Abstract
Arbutoid mycorrhizal plants are commonly found as understory vegetation in forests worldwide where ectomycorrhiza-forming trees occur. Comarostaphylis arbutoides (Ericaceae) is a tropical woody plant and common in tropical Central America. This plant forms arbutoid mycorrhiza, whereas only associations with Leccinum monticola as well as Sebacina sp. are described so far. We collected arbutoid mycorrhizas of C. arbutoides from the Cerro de la Muerte (Cordillera de Talamanca), Costa Rica, where this plant species grows together with Quercus costaricensis. We provide here the first evidence of mycorrhizal status for the Ascomycete Leotia cf. lubrica (Helotiales) that was so far under discussion as saprophyte or mycorrhizal. This fungus formed arbutoid mycorrhiza with C. arbutoides. The morphotype was described morphologically and anatomically. Leotia cf. lubrica was identified using molecular methods, such as sequencing the internal-transcribed spacer (ITS) and the large subunit (LSU) ribosomal DNA regions, as well as phylogenetic analyses. Specific plant primers were used to confirm C. arbutoides as the host plant of the leotioid mycorrhiza.
Keywords: Arbutoid mycorrhiza, Anatomy, Morphology, Phylogeny, Costa Rica, Leotiomycetes
Introduction
The tropical woody plant Comarostaphylis arbutoides Lindl. (Ericaceae) is common in tropical Central America (c. 2,500–3,430 m height above sea level (a.s.l.)), where it can form extensive thickets. Based on fruit and flower morphology, anatomy, and phytochemistry, this plant is a member of the subfamily Arbutoideae (Hileman et al. 2001; Fig. 1a). The Arbutoideae also include the circumboreal Arctostaphylos uva-ursi and species of Arbutus that are all known to form arbutoid mycorrhiza (Molina and Trappe 1982a; Münzenberger et al. 1992; Osmundson et al. 2007). The mycorrhizal fungus induces the branching of the lateral roots to form mostly, with some exceptions (Molina and Trappe 1982a), a pinnate-cruciate branching pattern that is typical for this mycorrhizal type (Massicotte et al. 1993). The arbutoid mycorrhiza is characterized by a hyphal mantle, a para-epidermal Hartig net and intracellular hyphae penetrating the living epidermal cells of the host (Münzenberger et al. 1992; Selosse et al. 2007). Suberization of the outer cortical layer leads to the formation of an exodermis and prevents penetration of the fungus into deeper root cell layers (Münzenberger et al. 1992; Massicotte et al. 1993).
Fig. 1.

a Leafs and flowers of the Ericaceae Comarostaphylis arbutoides. Photo by Roy E. Halling. b Two fruit bodies and one initial (arrowhead) of Leotia lubrica found at Estación Biologíca de la Muerte (site I), Costa Rica. Ascomata, 10–40 mm. Photo by Katja Kühdorf. c Habit of the mycorrhiza Leotia cf. lubrica-Comarostaphylis arbutoides. Mycorrhiza arbutoid ramified, mantle smooth to moderately hairy, and transparent with foreign brown hyphae; bar, 0.5 mm
C. arbutoides is a refuge plant for ectomycorrhizal fungi as it shares these fungi with ectomycorrhizal tropical trees such as Quercus costaricensis (Halling and Mueller 2003; Kühdorf et al. 2014). After forest clearcutting of the economically important forest trees, arbutoid mycorrhizal plants host the ectomycorrhizal fungi that contribute to forest recovery later on (Dahlberg 1990; Visser 1995; Molina et al. 1997; Horton et al. 1999; Hagerman et al. 2001). Ectomycorrhizal fungi of Arbutoideae show identical morphology (apart from, e.g., host-depending branching pattern and habit dimensions) and hyphal mantle anatomy as the ectomycorrhiza (ECM) of other hosts such as Pinus, Picea, Pseudotsuga, and Quercus (Zak 1976a, b; Molina and Trappe 1982b; Mühlmann and Göbl 2006; Kühdorf et al. 2014). The ericaceous hosts Arbutus menziesii and A. uva-ursi lack mycorrhizal specificity, which means numerous ectomycorrhizal fungi are able to colonize these ericads (Molina and Trappe 1982a; Acsai and Largent 1983a, b; Massicotte et al. 1994; Richard et al. 2005; Kennedy et al. 2012).
