Abstract
Shigella flexneri is one of the major etiologic causes of shigellosis in Guizhou Province, China. However, the genetic characteristics of circulating isolates are unknown. Phenotypic and molecular profiles of 60 S. flexneri isolates recovered in Guizhou between 1972 to 1982 and 2008 to 2010 were determined. Nine serotypes (1a, 2a, 3a, 1b, 2b, X, Y, 4av and Yv) were identified. Multi-locus sequence typing differentiated the isolates into 20 sequence types (STs); 18 were novel. Four STs, ST 129, ST 100, ST 126 and ST 18, were most abundant, accounting for 65% of the isolates. Thirty-nine NotI-pulsed field gel electrophoresis patterns (pulsotypes, PTs) were observed; eight PTs were represented by more than one isolate with six isolates sharing the PT 13 profile. Multi-locus variable-nucleotide tandem-repeat analysis recognized 44 different types (MTs); seven MTs were represented by more than one isolate and MT 1 was most commonly encountered. Correlation between genetic relationships and serotypes was observed among the isolates studied; the majority of isolates belonging to the same serotype from different years clustered together based on the molecular data. These clustered isolates were also from similar geographical origins. These results enhance our understanding of genetic relationships between S. flexneri in Guizhou Province and can be used to help understand the changing etiology of shigellosis in China.
Introduction
Shigellosis, gastroenteritis caused by Shigella spp., is a major public health problem in both developing and developed countries [1, 2]. Approximately 164.7 million cases of shigellosis occur annually worldwide, resulting in 1.1 million deaths, primarily among children aged < 5 years [3]. Shigellosis occurs mainly in developing countries due to poor hygiene and limited access to clean drinking water; in industrialized countries the disease mainly results from travel to developing countries and exposure to contaminated foods and/or food handlers [1]. In China, Shigella spp. is one of the most frequently isolated gastrointestinal pathogens [4], accounting for up to 1.7 million episodes of shigellosis annually, with up to 200,000 patients admitted to hospitals [5, 6].
Guizhou Province, with nearly 50 million people, is an under-developed province in the southwest of China. Shigellosis has been one of the primary bacterial diseases in Guizhou in past decades, and from 2007 to 2010, 48,222 cases of shigellosis were reported [7]. Four species of Shigella can cause shigellosis but Shigella flexneri is the predominant species in China. Although there has been an observed shift in prevalence from S. flexneri to Shigella sonnei in recent years, S. flexneri is still one of the major etiologic cause of shigellosis in Guizhou Province [7].
Although Shigella has been a major source of disease over the past decades, information on the genetic relationships of circulating S. flexneri isolates from Guizhou Province is lacking. In this study, the molecular techniques multi-locus sequence typing (MLST), pulsed field gel electrophoresis (PFGE) and multi-locus variable-nucleotide tandem-repeat analysis (MLVA) were used to analyze the relationships between S. flexneri isolates recovered from Guizhou during the periods 1972 to 1982 and 2008 to 2010.
Material and Methods
Bacterial isolates and serotyping
Sixty isolates of S. flexneri, including 30 isolates recovered from 1972 to 1982 and 30 recovered from 2008 to 2010 in Guizhou Province, were analyzed (Table 1). The isolates were from Guiyang, Anshun, Qianxinan, Qiandongnan, Qiannan, Tongren and Zunyi Prefectures (seven of the nine prefectures making up Guizhou Province). All S. flexneri isolates were serotyped by slide agglutination using a commercially available monovalent antisera kit (Denka Seiken, Tokyo, Japan) and monoclonal antibody reagents (Reagensia AB, Sweden) per the manufacturer’s instructions [8]. S. flexneri isolates were routinly cultured in a 37°C incubator on Luria-Burtani (LB) agar plates or in an orbital shaker in LB broth.
Table 1. Isolation location, year and serotyping results of 60 S. flexneri isolates, Guizhou, 1972 to 1982 and 2008 to 2010.
| No. | Isolate No. | Prefecture | County | Year | Serotype |
|---|---|---|---|---|---|
| F01 | 1972GZ01 | Tongren | Songtao | 1972 | 2a |
| F02 | 1972GZ02 | Tongren | Songtao | 1972 | 2a |
| F03 | 1973GZ01 | Guiyang | Guiyang | 1973 | y |
| F04 | 1973GZ03 | Guiyang | Guiyang | 1973 | 4av |
| F05 | 1973GZ02 | Guiyang | Guiyang | 1973 | 2a |
| F06 | 1973GZ04 | Guiyang | Guiyang | 1973 | 3a |
| F07 | 1973GZ05 | Guiyang | Guiyang | 1973 | 3a |
| F08 | 1973GZ06 | Guiyang | Guiyang | 1973 | 3a |
| F09 | 1973GZ07 | Guiyang | Guiyang | 1973 | 3a |
| F10 | 1973GZ08 | Guiyang | Guiyang | 1973 | 2a |
| F11 | 1978GZ01 | Qiandongnan | Kaili | 1978 | Yv |
| F12 | 1981GZ01 | Zunyi | Qiannan | 1981 | 1a |
| F13 | 1981GZ02 | Zunyi | Qiannan | 1981 | 1a |
| F14 | 1982GZ01 | Qianxinan | Qinglong | 1982 | 3a |
| F15 | 1982GZ02 | Qianxinan | Puan | 1982 | 1a |
| F16 | 1982GZ03 | Qianxinan | Ceheng | 1982 | 2a |
| F17 | 1982GZ04 | Qianxinan | Zengfeng | 1982 | 1a |
| F18 | 1982GZ05 | Guiyang | Guiyang | 1982 | 1a |
| F19 | 1982GZ06 | Guiyang | Guiyang | 1982 | 1a |
