheritance and intragenomic conflict. J Theor Biol. 1981;89:83–129. doi: 10.1016/0022-5193(81)90181-8. [DOI] [PubMed] [Google Scholar]
- Eberhard WG. Evolutionary consequences of intracellular organelle competition. Q Rev Biol. 1980;55:231–49. doi: 10.1086/411855. [DOI] [PubMed] [Google Scholar]
- Hurst LD. Why are there only two sexes. Proc R Soc Lond B. 1996;263:415–22. [Google Scholar]
- Hurst LD, Hamilton WD. Cytoplasmic fusion and the nature of sexes. Proc R Soc Lond B. 1992;247:189–94. [Google Scholar]
- Billiard S, López-Villavicencio M, Devier B, Hood ME. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol Rev. 2011;86:421–42. doi: 10.1111/j.1469-185X.2010.00153.x. [DOI] [PubMed] [Google Scholar]
- Grun P. Cytoplasmic Genetics and Evolution. New York: Columbia University Press; 1976. [Google Scholar]
- Law R, Hutson V. Intracellular symbionts and the evolution of uniparental cytoplasmic inheritance. Proc R Soc Lond B. 1992;248:69–77. doi: 10.1098/rspb.1992.0044. [DOI] [PubMed] [Google Scholar]
- Randerson JP, Hurst LD. Small sperm, uniparental inheritance and selfish cytoplasmic elements: a comparison of two models. J Evol Biol. 1999;12:1110–24. [Google Scholar]
- Hastings IM. Population genetic aspects of deleterious cytoplasmic genomes and their effect on the evolution of sexual reproduction. Genet Res. 1992;59:215–25. doi: 10.1017/s0016672300030500. [DOI] [PubMed] [Google Scholar]
- Hoekstra RF. Evolution of uniparental inheritance of cytoplasmic DNA. In: Smith MJ, Vida J, editors. Organizational Constrains of the Dynamics of Evolution. Manchester: Manchester University Press; 1990. pp. 269–78. [Google Scholar]
- Hurst LD. Parasite diversity and the evolution of diploidy, multicellularity and anisogamy. J Theor Biol. 1990;144:429–43. doi: 10.1016/s0022-5193(05)80085-2. [DOI] [PubMed] [Google Scholar]
- Frank SA. Host-symbiont confict over the mixing of symbiotic lineages. Proc R Soc Lond B. 1996;263:339–44. doi: 10.1098/rspb.1996.0052. [DOI] [PubMed] [Google Scholar]
- Vautrin E, Vavre F. Interactions between vertically transmitted symbionts: cooperation or conflict. Trends Microbiol. 2009;17:95–9. doi: 10.1016/j.tim.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Card S, Pearson M, Clover G. Plant pathogens transmitted by pollen. Australas Plant Path. 2007;36:455–61. [Google Scholar]
- Charlesworth B. Reproductive evolution: mating types and uniparental transmission of chloroplast genes. Nature. 1983;304:211. [Google Scholar]
- Sharpley MS, Marciniak C, Eckel-Mahan K, McManus M. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell. 2012;151:333–43. doi: 10.1016/j.cell.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N. The problem with mixing mitochondria. Cell. 2012;151:246–8. doi: 10.1016/j.cell.2012.09.028. [DOI] [PubMed] [Google Scholar]
- Hill JH, Chen Z, Xu H. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet. 2014;46:389–92. doi: 10.1038/ng.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Xu H, O'Farrell PH. Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nat Genet. 2014;46:393–7. doi: 10.1038/ng.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom CT, Pritchard J. Germline bottlenecks and the evolutionary maintenance of mitochondrial genomes. Genetics. 1998;149:2135–46. doi: 10.1093/genetics/149.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze D, Rousset Fo, Michalakis Y. Germline bottlenecks, biparental Inheritance and selection on mitochondrial variants. Genetics. 2005;170:1385–99. doi: 10.1534/genetics.104.039495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc R Soc Lond B. 2012;279:1865–72. doi: 10.1098/rspb.2011.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R, Timmis JN. Reconstructing evolution: gene transfer from plastids to the nucleus. BioEssays. 2008;30:556–66. doi: 10.1002/bies.20761. [DOI] [PubMed] [Google Scholar]