Osmundson et al. (2007) were the first to describe the arbutoid mycorrhiza of C. arbutoides and Leccinum monticola anatomically and molecularly. To date, only one more morphotype has been described in literature (Kühdorf et al. 2014). However, Halling and Mueller (2004) mention that species of other mycorrhizal genera such as Tricholoma, Cortinarius, Russula, and Laccaria are likely associates of C. arbutoides.
The ecology of the helotialean fungi is diverse. Members of the Helotiales have been described as plant pathogens, ECM and ericoid mycorrhiza formers, terrestrial and aquatic saprobes, and wood rot fungi (Wang et al. 2006a). The genus Leotia (Helotiales) is globally distributed (Wang et al. 2006a, b), whereas the ecological role of the Ascomycete is not clear. Species of Leotia and Microglossum are found usually on humus-rich soil, sometimes on decaying wood, but rarely on leaf litter (Arora 1986). Leotia was already recorded as mycorrhiza-forming fungus by Molina et al. (1992). This status was later speculated to be saprotrophic by Rinaldi et al. (2008) based on insufficient evidence. Both Zeller et al. (2007) and Seitzman et al. (2011) later found the 13C signature of the species L. lubrica comparable with those of ectomycorrhizal fungi and therefore, conclude that this fungus should be mycorrhizal. Furthermore, L. lubrica was reported to form ECM with diverse plant species such as Quercus rotundifolia, Polygonum sp., and Nothofagus menziesii (Branco and Ree 2010; Gao and Yang 2010; Orlovich et al. 2013). However, these records have been proven by DNA sequencing only and were not further described.
The genus Leotia contains three species of inoperculate discomycetes with stipitate-capitate ascomata. They are constructed, at least in part, of tissues built-up of hyphae imbedded in a gelatinous matrix (Zhong and Pfister 2004). They are distinguished by the color of their fresh ascomata: species with a yellow stipe and yellow hymenia are assigned to Leotia lubrica, those with an entirely green ascomata to Leotia atrovirens. Species of Leotia viscosa have a yellow stalk and green hymenia (Durand 1908; Mains 1956; Grund and Harrison 1967).
Here, we describe arbutoid mycorrhizal systems morphologically and anatomically according to Agerer (1991), formed by L. cf. lubrica and C. arbutoides from the Cerro de la Muerte (Cordillera de Talamanca), Costa Rica. L. cf. lubrica was identified using molecular methods such as internal-transcribed spacer (ITS) and large subunit (LSU) sequencing as well as phylogenetic analysis. Plant primers were used to sequence the ITS region of C. arbutoides from the same arbutoid mycorrhiza as used for fungal analysis.
Materials and methods
Site description and sampling
Sampling was conducted at two forest sites around the Mountain Cerro de la Muerte (3,491 m a.s.l.) in the Cordillera de Talamanca of Costa Rica, 54 km southeast of the capital San José. Both sites are secondary cloud forests and about 1.4 km apart from each other: Estación Biologíca de la Muerte (site I; 3,100 m a.s.l.; 9° 33′ N, 83° 45′ W) and Reserva Forestal Los Santos (site II; 3,300 m a.s.l.; 9° 34′ N, 83° 45′ W). Site I is dominated by Q. costaricensis mixed with solitary individuals of C. arbutoides. At sampling site II, C. arbutoides itself is the dominating species, mixed with a few individuals of Q. costaricensis. The understorey species consist of members of the families Araliaceae (Schefflera and Oreopanax), Cunoniaceae (Weinmannia), Ericaceae (Cavendishia, Disterigma, and Vaccinium), Poaceae (Chusquea), Primulaceae (Myrsine), and Winteraceae (Drimys).
Fine root systems of C. arbutoides were collected during the rainy seasons in October 2010 and 2011. For this, a soil corer (diameter, 3 cm; length, 40 cm) was used at distances of 50 and 100 cm from the trunk. Within these 2 years, a total of 60 soil cores were taken and analyzed. At the University of Costa Rica, turgid and apparently healthy morphotypes were sorted out using a stereomicroscope. Systems with the same morphological features (e.g., color, hydrophobicity presence, emanating elements, and rhizomorphs) were assigned to one morphotype. For further analyses, the morphotypes were preserved in 2 % glutaraldehyde with 0.1 M sodium cacodylate buffer (Münzenberger et al. 2009) for light microscopy or dried on silica gel for DNA extraction, respectively. Identification of each morphotype is based on their respective sequence type. The genus Leotia was proven genetically in six soil cores.