| F20 | 1982GZ07 | Guiyang | Guiyang | 1982 | 1a |
| F22 | 1982GZ09 | Guiyang | Guiyang | 1982 | y |
| F23 | 1982GZ10 | Zunyi | Zunyi | 1982 | 1a |
| F24 | 1982GZ11 | Guiyang | Guiyang | 1982 | 1a |
| F25 | 1982GZ12 | Guiyang | Guiyang | 1982 | 1a |
| F26 | 1982GZ13 | Zunyi | Zunyi | 1982 | 1a |
| F27 | 1982GZ14 | Zunyi | Zunyi | 1982 | 1a |
| F28 | 1982GZ15 | Zunyi | Zunyi | 1982 | 1a |
| F29 | 1982GZ16 | Zunyi | Zunyi | 1982 | 1a |
| F30 | 1982GZ17 | Zunyi | Zunyi | 1982 | 1b |
| F31 | 1982GZ18 | Zunyi | Zunyi | 1982 | 1a |
| F32 | 2008GZ01 | Guiyang | Kaiyang | 2008 | 2a |
| F33 | 2008GZ02 | Guiyang | Kaiyang | 2008 | 2a |
| F34 | 2008GZ03 | Anshun | Ziyun | 2008 | 3a |
| F35 | 2008GZ07 | Anshun | Pingba | 2008 | 2a |
| F36 | 2008GZ08 | Anshun | Pingba | 2008 | 2a |
| F37 | 2008GZ11 | Guiyang | Kaiyang | 2008 | 3a |
| F38 | 2008GZ12 | Guiyang | Kaiyang | 2008 | 3a |
| F39 | 2008GZ13 | Guiyang | Kaiyang | 2008 | 3a |
| F40 | 2008GZ14 | Guiyang | Kaiyang | 2008 | 3a |
| F41 | 2008GZ15 | Guiyang | Kaiyang | 2008 | 3a |
| F42 | 2008GZ16 | Guiyang | Kaiyang | 2008 | 3a |
| F43 | 2008GZ17 | Guiyang | Kaiyang | 2008 | 3a |
| F44 | 2009GZ01 | Guiyang | Kaiyang | 2009 | 2a |
| F45 | 2009GZ02 | Guiyang | Kaiyang | 2009 | 2a |
| F46 | 2009GZ03 | Anshun | Pingba | 2009 | 2a |
| F47 | 2009GZ04 | Anshun | Pingba | 2009 | 2b |
| F48 | 2009GZ05 | Anshun | Pingba | 2009 | 2a |
| F49 | 2009GZ06 | Anshun | Pingba | 2009 | 2a |
| F50 | 2009GZ23 | Anshun | Ziyun | 2009 | 3a |
| F51 | 2009GZ25 | Anshun | Ziyun | 2009 | 2a |
| F52 | 2009GZ28 | Guiyang | Kaiyang | 2009 | 2a |
| F53 | 2009GZ29 | Guiyang | Kaiyang | 2009 | 2a |
| F54 | 2009GZ60 | Guiyang | Kaiyang | 2009 | 2a |
| F55 | 2009GZ68 | Anshun | Pingba | 2009 | 1a |
| F56 | 2010GZ01 | Anshun | Ziyun | 2010 | 2a |
| F57 | 2010GZ02 | Anshun | Ziyun | 2010 | 2a |
| F58 | 2010GZ03 | Anshun | Ziyun | 2010 | x |
| F59 | 2010GZ04 | Anshun | Ziyun | 2010 | x |
| F60 | 2010GZ05 | Anshun | Ziyun | 2010 | 2a |
| F61 | 2010GZ06 | Anshun | Ziyun | 2010 | 2a |
Preparation of DNA
Genomic DNA for PCR was prepared directly from bacterial colonies by the lysis by boiling method [8]. Briefly, a single colony from an overnight culture at 37°C on LB agar was suspended in 30 μl of distilled water and boiled at 100°C for 10 min. The sample was immediately cooled on ice for 5 min and centrifuged at 13,000 × g at 4°C for 10 min. The supernatant, containing DNA, was used as the template for PCR amplification.
MLST
MLST analysis of 15 housekeeping genes was performed as described on the EcMLST website (http://www.shigatox.net/ecmlst). PCR products were sequenced bi-directionally.
Each unique allele was assigned a different number and the allelic profile (string of fifteen allelic loci) was used to define each isolate’s sequence type (ST). New allele numbers and STs were submitted to the EcMLST curator for confirmation and allocation of a unique identifier. Clustering and minimum spanning tree (MST) analysis was used to infer relationships among the isolates using the fingerprint analysis software BioNumerics version 4.5 (Applied Maths, Kortrijk, Belgium) [9].
PFGE
PFGE analysis was performed using the method described by Ye et al. [9] using the restriction enzyme NotI. PFGE images were analyzed using BioNumerics. A PFGE pulsotype (PT) was defined as a pattern with one or more DNA bands different from other patterns. A dendrogram constructed using PFGE patterns was generated using the UPGMA algorithm with Dice-predicted similarity value of two patterns set at 1.0% pattern optimization and 0.8% band position tolerance.
MLVA
MLVA was performed as previously described [10]. Eight VNTR loci (SF3, SF4, SF6, SF7, SF8, SF9, SF10 and SF25) were selected. The forward primer for each primer set was labeled at its 5′ end with a compatible HEX, FAM, TAMRA, and ROX dye, respectively. The loci were amplified individually, with each 20 μl PCR mixture containing 1 μl each primer, 1 μl DNA template, 10 μl 2 × Taq MasterMix (Cowin Biotech, Beijing, China) and deionized water used to make up volume differences to 20 μl. PCR products were analyzed by capillary electrophoresis on an ABI 3730XL sequencer with GeneScan 500 LIZ Size Standard (Applied Biosystems Incorporated, Carlsbad, CA, USA) as described [11]. The copy number of each VNTR locus was incorporated into BioNumerics software and analyzed as described previously [12]. Each unique allelic string was designated a unique MLVA type (MT). A dendrogram was constructed by UPGMA clustering based on categorical coefficient analysis.
Results
Distribution of serotypes
The 60 S. flexneri isolates were grouped into nine serotypes (1a, 2a, 1b, 2b, 3a, X, Y, 4av and Yv) (Table 1). Three serotypes, 1a, 2a and 3a, were predominant. Serotype 1a was the most frequently identified serotype (50%, 15/30) among isolates from 1972 to 1982, however 2a was dominated from 2008 to 2010 (56.7%, 17/30) of isolates. Further, 93.8% (15/16) of the 1a isolates were from 1972 to1982, and 72.3% (17/22) of the 2a isolates were isolated during 2008 to 2010; 3a isolates were almost equally distributed across both time periods. Isolate expressing serotype 4av (1973GZ03) and Yv (1978GZ01), recently described by Sun et al [13–15], were recovered from diarrheal cases in 1973 and 1978, respectively.