- Lessells CM, Snook RR, Hosken DJ. The evolutionary origin and maintenance of sperm: selection for a small, motile gamete mating type. In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm Biology: An Evolutionary Perspective. Burlington: Academic Press; 2009. pp. 43–67. [Google Scholar]
- Hutson V, Law R. Four steps to two sexes. Proc R Soc Lond B. 1993;253:43–51. doi: 10.1098/rspb.1993.0080. [DOI] [PubMed] [Google Scholar]
- Sears BB. Elimination of plastids during spermatogenesis and fertilization in the plant kingdom. Plasmid. 1980;4:233–55. doi: 10.1016/0147-619x(80)90063-3. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T. Review of cytological studies on cellular and molecular mechanisms of uniparental (maternal or paternal) inheritance of plastid and mitochondrial genomes induced by active digestion of organelle nuclei (nucleoids) J Plant Res. 2010;123:207–30. doi: 10.1007/s10265-009-0306-9. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K. Maternal inheritance of mitochondrial DNA by diverse mechanisms to eliminate paternal mitochondrial DNA. Biochim Biophys Acta. 2013;1833:1979–84. doi: 10.1016/j.bbamcr.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Mogensen HL. The hows and whys of cytoplasmic inheritance in seed plants. Am J Bot. 1996;83:383–404. [Google Scholar]
- Sreedharan V, Shpak M. Selection for male-enforced uniparental cytoplasmic inheritance. Theor Biosci. 2010;129:295–306. doi: 10.1007/s12064-010-0113-9. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 1987;84:9054–8. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogen Evol. 2008;49:827–31. doi: 10.1016/j.ympev.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Hagemann R. The sexual inheritance of plant organelles. In: Daniell H, Chase CD, editors. Molecular Biology and Biotechnology of Plant Organelles – Chloroplasts and Mitochondria. Berlin, Heidelberg, New York: Springer; 2004. pp. 93–114. [Google Scholar]
- Nagata N. Mechanisms for independent cytoplasmic inheritance of mitochondria and plastids in angiosperms. J Plant Res. 2010;123:193–9. doi: 10.1007/s10265-009-0293-x. [DOI] [PubMed] [Google Scholar]
- Birky CW. Relaxed cellular controls and organelle heredity. Science. 1983;222:468–75. doi: 10.1126/science.6353578. [DOI] [PubMed] [Google Scholar]
- Allen JF. Separate sexes and the mitochondrial theory of ageing. J Theor Biol. 1996;180:135–40. doi: 10.1006/jtbi.1996.0089. [DOI] [PubMed] [Google Scholar]
- Allen JF, de Paula WBM. Mitochondrial genome function and maternal inheritance. Biochem Soc Trans. 2013;41:1298–304. doi: 10.1042/BST20130106. [DOI] [PubMed] [Google Scholar]
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y. Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature. 2004;432:779–82. doi: 10.1038/nature03145. [DOI] [PubMed] [Google Scholar]
- Whittle C-A, Johnston MO. Male-driven evolution of mitochondrial and chloroplastidial DNA sequences in plants. Mol Biol Evol. 2002;19:938–49. doi: 10.1093/oxfordjournals.molbev.a004151. [DOI] [PubMed] [Google Scholar]
- Crosby K, Smith DR. Does the mode of plastid inheritance influence plastid genome architecture. PLoS One. 2012;7:e46260. doi: 10.1371/journal.pone.0046260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D-Y, Zhang Q, Liu Y, Lin Z-F. The levels of male gametic mitochondrial DNA are highly regulated in angiosperms with regard to mitochondrial inheritance. Plant Cell. 2010;22:2402–16. doi: 10.1105/tpc.109.071902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman M, Taylor DR. The causes of mutation accumulation in mitochondrial genomes. Proc R Soc Lond B. 2009;276:1201–9. doi: 10.1098/rspb.2008.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N. Mitonuclear match: optimizing fitness and fertility over generations drives ageing within generations. BioEssays. 2011;33:860–9. doi: 10.1002/bies.201100051. [DOI] [PubMed] [Google Scholar]