DNA extraction, PCR, and sequencing
One unramified root tip per morphotype was used for DNA extraction using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s recommendations. For phylogenetic analysis at family and species level, the ribosomal nuclear LSU and the ITS region from the ribosomal DNA (rDNA) were amplified. For this purpose, the primer combinations LR0R/LR5 (Moncalvo et al. 2000) and ITS1F/ITS4 (Gardes and Bruns 1993; White et al. 1990) were used. To identify the plant from mycorrhizal roots without co-amplifying fungal DNA, the angiosperm-specific ITS primer pair ITS-5A/ITS-241r was amplified (Osmundson et al. 2007). Direct sequencing of PCR products was performed using the PCR primers as sequencing primers. Sequencing service was facilitated by GATC Biotech AG (Konstanz, Germany).
Identification and phylogenetic analysis
All fungal sequences obtained for ITS and LSU rDNA were analyzed and edited using Chromas Lite v2.01 software (http://technelysium.com.au). Sequence comparisons were performed in the NCBI database (http://www.ncbi.nlm.nih.gov/) using Megablast, and the database UNITE (Kõljalg et al. 2005; http://unite.ut.ee/) using blastn. To calculate the phylogenetic tree of the ITS region, the 100 most similar sequences for each reference sequence in NCBI database were downloaded and complemented with an additional search in the nucleotide database and sequences of other publications as well. Alignment was performed with the program MAFFT v7 (Katoh et al. 2002) using the FFT-NS-2 alignment algorithm. To estimate phylogenetic relationships, we used maximum likelihood and Bayesian approaches. Maximum likelihood analyses was performed using RAxML (v7.3.2; Stamatakis 2006; Stamatakis et al. 2008) in a parallelized version supplied by Bioportal (http://bioportal.uio.no/) with eight parallel processors and trees inferred from 10,000 rapid bootstrap analyses as starting trees in a heuristic search for the tree with the highest likelihood. GTRCAT was used in the heuristic search and the final evaluation of the best tree found was based on the GTR + Gamma model. The Bayesian analyses were performed using MrBayes v3.2.1 (Ronquist et al. 2012) on an iMac (2.9 GHz Quad-Core Intel Core i5). The GTR + Gamma model was in effect, and four chains in two parallel runs were performed for 2,000,000 generations. The first 50,000 trees were discarded before calculating the posterior probabilities.
Microscopy
The morphological and anatomical description of the mycorrhizas was carried out according to Agerer (1987–2012, 1991), and using the online key of DEEMY (Agerer and Rambold 2004–2014). The iodine reaction specially adapted to Ascomycetes was done after Baral (1987, 2009). Anatomical studies are based on 15 mycorrhizal systems. Drawings were performed by using an interference contrast microscope (BX50F-3, Olympus Corporation, Tokyo, Japan) connected with a drawing tube. All drawings were made at a thousand-fold magnification.
For semi-thin sections, the mycorrhizas were fixed with 2 % glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) at room temperature until further processing. Thereafter, six washes in 0.1 M sodium cacodylate buffer were performed. Samples were postfixed in 1 % osmium tetroxide in the same buffer for 1 h under light exclusion at room temperature. After six washes in double-distilled water, samples were dehydrated by immersion for 15 min each in 25, 50, 70, and 95 % acetone and three times for 1 h in 100 % acetone (Münzenberger et al. 2009). The mycorrhizal tips were embedded in Spurr’s plastic (Spurr 1969) and sectioned with a diamond knife on an Ultracut Reichert Ultramicrotome (W. Reichert-LABTEC, Wolfratshausen, Germany). The sections (0.5 μm thick) were stained with crystal violet and investigated by use of a light microscope (Zeiss Axioskop 50, Oberkochen, Germany).