MLST based genotypes
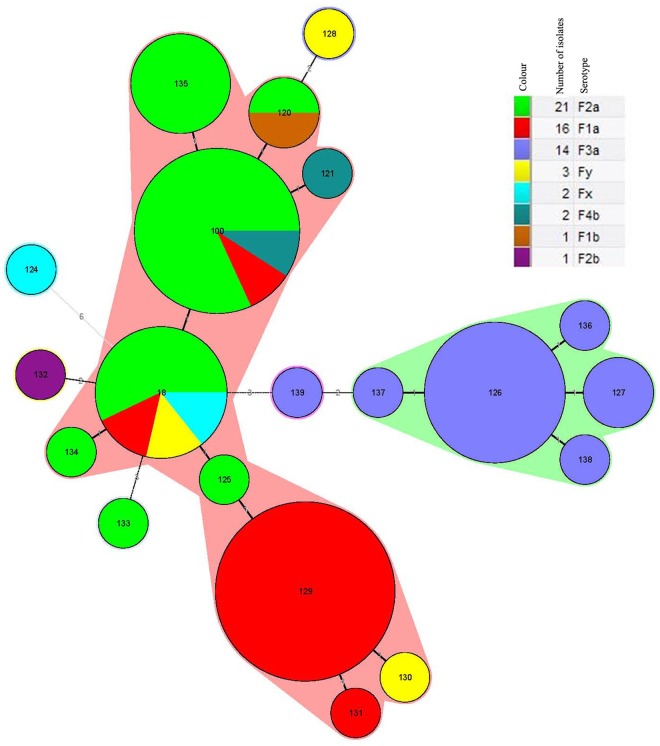
The 60 isolates were divided into 20 STs, among which 2 STs (ST 18 and ST 100) have been previously reported; the remaining 18 STs (ST 120, ST 121, ST 124—ST 139) were unique (Table 2). The most common STs identified were ST 129 (22%), including isolates of serotype 1a; ST 100 (18%) including isolates of serotype 1a, 2a and X; ST 126 (13%) including all 3a isolates and ST 18 (12%) including isolates of serotype 1a, 2a, and Y. Among the most common STs, ST 18 (except for isolate 2008GZ02) and ST 129 isolates were recovered from 1972 to 1982; ST 100 isolates were only recovered from 2009 to 2010; and ST 126 isolates were recovered from both time periods, respectively. Eleven STs (18%) were singletons (Fig. 1). The predominant ST from 1972 to 1982 was ST 129 (43.3%, 13/30), while ST 100 was the predominant ST (36.7%, 11/30) from 2008 to 2010. A MLST cluster tree of the isolates showed they were divided into two clusters, designated A and B, with an overall coefficient of similarity of 50% (Fig. 1). ST 124 (4av, isolate 1973GZ03 isolated in 1973) was the only isolate within cluster B, while the remaining 19 STs formed cluster A. Cluster A was further divided in to subclusters A1 (15 isolates) and A2 (44 isolates); all the isolates in cluster A1 belonged to serotype 3a with the exception of one serotype 2a (2008GZ01) isolate. The STs in cluster A1 included ST 126, ST 127, ST 133 and ST 136–139. Cluster A2 was further divided into two distinct branches A2a (28 isolates) and A2b (16 isolates); A2a included 20 of the 22 S. flexneri 2a isolates, 2 1a, 2 X, and one each of 1b, 2b, Y and Yv. Branch A2b contained 14 of the 16 S. flexneri 1a isolates and a single 2a and Y isolate. The cluster tree indicated that isolates belonging to the same serotype closely clustered based on the time of isolation. A minimum spanning tree (MST), based on the 20 STs indicated that 20 STs were divided into 2 clonal complexes (CCs) (CC 18 and CC 126) and four singletons; CC18 contains isolates expressing serotypes 1a, 2a, 2b, 4b, X and Y, and included 10 STs. In contrast all isolates in CC 126 were serotype 3a and included five STs; ST 124, ST 132, ST 133 and ST 139 were singletons (Fig. 2).
Table 2. S. flexneri MLST allelic profiles and ST designation, Guizhou, 1972 to 1982 and 2008 to 2010.
| IsolatesName | Alle profile | ST | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| arcA | aroE | aspC | clpX | cyaA | dNaG | fadD | grpE | icdA | lysP | mdh | mtlD | mutS | rpoS | uidA | ||
| 1972GZ01 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 18 |
| 1972GZ02 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 18 |
| 1973GZ01 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 18 |
| 1973GZ02 | 8 | 33 | 159 | 16 | 33 | 11 | 14 | 10 | 16 | 11 | 179 | 15 | 37 | 15 | 236 | 124 |
| 1973GZ03 | 8 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 125 |
| 1973GZ04 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 30 | 11 | 193 | 15 | 14 | 73 | 14 | 126 |
| 1973GZ05 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 207 | 11 | 193 | 15 | 14 | 73 | 14 | 127 |
| 1973GZ06 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 30 | 11 | 193 | 15 | 14 | 73 | 14 | 126 |
| 1973GZ07 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 30 | 11 | 193 | 15 | 14 | 73 | 14 | 126 |
| 1973GZ08 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 18 |
| 1978GZ-01 | 8 | 10 | 15 | 16 | 10 | 11 | 229 | 10 | 16 | 127 | 19 | 15 | 14 | 15 | 14 | 128 |
| 1981GZ01 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1981GZ02 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1982GZ01 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 30 | 11 | 193 | 15 | 14 | 73 | 14 | 126 |
| 1982GZ02 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1982GZ03 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 18 |