- Greiner S. Plastome mutants of higher plants. In: Bock R, Knoop V, editors. Genomics of Chloroplasts and Mitochondria. Dordrecht, Heidelberg, New York, London: Springer; 2012. pp. 237–66. [Google Scholar]
- Rose RJ, Thomas MR, Fitter JT. The transfer of cytoplasmic and nuclear genomes by somatic hybridisation. Aust J Plant Physiol. 1990;17:303–21. [Google Scholar]
- Ziegler ML, Davidson RL. Elimination of mitochondrial elements and improved viability in hybrid cells. Somatic Cell Mol Genet. 1981;7:73–88. doi: 10.1007/BF01544749. [DOI] [PubMed] [Google Scholar]
- Day A, Madesis P. DNA replication, recombination, and repair in plastids. In: Bock R, editor. Cell and Molecular Biology of Plastids. Berlin, Heidelberg, New York: Springer; 2007. pp. 65–119. [Google Scholar]
- Nishimura Y, Stern DB. Differential replication of two chloroplast genome forms in heteroplasmic Chlamydomonas reinhardtii gametes contributes to alternative inheritance patterns. Genetics. 2010;185:1167–81. doi: 10.1534/genetics.110.118265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montier LLC, Deng JJ, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genom. 2009;36:125–31. doi: 10.1016/S1673-8527(08)60099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuten T, Cincu E, Fuchs J, Zoschke R. Fewer genes than organelles: extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 2010;64:948–59. doi: 10.1111/j.1365-313X.2010.04389.x. [DOI] [PubMed] [Google Scholar]
- Golczyk H, Greiner S, Wanner G, Weihe A. Chloroplast DNA in mature and senescing leaves: a reappraisal. Plant Cell. 2014;26:847–54. doi: 10.1105/tpc.113.117465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D. The curious history of yeast mitochondrial DNA. Nat Rev Genet. 2002;3:475–81. doi: 10.1038/nrg814. [DOI] [PubMed] [Google Scholar]
- Chiu W-L, Stubbe W, Sears BB. Plastid inheritance in Oenothera: organelle genome modifies the extent of biparental plastid transmission. Curr Genet. 1988;13:181–9. [Google Scholar]
- Chiu W-L, Sears BB. Plastome-genome interactions affect plastid transmission in Oenothera. Genetics. 1993;133:989–97. doi: 10.1093/genetics/133.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhlova O, Bock R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006;46:85–94. doi: 10.1111/j.1365-313X.2006.02673.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Morgan MT, Charlesworth B. Mutation accumulation in finite outbreeding and inbreeding populations. Genet Res. 1993;61:39–56. [Google Scholar]
- Svab Z, Maliga P. Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc Natl Acad Sci USA. 2007;104:7003–8. doi: 10.1073/pnas.0700063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky CW, Maruyama T, Fuerst P. An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics. 1983;103:513–27. doi: 10.1093/genetics/103.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S, Karcher D, Bock R. Determining the transgene containment level provided by chloroplast transformation. Proc Natl Acad Sci USA. 2007;104:6998–7002. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann R. Indications for nuclear influences on the mode of plastid inheritance. Endocytobiosis Cell Res. 2013;24:16–22. [Google Scholar]
- Derepas A, Dulieu H. Inheritance of the capacity to transfer plastids by the pollen parent in Petunia hybrida Hort. J Hered. 1992;83:6–10. [Google Scholar]
- Dietrich W, Wagner WL, Raven PH. Systematics of Oenothera section Oenothera subsection Oenothera (Onagraceae) In: Anderson C, editor. Systematic Botany Monographs. Laramie: The American Society of Plant Taxonomists; 1997. [Google Scholar]
- Cleland RE. Oenothera – cytogenetics and evolution. In: Sutcliffe JF, Mahlberg P, editors. Experimental Botany. London, New York: Academic Press, Inc; 1972. [Google Scholar]
- Miyamura S. Cytoplasmic inheritance in green algae: patterns, mechanisms and relation to sex type. J Plant Res. 2010;123:171–84. doi: 10.1007/s10265-010-0309-6. [DOI] [PubMed] [Google Scholar]