Fruit bodies of L. lubrica
During 2 years of sampling, fruit bodies of L. lubrica were found only once and were coincidentally documented. For that reason, no further genetic investigations were carried out to clearly identify the fungus. As the three species of the genus Leotia are easy to distinguish morphologically, the ascomata (Fig. 1b) were preliminary identified as L. lubrica, based on the yellow color of both the stipe and the hymenium.
Results
Morpho-anatomical description of the mycorrhiza Leotia cf. lubrica-C. arbutoides
Morphological characters
(Fig. 1c) Mycorrhizal systems arbutoid, with 0–1 (4) orders of ramification, solitary or in small numbers, up to 2.8 mm long, main axis at 0.3 mm diameter; mantle surface hydrophilic and smooth to moderately hairy, of contact exploration type or short-distance exploration type. Unramified ends straight to slightly bent, cylindric, not inflated, 0.1–0.7 (1.4) μm long, 0.2–0.3 (0.1) μm diameter, mantle consistently transparent and yellowish to light orange, older parts dark orange to ochre. Surface of unramified ends smooth to occasionally hairy, cortical cells (correspond to epidermal cells) visible. Cystidia not distinct under stereomicroscope magnification. Rhizomorphs not found. Sclerotia not observed.
Anatomical characters of the mantle in plan views
(Figs. 2 and 3a–d) Mantle plectenchymatous throughout, all hyphae clampless and smooth; laticifers are lacking. Outer mantle layers (Figs. 2a and 3a) hyphae arranged net-like; hyphae frequently branched, often with merged hyphal tips, matrix lacking (mantle type E; Agerer 1991); hyphae septate, even and straight, not constricted at septa; hyphae with closed anastomoses, anastomosal bridge long, mostly thinner than hyphae; hyphae with numerous oily droplets, droplets light yellow to light orange; cytoplasm colorless; hyphae 10–90 μm long, 0.7–3.1 μm diameter; cell walls 0.2–0.4 μm thick; septa as thick as cell walls and often difficult to discern due to frequent droplets. Middle mantle layers (Figs. 2d, e and 3b) contain a nongelatinous matrix, hyphae irregularly interwoven, repeatedly branched and septate, colorless, no discernible pattern; frequently merged hyphal tips, hyphae at distal end simple; hyphae 15–50 (90) μm long, 0.9–2.4 μm diameter; cell walls 0.2 μm thick; septa as thick as cell walls; anastomoses frequently and very variable in shape; anastomoses open or closed by a simple septum; anastomosal bridge long, short, or almost lacking; bridge thinner or thicker than hyphae or as thick as hyphae; cell walls of anastomoses as thick as remaining wall; anastomoses close to hyphal tips not found. Inner mantle layers (Figs. 2f, h and 3c) ring-like arrangement of hyphal bundles; hyphae even or irregularly inflated, at distal end simple or slightly inflated; hyphae rarely septate, colorless; hyphae (7) 20–130 μm long, 1.5–3.9 μm diameter; cell walls 0.2–0.3 μm thick, septa as thick as cell walls; anastomoses frequently, anastomoses open, with short bridge or bridge almost lacking, bridge thinner or thicker than hyphae, or as thick as hyphae; cell walls of anastomoses as thick as remaining walls; anastomoses close to hyphal tips also present. Very tip (Figs. 2g, h and 3d) inner mantle layers similar to remaining part but without hyphal bundles; hyphae often irregularly inflated, many hyphae with hyphal tips; outer and middle mantle layers organized as older parts of the mantle.
Fig. 2.