| 1982GZ04 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1982GZ05 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1982GZ06 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1982GZ07 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1982GZ09 | 28 | 10 | 13 | 189 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 130 |
| 1982GZ10 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1982GZ11 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 72 | 14 | 131 |
| 1982GZ12 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 18 |
| 1982GZ13 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1982GZ14 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1982GZ15 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1982GZ16 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 1982GZ17 | 8 | 34 | 13 | 189 | 10 | 11 | 14 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 132 |
| 1982GZ18 | 28 | 10 | 13 | 16 | 10 | 11 | 230 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 129 |
| 2008GZ01 | 8 | 10 | 15 | 16 | 10 | 11 | 14 | 10 | 207 | 11 | 19 | 15 | 14 | 73 | 14 | 133 |
| 2008GZ02 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 18 |
| 2008GZ03 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 207 | 11 | 193 | 15 | 14 | 73 | 14 | 127 |
| 2008GZ07 | 8 | 10 | 171 | 16 | 10 | 11 | 14 | 10 | 16 | 11 | 19 | 15 | 14 | 15 | 14 | 134 |
| 2008GZ08 | 8 | 10 | 171 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 135 |
| 2008GZ11 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 30 | 23 | 193 | 15 | 14 | 73 | 14 | 136 |
| 2008GZ12 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 30 | 11 | 193 | 15 | 14 | 73 | 14 | 126 |
| 2008GZ13 | 8 | 35 | 13 | 16 | 10 | 11 | 14 | 30 | 30 | 11 | 193 | 15 | 14 | 73 | 14 | 137 |
| 2008GZ14 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 30 | 11 | 193 | 15 | 17 | 73 | 14 | 138 |
| 2008GZ15 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 30 | 11 | 193 | 15 | 14 | 73 | 14 | 126 |
| 2008GZ16 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 30 | 11 | 193 | 15 | 14 | 73 | 14 | 126 |
| 2008GZ17 | 8 | 35 | 13 | 16 | 10 | 11 | 14 | 30 | 30 | 11 | 19 | 15 | 14 | 15 | 14 | 139 |
| 2009GZ01 | 8 | 10 | 15 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 120 |
| 2009GZ02 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 100 |
| 2009GZ03 | 8 | 10 | 171 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 135 |
| 2009GZ04 | 8 | 10 | 15 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 120 |
| 2009GZ05 | 8 | 10 | 171 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 135 |
| 2009GZ06 | 8 | 10 | 171 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 135 |
| 2009GZ23 | 8 | 35 | 15 | 16 | 10 | 11 | 14 | 30 | 30 | 11 | 193 | 15 | 14 | 73 | 14 | 126 |
| 2009GZ25 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 100 |
| 2009GZ28 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 100 |
| 2009GZ29 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 100 |
| 2009GZ60 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 100 |
| 2009GZ68 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 100 |
| 2010GZ01 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 100 |
| 2010GZ02 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 100 |
| 2010GZ03 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 100 |
| 2010GZ04 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 17 | 14 | 121 |
| 2010GZ05 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 100 |
| 2010GZ06 | 8 | 10 | 13 | 16 | 10 | 11 | 14 | 10 | 16 | 23 | 19 | 15 | 14 | 15 | 14 | 100 |
Fig 1. MLST clustering tree of S. flexneri isolates, Guizhou, from 1972 to 1982 and 2008 to 2010.
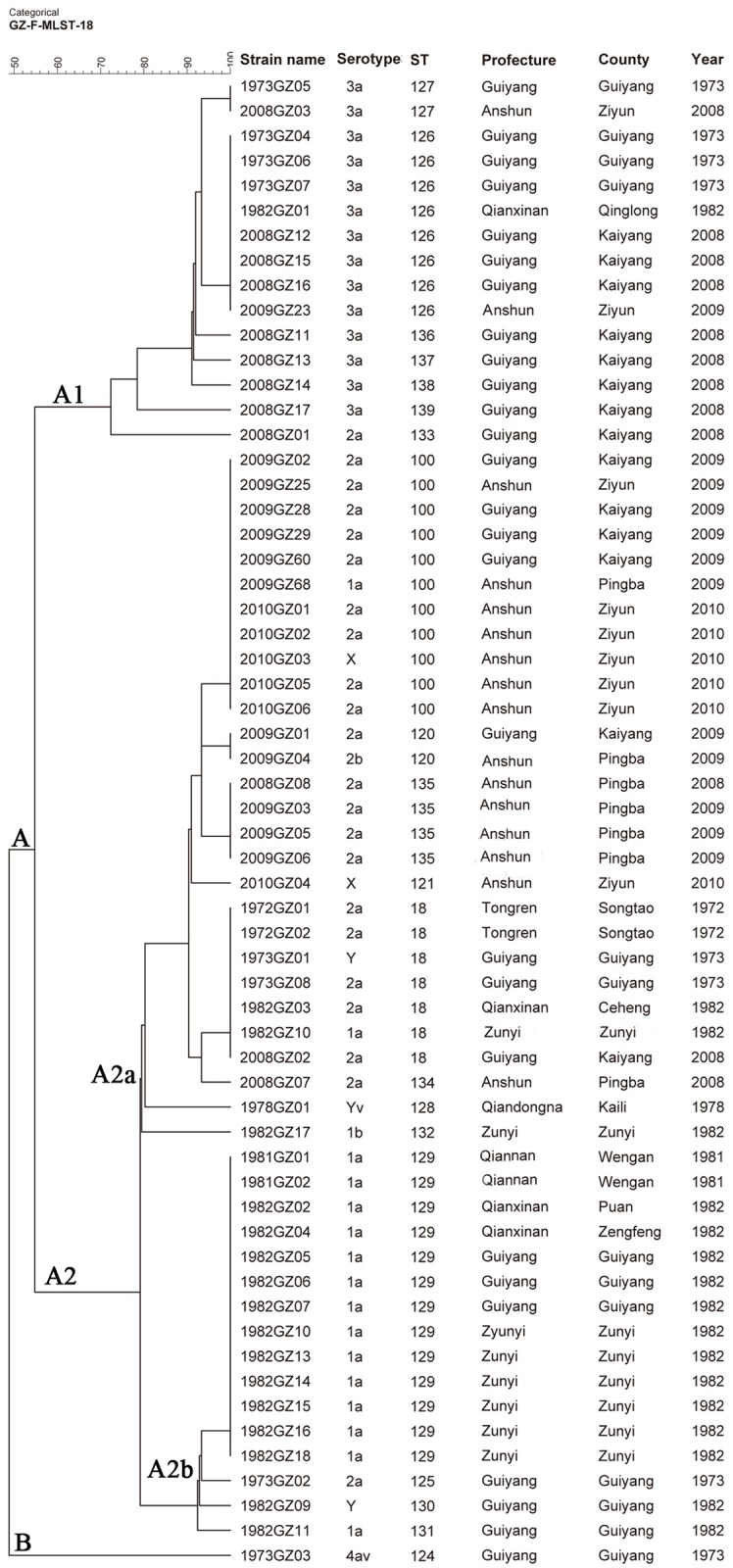
The 60 isolates from Guizhou province were analyzed using a 15 allele MLST as described in the Materials and Methods.