- Motomura T, Nagasato C, Kimura K. Cytoplasmic inheritance of organelles in brown algae. J Plant Res. 2010;123:185–92. doi: 10.1007/s10265-010-0313-x. [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhang Q, Rao G, Sodmergen Occurrence of plastids in the sperm cells of Caprifoliaceae: biparental plastid inheritance in angiosperms is unilaterally derived from maternal inheritance. Plant Cell Physiol. 2008;49:958–68. doi: 10.1093/pcp/pcn069. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Liu Y, Sodmergen Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 2003;44:941–51. doi: 10.1093/pcp/pcg121. [DOI] [PubMed] [Google Scholar]
- Kmiec B, Woloszynska M, Janska H. Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr Genet. 2006;50:149–59. doi: 10.1007/s00294-006-0082-1. [DOI] [PubMed] [Google Scholar]
- Wolfe AD, Randle CP. Recombination, heteroplasmy, haplotype polymorphism, and paralogy in plastid genes: implications for plant molecular systematics. Syst Bot. 2004;29:1011–20. [Google Scholar]
- Ellis JR, Bentley KE, McCauley DE. Detection of rare paternal chloroplast inheritance in controlled crosses of the endangered sunflower Helianthus verticillatus. Heredity. 2008;100:574–80. doi: 10.1038/hdy.2008.11. [DOI] [PubMed] [Google Scholar]
- Nunes MDS, Dolezal M, Schlötterer C. Extensive paternal mtDNA leakage in natural populations of Drosophila melanogaster. Mol Ecol. 2013;22:2106–17. doi: 10.1111/mec.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhagiri AK, Maliga P. Exceptional paternal inheritance of plastids in Arabidopsis suggests that low-frequency leakage of plastids via pollen may be universal in plants. Plant J. 2007;52:817–23. doi: 10.1111/j.1365-313X.2007.03278.x. [DOI] [PubMed] [Google Scholar]
- Schwemmle J, Haustein E, Sturm A, Binder M. Genetische und zytologische Untersuchungen an Eu-Oenotheren: Teil I bis VI. Z Indukt Abstamm Vererbungsl. 1938;75:358–800. [Google Scholar]
- Smith SE. Influence of parental genotype on plastid Inheritance in Medicago sativa. J Hered. 1989;80:214–7. doi: 10.1093/oxfordjournals.jhered.a110838. [DOI] [PubMed] [Google Scholar]
- Zhu T, Mogensen HL, Smith S. Quantitative, three-dimensional analysis of alfalfa egg cells in two genotypes: implications for biparental plastid inheritance. Planta. 1993;190:143–50. [Google Scholar]
- Hansen AK, Escobar LK, Gilbert LE, Jansen RK. Paternal, maternal, and biparental inheritance of the chloroplast genome in Passiflora (Passifloraceae): implications for phylogeneitc studies. Am J Bot. 2007;94:42–6. doi: 10.3732/ajb.94.1.42. [DOI] [PubMed] [Google Scholar]
- Bogdanova V, Galieva E, Kosterin O. Genetic analysis of nuclear-cytoplasmic incompatibility in pea associated with cytoplasm of an accession of wild subspecies Pisum sativum subsp. elatius (Bieb.) Schmahl. Theor Appl Genet. 2009;118:801–9. doi: 10.1007/s00122-008-0940-y. [DOI] [PubMed] [Google Scholar]
- Li D, Qi X, Li X, Li L. Maternal inheritance of mitochondrial genomes and complex inheritance of chloroplast genomes in Actinidia Lind.: evidences from interspecific crosses. Mol Genet Genom. 2013;288:101–10. doi: 10.1007/s00438-012-0732-6. [DOI] [PubMed] [Google Scholar]
- Bentley KE, Mandel JR, McCauley DE. Paternal leakage and heteroplasmy of mitochondrial genomes in Silene vulgaris: evidence from experimental crosses. Genetics. 2010;185:961–8. doi: 10.1534/genetics.110.115360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Mitochondrial genetics and functions. In: Strathern JN, Jones EW, Broach JR, editors. Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1981. pp. 505–635. [Google Scholar]
- Sears BB. Replication, recombination, and repair in the chloroplast genetic system of Chlamydomonas. In: Rochaix JD, Goldschmidt-Clermont M, Merchant S, editors. The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. New York Bosten, Dordrecht, London, Moscow: Kluwer Academic Publishers; 1998. pp. 115–38. [Google Scholar]