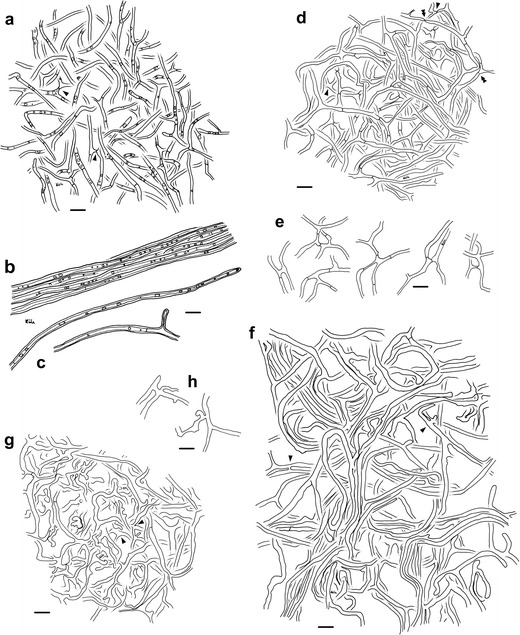
Arbutoid mycorrhiza of Leotia cf. lubrica-Comarostaphylis arbutoides. a–f Plan view of different mantle layers, emanating hyphae and anastomoses; bars, 10 μm: a outer mantle layer with ramified hyphae, irregularly arranged; hyphae with numerous oily droplets; anastomoses with closed long bridge (arrowheads); b bundle of emanating hyphae, hyphae with oily droplets; c simple and branched emanating hyphae; d middle mantle layer with repeatedly branched hyphae, densely arranged; anastomoses with short or long bridge, bridge closed or open (single arrowheads); merged hyphal tips, some with remnants of a partly solved septum (double arrowheads); e different anastomoses of the middle mantle layer; f inner mantle layer with ring-like arrangement of hyphal bundles, some hyphae irregularly inflated; anastomoses (arrowheads); g inner mantle layer close to the very tip with many irregularly inflated hyphae, many hyphae with hyphal tips; different anastomoses (arrowheads); h anastomoses of both the inner mantle layer and the inner mantle layer close to very tip, anastomosis close to the hyphal tip
Fig. 3.
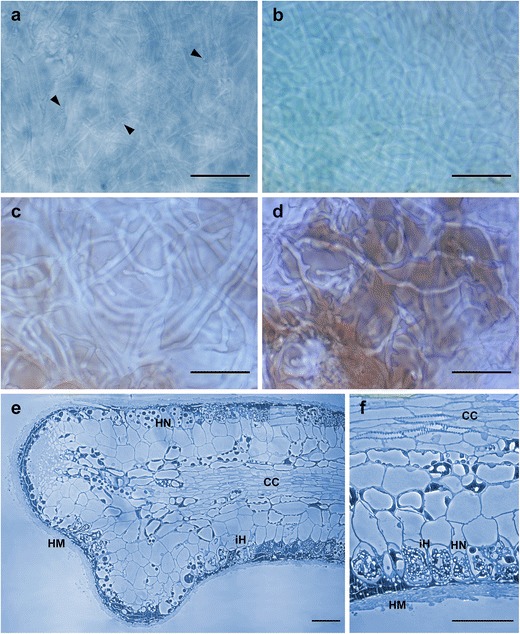
a–d Interference contrast of the three mantle layers of leotioid mycorrhiza; bars, 20 μm: a outer mantle layer, hyphae containing numerous oily droplets (arrowheads). b Middle mantle layer. c Inner mantle layer. d Inner mantle layer close to very tip with brownish matrix (remnants of root cap cells). e–f Semi-thin sections of the arbutoid mycorrhiza Leotia cf. lubrica-Comarostaphylis arbutoides; bars, 50 μm: e hyphal mantle (HM), Hartig net (HN), intracellular hyphae (iH), and central cylinder (CC). f Anatomical features in detail
Anatomical characters of emanating elements
(Fig. 2b, c) Rhizomorphs lacking. Emanating hyphae rare to frequently, not specifically distributed; hyphae even, not striking, sometimes one-side branched, sometimes with open anastomoses; hyphal tips sometimes merged with hyphae; some hyphae in bundles; hyphae at distal end simple; hyphae straight and even, up to 500 μm long, maybe longer since often observed without hyphal tip, 2.3–4.2 μm diameter; cell walls 0.5–1.2 μm thick, not constricted at septa, septa 0.2–0.3 (0.6) μm thick, septa often difficult to discern due to frequent droplets; cell wall light yellow to light orange, cytoplasm light dirty blue, oily droplets same color as cell wall; lacking are clamps, elbow-like protrusions, and intrahyphal hyphae. Cystidia not found.
Anatomical characters of longitudinal section
(Fig. 3e, f) Mantle plectenchymatous, 5–17 μm thick. Mantle of very tip plectenchymatous, 8–20 μm thick. Epidermal layer with intracellular hyphae, epidermal cells radially oval to eliptic; Hartig net around epidermal cells para-epidermal in one row; hyphal cells roundish to cylindrical. Tannin cells lacking.