Fig 2. Genetic relationships of S. flexneri isolates recovered from Guizhou Province based on MLST.
The minimum spanning tree was constructed using the 20 identified STs obtained from the 60 Guizhou Province isolates. Each circle corresponds to a single ST. The shadow zones in different color correspond to different clonal complexes. The size of the circle is proportional to the number of the isolates, and the color within the cycles represents the serotypes of the isolates. The corresponding color, serotype, number of isolates and back ground information are shown on the right of the minimum spanning tree.
PFGE based Genotypes
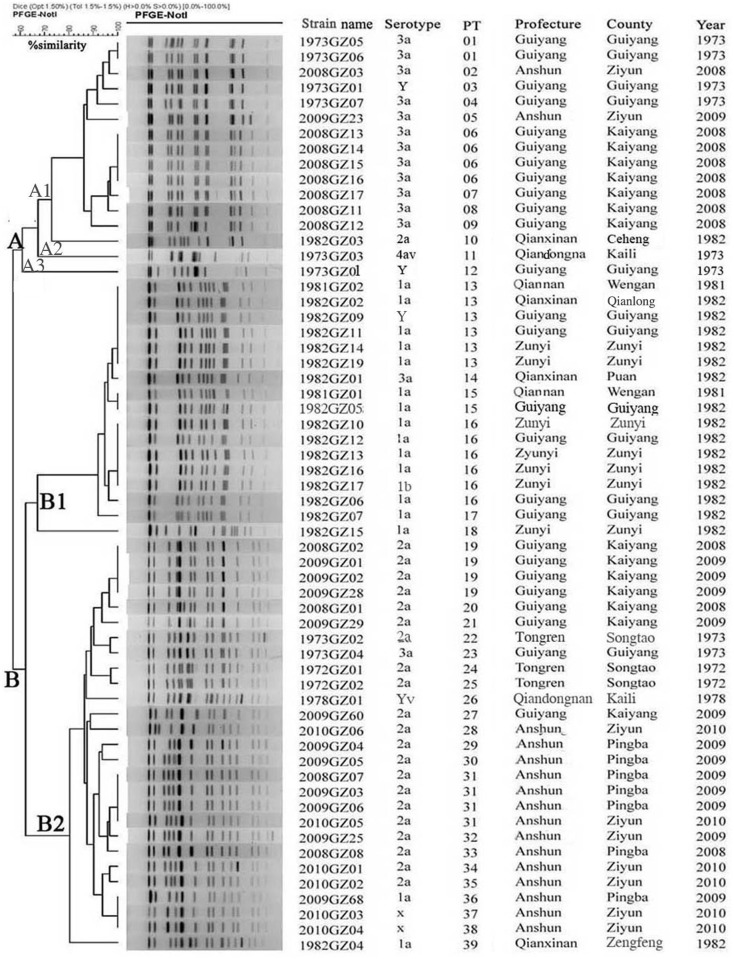
The genotypes and genetic relatedness of the Guizhou S. flexneri isolates were also determined using PFGE. NotI-digested S. flexneri DNA generated 39 reproducible unique PTs, each with 12–17 bands. Eight patterns were represented by more than one isolate with PT 13 containing the greatest number of isolates, followed by PT 16. Among the isolates from 1972 to 1982, the predominant PT was PT 13 or PT 16, each representing 20.0% (6/30) of the total, while the predominant PT among isolates from 2008 to 2010 was PT 06, PT 19 or PT 31, each representing 13.3% (4/30) of the total. All 60 isolates were related at a coefficient of similarity of 60%, but two main clusters could be distinguished at a 62% similarity value (cluster A and B; Fig. 3). Cluster A was split into two additional broad subgroups, A1 (15 isolates) and A2 (1 isolate). The majority (12 of 14) serotype 3a isolates, with the exception of isolates 1973GZ04 and 1982GZ01, grouped together in A1; single isolates of serotype 2a, 4av and Y complete this group. Subgroup A2 contained a single Y serotype isolate. Cluster B split into two subgroups as well, subgroup B1 (17 isolates) and B2 (27 isolates). The majority of serotype 1a isolates (14 of 16) were found in subgroup B1; the three remaining isolates expressed 1b, 3a and Y. Subgroup 2B contained 21 of the 22 serotype 2a isolates, 2 isolates of 1a and X and single isolates of 3a and Yv. Similar to MLST, the majority of isolates expressing the same serotype were closely clustered and this was also related to the year of isolation. Additionally, the isolate (isolate 1973GZ03 recovered in 1973) of serotype 4av, was formed cluster A3, while the serotype Yv isolate, shared a unique but similar PT and was included in subgroup B2.
Fig 3. Relationship of S. flexneri isolates recovered from Guizhou based on NotI-PFGE analysis.
The dendrogram were constructed using UPGMA. The corresponding PFGE pattern, serotype and background information are shown to the right of the dendrogram.