- Remacle C, Colin M, Matagne RF. Genetic mapping of mitochondrial markers by recombinational analysis in Chlamydomonas reinhardtii. Mol Gen Genet. 1995;249:185–90. doi: 10.1007/BF00290365. [DOI] [PubMed] [Google Scholar]
- Sager R, Ramanis Z. Chloroplast genetics of Chlamydomonas: II. Mapping by cosegregation frequency analysis. Genetics. 1976;83:323–40. doi: 10.1093/genetics/83.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillham NW. Organelle Genes and Genomes. New York, Oxford: Oxford University Press; 1994. [Google Scholar]
- Linnane AW, Nagley P. Mitochondrial genetics in perspective: the derivation of a genetic and physical map of the yeast mitochondrial genome. Plasmid. 1978;1:324–45. doi: 10.1016/0147-619x(78)90049-5. [DOI] [PubMed] [Google Scholar]
- Harris EH, Burkhart BD, Gillham NW, Boynton JE. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics. 1989;123:281–92. doi: 10.1093/genetics/123.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SM, Harris EH, Johnson AM, Boynton JE. Nonrandom distribution of chloroplast recombination events in Chlamydomonas reinhardtii: evidence for a hotspot and an adjacent cold region. Genetics. 1992;132:413–29. doi: 10.1093/genetics/132.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton JE, Gillham NW, Newman SM, Harris EH. Organelle genetics and transformation of Chlamydomonas. In: Herrmann RG, editor. Cell Organelles. Vienna, New York: Springer; 1992. pp. 3–64. [Google Scholar]
- VanWinkle-Swift KP, Birky CWJ. The non-reciprocality of organelle gene recombination in Chlamydomonas reinhardtii and Saccharomyces cerevisiae. Mol Gen Genet. 1978;166:193–209. doi: 10.1007/BF00285922. [DOI] [PubMed] [Google Scholar]
- Chiu W-L, Sears BB. Recombination between chloroplast DNAs does not occur in sexual crosses of Oenothera. Mol Gen Genet. 1985;198:525–8. doi: 10.1007/BF00332951. [DOI] [PubMed] [Google Scholar]
- Medgyesy P, Fejes E, Maliga P. Interspecific chloroplast recombination in a Nicotiana somatic hybrid (protoplast fusion/chloroplast DNA/physical mapping) Proc Natl Acad Sci USA. 1985;82:6960–4. doi: 10.1073/pnas.82.20.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh N, Medgyesy P. Limited chloroplast gene transfer via recombination overcomes plastome-genome incompatibility between Nicotiana tabacum and Solanum tuberosum. Plant Mol Biol. 1989;12:87–93. doi: 10.1007/BF00017450. [DOI] [PubMed] [Google Scholar]
- Gillham NW, Boynton JE, Harris EH. Transmission of plastid genes. In: Bogorad L, Vasil IK, editors. Cell Culture and Somatic Cell Genetics of Plants. San Diego, New York: Academic Press; 1991. pp. 55–92. [Google Scholar]
- Apitz J, Weihe A, Pohlheim F, Börner T. Biparental inheritance of organelles in Pelargonium: evidence for intergenomic recombination of mitochondrial DNA. Planta. 2013;237:509–15. doi: 10.1007/s00425-012-1768-x. [DOI] [PubMed] [Google Scholar]
- Maréchal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186:299–317. doi: 10.1111/j.1469-8137.2010.03195.x. [DOI] [PubMed] [Google Scholar]
- Scott I, Logan DC. Mitochondrial dynamics. In: Kempken F, editor. Plant Mitochondria. New York, Dordrecht, Heidelberg, London: Springer; 2011. pp. 31–63. [Google Scholar]
- Kraytsberg Y, Schwartz M, Brown TA, Ebralidse K. Recombination of human mitochondrial DNA. Science. 2004;304:981. doi: 10.1126/science.1096342. [DOI] [PubMed] [Google Scholar]
- Rokas A, Ladoukakis E, Zouros E. Animal mitochondrial DNA recombination revisited. Trends Ecol Evol. 2003;18:411–7. [Google Scholar]
- Smith ML, Duchesne LC, Bruhn JN, Anderson JB. Mitochondrial genetics in a natural population of the plant pathogen armillaria. Genetics. 1990;126:575–82. doi: 10.1093/genetics/126.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. Structure, function, and inheritance of plastid genomes. In: Bock R, editor. Cell and Molecular Biology of Plastids. Berlin, Heidelberg, New York: Springer; 2007. pp. 29–63. [Google Scholar]