Color reactions with different reagents (mantle preparations and emanating hyphae)
Acetic acid: no reaction; congo red: no reaction; cotton blue: hyphae of outer mantle layer with blue cytoplasm and blue (sometimes reddish) oily droplets, emanating hyphae with violet (sometimes reddish) cell walls, blue cytoplasm, and red to red brown oily droplets; ethanol 70 %: no reaction; Fe(II)SO4: no reaction; guaiac: no reaction; KOH 10 %: no reaction; lactic acid: no reaction; Lugol’s solution: no reaction; Melzer’s reagent: no reaction; NH4OH: no reaction; sulpho-vanillin: no reaction; H2SO4 concentration: no reaction; toluidine blue: hyphae of outer mantle layer with patchy pale blue to blue cytoplasm and red oily droplets, emanating hyphae with violet cell walls, patchy pale blue to blue cytoplasm, and red oily droplets.
Reference specimen
Costa Rica, province of San José, canton of Pérez Zeledón, at mountain Cerro de la Muerte, Reserva Forestal Los Santos (3,300 m a.s.l.; precipitation c. 2,812 mm/year; inceptisol (USDA)), in a secondary cloud forest with Q. costaricensis, soil core exc., myc. isol. Katja Kühdorf; KKM 337 and KKM 348, 18 October 2011; mycorrhiza deposited by B. Münzenberger (ZALF Müncheberg, Germany). Further material studied same location, soil core exc., myc. isol. Katja Kühdorf; KKM 334 and KKM 347, 18 October 2011; mycorrhiza deposited by B. Münzenberger (ZALF Müncheberg, Germany).
Phylogenetic analyses
A total of 399 root tips were analyzed genetically, of which ten were identified as Leotia cf. lubrica. All leotioid sequences were deposited in NCBI GenBank under the accession numbers KF836622-KF836631 (LSU) and KF836612-KF836621 (ITS), respectively. In all samples, C. arbutoides (KF419121) was proven as host tree.
The sequenced LSU rDNA region of the ten arbutoid mycorrhizas of C. arbutoides resulted in sequences with a length of 890–913 bp (KF836622–KF836631), in which the overlapping area was different in five positions within these samples. Sequence comparison with BLASTn in NCBI database resulted in matches mainly belonging to the order Helotiales, where the Leotiaceae L. lubrica (AY789359) showed the highest similarity values (Table 1). In UNITE, best values were reached with the Helotiaceae Unguiculariopsis thallophila (UDB016232), which also belongs to the Helotiales. In the sequenced 563–616 bp long ITS region (KF836612-KF836621) of the arbutoid mycorrhizas of C. arbutoides, 30 positions were different in the overlapping area. Sequences obtained from NCBI and UNITE comparison with ITS sequences belong to members of the genus Leotia (Table 1), whereby UNITE provides lower similarity values.
Table 1.
Comparison of ITS and LSU sequences with NCBI and UNITE database obtained from ten mycobionts of Comarostaphylis arbutoides
| Samples with accession numbers (ITS; LSU) | NCBI (ITS) | UNITE (ITS) | NCBI (LSU) | UNITE (LSU) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Closest matcha | Highest maximum score | E value/query coverage (%) | Identity (%) | Closest matcha | Highest bit-score | E value/query coverage (%) | Identity (%) | Closest matcha | Highest maximum score | E value/query coverage (%) | Identity (%) | Closest matcha | Highest bit-score | E value/query coverage (%) | Identity (%) | |
| KKM 145 (KF836612; KF836622) | Leotia lubrica (EU819412) | 1,072 | 0.0/99 | 99 | Leotia sp. (UDB013464) | 504 | e − 143/56 | 94 | Leotia lubrica (AY789359) | 1,650 | 0.0/100 | 99 | Unguiculariopsis thallophila (UDB016232) | 985 | 0.0/–a | –a |