MLVA typing
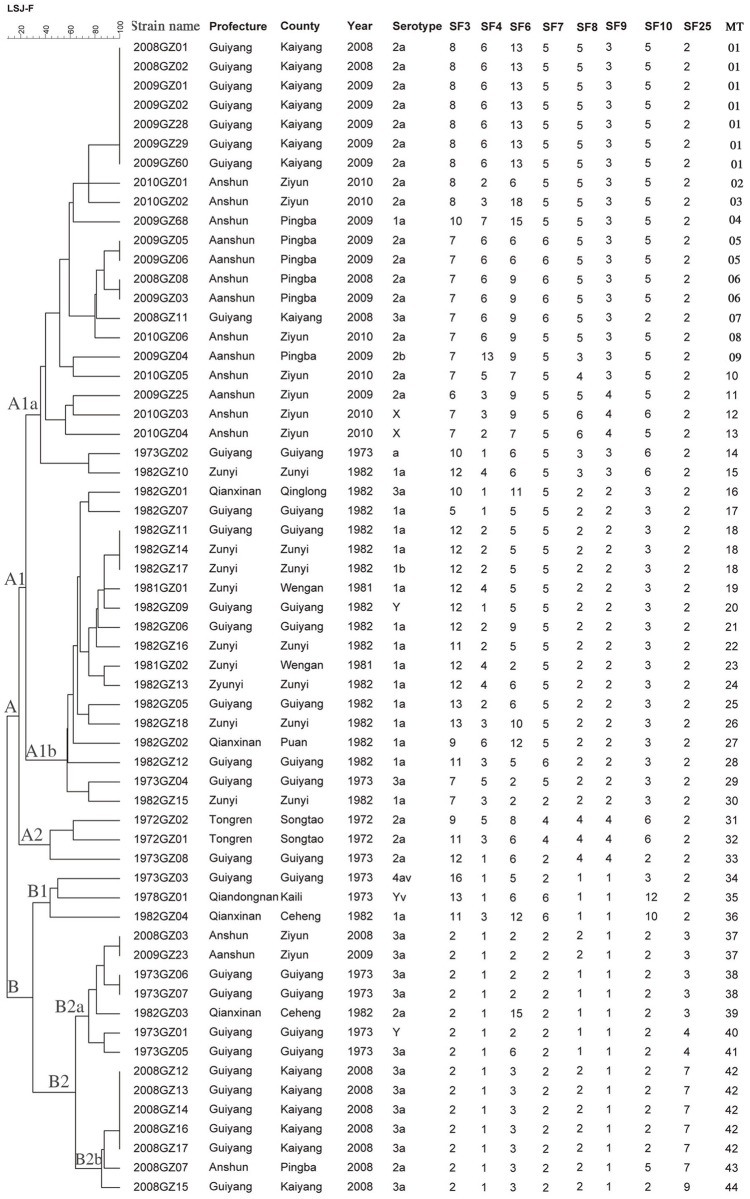
Using MLVA, the 60 S. flexneri isolates grouped to 44 different MTs (Fig. 4). Seven MTs were represented by more than one isolate with MT 1 occurring most frequently (n = 7) followed by MT 42 (n = 5). The predominant MT among isolates recovered from 2008 to 2010 was MT 1 (23.3%; 7/30), however the predominant MT among isolates from 1972 to 1982 was not obvious. MLVA analysis showed the greatest diversity among the 60 S. flexneri isolates resulting in an extensively branched tree. Like MLST and PFGE, two main clusters, A and B, were observed (Fig. 4). The majority of serotype 1a and 2a isolates were assigned in cluster A, in which the 1a and 2a isolates were further grouped in cluster A1 and A2, respectively, whereas most of the serotype 3a isolates were grouped in cluster B. Isolates of serotype X assigned to cluster A showed relatively close relationship to serotype 2a, while the serotype Y isolates were closely related to serotype 1a or 3a isolates. Isolates belonging to the same serotype but recovered from different years showed clear relatedness, indicated by grouping in the same clusters. For example, serotype 3a isolates recovered from 1972 to 1982 and 2008 to 2010 clustered together, and similar characteristics were also observed for isolates belonging to serotype 1a and 2a (Fig. 4). Serotype 4av and Yv isolates were closely grouped in cluster B1. Additionally, isolates belonging to the same serotype and within a close time span clustered together based on geographical origin. For example, 2a isolates recovered in Guiyang, Ziyun and Pingba shared very similar patterns.
Fig 4. Relationship of S. flexneri isolates recovered from Guizhou based on MLVA.
Isolates were analyzed using an eight VNTR loci MLVA scheme. The dendrogram was constructed using UPGMA. The corresponding MLVA type with copy numbers for the eight VNTRs, serotype, and background information are shown to the right of the dendrogram.
Discussion
In the present study, the genetic characteristics of 60 isolates of S. flexneri recovered from Guizhou Province between 1972 to 1982 and 2008 to 2010 were systematically studied. Thirty of the S. flexneri isolates were from shigellosis cases from 1972 to 1982 and the remaining isolates were recovered from patients with shigellosis from 2008 to 2010. The serotypes of the isolates used in this study included 1a, 2a, 3a 1b, 2b, X, Y, 4av and Yv; isolates were also from seven of the nine prefectures in Guizhou Province. Serotyping results indicated that S. flexneri 1a and 2a were the predominant serotypes from 1972 to 1982 (94%) and 2008 to 2010 (72%), respectively, while 3a isolates were almost equally recovered in both periods. The predominant serotype (2a) in Guizhou Province recovered from 2008 to 2010 are consistent with isolates from Suzhou of Jiangsu, Henan and Shanxi Province [16–18], but are different from isolates of other provinces such as Beijing (4a and 4b) and Jiading of Shanghai Province [19, 20]. It is noteworthy that isolates of serotype 4av and Yv were recovered as early as 1973 and 1978, respectively, indicating the early emergence of these serotypes in China.
Recently a number of genotyping methods with higher discriminatory power than serotyping or biochemical testing such as MLST [21, 22], PFGE [5, 23] and MLVA [10, 11, 24] were introduced to characterize Shigella isolates. These methods are based primarily based on changes in isolate genotype, permitting analysis of phylogenetic relationships. Analysis of the isolates can be helpful for clinical diagnosis, treatment, prevention and control of shigellosis. Choi et al. [22] showed that S. flexneri serotypes 1–5, X and Y clustered together in a group containing many allelic variants while serotype 6 formed a distinct group, as previously established [25, 26]. Wang et al.[10] reported that phylogenetic groupings of 242 S. flexneri isolates recovered from shigellosis cases in Taiwan between 1995 to 2008, based on PFGE and MLVA profiles, correlated with serotype and isolate origin. Two distinct clusters for isolates of serotype 3 were shown but only one distinct cluster for each of the serotype groups 1a/1b/NT, 2a/2b/X/NT, 4a/Y, and 6 were revealed. Serologically different isolates including serotype Y and subserotype 4a; serotype X and subserotype 2b; subserotypes 1a and 1b, and subserotypes 3a and 3b, were genetically more closely related than indicated by serotyping alone.
Ye et al. [9] previously analyzed 37 serotype X and 69 serotype 1a, 2a, 2b, 3a, 4a, 5b, and Y isolates from China; all belonged to ST91 (later renamed ST 100), and concluded that S. flexneri epidemics in China have been caused by a single epidemic clone, ST 100. In this study, 60 isolates of S. flexneri from Guizhou Province separated into 20 STs based on a 15 loci MLST scheme; 18 of the STs were novel. The most common STs from 1972 to 1982 were ST 18, ST 126 and ST 129, however, ST 100 and ST 126 appeared between 2008 to 2010. Our results suggested that the predominant ST was ST 129 from 1972 to 1982, while ST 100 was the the predominant ST during 2008 to 2010, and the predominant ST is consistent with ST of isolates from other provinces of China [9]. Isolates belonging to the same serotype clustered in accordance with the year of isolation using all three genotyping approaches. MST indicated that the 20 STs were divided into 2 CCs, CC 18 and CC 126, and 4 singletons. CC 18 contained isolates expressing serotypes 1a, 2a, 2b, 4b, X and Y, while all the isolates in CC 126 belonged to serotype 3a. In addition, both the cluster tree and the MST, based on MLST data, showed that the isolates of serotype 4av (1973GZ03) was distant from the isolates belonging to other serotypes.
PFGE is a broadly applicable typing method with a high degree of intra- and inter laboratory reproducibility when standardized protocols are followed [23]. It has been shown to be a powerful tool in the laboratory for discriminating Shigella isolates during an outbreak [27]. In this study, PFGE discriminated the 60 isolates of S. flexneri into 39 unique PFGE patterns. Isolates belonging to the same serotypes mainly clustered together; the most closely related isolates were temporally associated with one another was well, suggesting that some drift was associated within each serotype over time. For instance, isolates of serotype 2a in cluster B2 isolated during the period of 1972 to 1982 and 2008 to 2010 were closely clustered, respectively, and similar clustering characteristics were observed for the isolates belonging to serotype 1a (cluster B1) and 3a (cluster A1), but some isolates, such as 2008GZ03, 1982GZ03 and 2009GZ23, were assigned irrespective their isolation time. Additionally, isolates with similar geographic origin were also often grouped by PFGE together as they tended to express the same serotypes.
MLVA is a prominent typing tool which has been used for characterizing S. flexneri; it has also been a useful tool for phylogenetic analysis [10]. In the present study, the S. flexneri isolates were discriminated into 44 different MTs and showed a low (approximately 20%) coefficient of similarity, indicating the high discriminatory power of the MLVA method. This finding is consistent with a previous study showing MLVA exhibited a discriminatory power greater than PFGE [10]. For most of isolates belonging to serotypes 1a, 2a and 3a, MLVA results correlated with serotyping. However, isolates of serotype X and Y were associated with serotype 1a and 2a isolates, respectively, and the serotype 4av isolate was closely related to serotype 3a isolates; this is similar to results observed previously [10]. Similar to PFGE, the majority of isolates belonging to the same serotype were temporally and geographically related.
In this study, isolates 1973GZ03 and 1978GZ01were serologically identified as serotype 4av and Yv respectively. These serotypes differ from 4a and Y because they react with monoclonal antibody MASF IV-1 [13–15]. In theory, serotype 4av and Yv should have originated from serotype 4a and Y, primarily differing only in the acquisition of a 6.8k plasmid carrying a phospheantransferase gene (opt) responding for the MASF IV-1 antigenic determinant in these isolates [13–15]. In this study, isolates of serotype of 4av (1973GZ03) and Yv (1978GZ01) were typed as ST 124 and ST 128 with MLST, respectively. MLST showed that 4av isolate were genetically distinct from isolates belonging to other serotypes, while Yv isolates were relatively close to isolates of serotype 2a. Molecular analysis indicates that isolate 1978GZ01 carries a dysfunctional gtrII gene within genome, and hence it is genetically similar to isolates from serotype 2a (unpublished data). This would explain the observed similarity between isolate 1978GZ01 and the majority of serotype 2a isolates.
Conclusions
In conclusion, phenotypic and molecular profiles of 60 S. flexneri isolates recovered in Guizhou between 1972 to 1982 and 2008 to 2010 were analysed. Nine serotypes (1a, 2a, 3a, 1b, 2b, X, Y, 4av and Yv) were identified, and the predominant serotype has changed from 1a to 2a in Guizhou Province. MLST differentiated the isolates into 20 sequence types (STs); 18 were novel. Four STs, ST 129, ST 100, ST 126 and ST 18, were most abundant, accounting for 65% of the isolates. The predominant ST was ST 129 in 1972 to 1982, while ST 100 was the predominant ST during 2008 to 2010. Thirty-nine NotI-PFGE (pulsotypes, PTs) were observed; eight PTs were represented by more than one isolate with six isolates sharing the PT 13 profile. MLVA analysis recognized 44 different types (MTs); seven MTs were represented by more than one isolate and MT 1 was most commonly encountered. Correlation between genetic relationships and serotypes was observed among the isolates studied; the majority of isolates belonging to the same serotype from different years clustered together based on the molecular data. These clustered isolates were also from similar geographical origins.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the grant of national science and technology major project of China (No. 2009ZX10602-12), Talent Base Funds for Infectious Disease Control and Prevention in Guizhou Provincial Government (No. Qian Ren Ling Fa [2013] 15), National Natural Science Foundation of China (No. 81271788 and 81290345), National Basic Research Priorities Program (No. 2011CB504901) and National Key Program for Infectious Diseases of China (No. 2013ZX10004221, 013ZX10004216-001-002 and 2ZX10004215. We acknowledge the use of the EcMLST database which is operated by the Microbial Evolution Laboratory at Michigan State University. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Niyogi SK (2005) Shigellosis. J Microbiol 43: 133–143. [PubMed] [Google Scholar]
- 2. Izumiya H, Tada Y, Ito K, Morita-Ishihara T, Ohnishi M, et al. (2009) Characterization of Shigella sonnei isolates from travel-associated cases in Japan. Journal of medical microbiology 58: 1486–1491. 10.1099/jmm.0.011809-0 [DOI] [PubMed] [Google Scholar]
- 3. von Seidlein L, Kim DR, Ali M, Lee H, Wang X, et al. (2006) A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS medicine 3: e353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qiu S, Wang Y, Xu X, Li P, Hao R, et al. (2013) Multidrug-resistant atypical variants of Shigella flexneri in China. Emerging infectious diseases 19: 1147–1150. 10.3201/eid1907.111221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xia S, Xu B, Huang L, Zhao JY, Ran L, et al. (2011) Prevalence and characterization of human Shigella infections in Henan Province, China, in 2006. Journal of clinical microbiology 49: 232–242. 10.1128/JCM.01508-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang XY, Tao F, Xiao D, Lee H, Deen J, et al. (2006) Trend and disease burden of bacillary dysentery in China (1991–2000). Bulletin of the World Health Organization 84: 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wei X, Tian K, You L, Ma Q, Liu Y, et al. (2012) Epidemic Characteristics and Etiological Analysis of Bacillary Ib’sentery in Guizhou Province During the Period of 2007–2010. Practical Preventive Medicine 19: 1185. [Google Scholar]
- 8. Sun Q, Lan R, Wang Y, Zhao A, Zhang S, et al. (2011) Development of a multiplex PCR assay targeting O-antigen modification genes for molecular serotyping of Shigella flexneri. Journal of clinical microbiology 49: 3766–3770. 10.1128/JCM.01259-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ye C, Lan R, Xia S, Zhang J, Sun Q, et al. (2010) Emergence of a new multidrug-resistant serotype X variant in an epidemic clone of Shigella flexneri. Journal of clinical microbiology 48: 419–426. 10.1128/JCM.00614-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang YW, Watanabe H, Phung DC, Tung SK, Lee YS, et al. (2009) Multilocus variable-number tandem repeat analysis for molecular typing and phylogenetic analysis of Shigella flexneri. BMC microbiology 9: 278 10.1186/1471-2180-9-278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chiou CS, Watanabe H, Wang YW, Wang WL, Terajima J, et al. (2009) Utility of multilocus variable-number tandem-repeat analysis as a molecular tool for phylogenetic analysis of Shigella sonnei. Journal of clinical microbiology 47: 1149–1154. 10.1128/JCM.01607-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hyytia-Trees E, Smole SC, Fields PA, Swaminathan B, Ribot EM (2006) Second generation subtyping: a proposed PulseNet protocol for multiple-locus variable-number tandem repeat analysis of Shiga toxin-producing Escherichia coli O157 (STEC O157). Foodborne pathogens and disease 3: 118–131. [DOI] [PubMed] [Google Scholar]
- 13. Sun Q, Lan R, Wang J, Xia S, Wang Y, et al. (2013) Identification and characterization of a novel Shigella flexneri serotype Yv in China. PloS one 8: e70238 10.1371/journal.pone.0070238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun Q, Knirel YA, Lan R, Wang J, Senchenkova SN, et al. (2012) A novel plasmid-encoded serotype conversion mechanism through addition of phosphoethanolamine to the O-antigen of Shigella flexneri. PloS one 7: e46095 10.1371/journal.pone.0046095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Knirel YA, Lan R, Senchenkova SN, Wang J, Shashkov AS, et al. (2013) O-antigen structure of Shigella flexneri serotype Yv and effect of the lpt-O gene variation on phosphoethanolamine modification of S. flexneri O-antigens. Glycobiology 23: 475–485. 10.1093/glycob/cws222 [DOI] [PubMed] [Google Scholar]
- 16. Zhu Li TQ, Zou W, Zhang M (2013) Analysis on serotype and antimicrobial resistance of Shigella during 2010–2011 in Suzhou. Jiangsu Journal of Preventive Medicine 24: 3. [Google Scholar]
- 17. Ru W HL, Zhao J (2011) Serotype distribution and drug resistance analysis of shigella in Henan province between 2001 and 2010. Chinese Journal of Practical Medicine 38: 4. [Google Scholar]
- 18.Zang Z LJ, Xu Y, Wu D (2009) Analysis of Community-acquired Shigella Infection and Drug Resistance in Xian Area 2004–2008
- 19. Journal of Modern Laboratory Medicine 24: 3. [Google Scholar]
- 20. Liu J MY, Wang L, Cui J, Wang Y, Ji Y (2011) Analysis on Change of Serum Types and Drug Resistance of Shigella in Xicheng District of Beijing from 2008 to 2010. Occupation and Health 27: 3. [Google Scholar]
- 21. Chen W WJ, Yu W, Yu Y (2006) Study on the distribution and the level of resistance to drugs of Shigella in Jiading District in 2004. Modern preventive medicine 33: 2. [Google Scholar]
- 22. Cao Y, Wei D, Kamara IL, Chen W (2012) Multi-Locus Sequence Typing (MLST) and Repetitive Extragenic Palindromic Polymerase Chain Reaction (REP-PCR), characterization of shigella spp. over two decades in Tianjin China. International journal of molecular epidemiology and genetics 3: 321–332. [PMC free article] [PubMed] [Google Scholar]
- 23. Choi SY, Jeon YS, Lee JH, Choi B, Moon SH, et al. (2007) Multilocus sequence typing analysis of Shigella flexneri isolates collected in Asian countries. Journal of medical microbiology 56: 1460–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, et al. (2006) Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne pathogens and disease 3: 59–67. [DOI] [PubMed] [Google Scholar]
- 25. Gorge O, Lopez S, Hilaire V, Lisanti O, Ramisse V, et al. (2008) Selection and validation of a multilocus variable-number tandem-repeat analysis panel for typing Shigella spp. Journal of clinical microbiology 46: 1026–1036. 10.1128/JCM.02027-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Islam MA, Huq M, Nabi A, Talukdar PK, Ahmed D, et al. (2013) Occurrence and characterization of multidrug-resistant New Delhi metallo-beta-lactamase-1-producing bacteria isolated between 2003 and 2010 in Bangladesh. Journal of medical microbiology 62: 62–68. 10.1099/jmm.0.048066-0 [DOI] [PubMed] [Google Scholar]
- 27. Ahmed AM, Shimamoto T (2014) Isolation and molecular characterization of Salmonella enterica, Escherichia coli O157:H7 and Shigella spp. from meat and dairy products in Egypt. International journal of food microbiology 168–169: 57–62. 10.1016/j.ijfoodmicro.2013.10.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.