| KKM 147 (KF836613; KF836623) | Leotia viscosa (AY144536) | 942 | 0.0/100 | 97 | Leotia sp. (UDB013464) | 478 | e − 135/60 | 94 | Leotia lubrica (AY789359) | 1,644 | 0.0/100 | 99 | Unguiculariopsis thallophila (UDB016232) | 979 | 0.0/–a | –a |
| KKM 152 (KF836614; KF836624) | Leotia lubrica (AY144544) | 1,064 | 0.0/99 | 99 | Leotia sp. (UDB013464) | 496 | e − 140/56 | 94 | Leotia lubrica (AY789359) | 1,644 | 0.0/100 | 99 | Unguiculariopsis thallophila (UDB016232) | 977 | 0.0/–a | –a |
| Leotia lubrica (AY144551) | 1,064 | 0.0/99 | 99 | |||||||||||||
| KKM 317 (KF836615; KF836625) | Leotia lubrica (AY144547) | 1,083 | 0.0/97 | 99 | Leotia sp. (UDB013464) | 533 | e − 152/55 | 95 | Leotia lubrica (AY789359) | 1,631 | 0.0/99 | 99 | Unguiculariopsis thallophila (UDB016232) | 971 | 0.0/–a | –a |
| Leotia lubrica (AY144548) | 1,083 | 0.0/97 | 99 | |||||||||||||
| KKM 334 (KF836616; KF836626) | Leotia lubrica (EU819412) | 1,092 | 0.0/99 | 99 | Leotia sp. (UDB013464) | 496 | e − 140/55 | 94 | Leotia lubrica (AY789359) | 1,639 | 0.0/99 | 99 | Unguiculariopsis thallophila (UDB016232) | 971 | 0.0/–a | –a |
| KKM 337 (KF836617; KF836627) | Leotia lubrica (EU819412) | 1,092 | 0.0/99 | 99 | Leotia sp. (UDB013464) | 496 | e − 140/55 | 94 | Leotia lubrica (AY789359) | 1,629 | 0.0/100 | 99 | Unguiculariopsis thallophila (UDB016232) | 961 | 0.0/–a | –a |
| KKM 347 (KF836618; KF836628) | Leotia lubrica (EU819412) | 1,022 | 0.0/99 | 99 | Leotia sp. (UDB013464) | 496 | e − 140/58 | 94 | Leotia lubrica (AY789359) | 1,604 | 0.0/100 | 99 | Unguiculariopsis thallophila (UDB016232) | 938 | 0.0/–a | –a |
| KKM 348 (KF836619; KF836629) | Leotia lubrica (EU819412) | 1,092 | 0.0/99 | 99 | Leotia sp. (UDB013464) | 496 | e − 140/55 | 94 | Leotia lubrica (AY789359) | 1,602 | 0.0/100 | 99 | Unguiculariopsis thallophila (UDB016232) | 938 | 0.0/–a | –a |
| KKM 349 (KF836620; KF836630) | Leotia lubrica (EU819412) | 1,024 | 0.0/99 | 99 | Leotia sp. (UDB013464) | 496 | e − 140/58 | 94 | Leotia lubrica (AY789359) | 1,602 | 0.0/100 | 99 | Unguiculariopsis thallophila (UDB016232) | 938 | 0.0/–a | –a |
| KKM 427 (KF836621; KF836631) | Leotia lubrica (EU819412) | 1,092 | 0.0/99 | 99 | Leotia sp. (UDB013464) | 496 | e − 140/55 | 94 | Leotia lubrica (AY789359) | 1,637 | 0.0/100 | 99 | Unguiculariopsis thallophila (UDB016232) | 969 | 0.0/–a | –a |
Closest match was chosen according to the highest maximum score or bit-score
aInformation locked by the reference author
The Bayesian and RAxML phylogenies, generated by ITS sequences are concordant. Both trees show the same grouping structure, supported by high posterior probabilities (PP) in the Bayesian analysis and by typically lower bootstraps (BS) in the RAxML analysis (Fig. 4). The phylogenetic analysis in Fig. 4 reveals that the three species of the genus Leotia are paraphyletic and split in several groups. Members of the species L. atrovirens can be found in two groups (III and IV), whereby both are highly supported (PP 1/BS 100, each). Samples of L. lubrica are also divided and can be found exclusively in group II (PP 1/BS 99) on the one hand, and as a large complex together with L. viscosa samples in group I (PP 1/BS 78) on the other. In this L. lubrica/viscosa complex, again several subgroups (a–f) are formed, containing exclusively L. lubrica (b, d, e, f; PP 1/BS 90-100) and L. viscosa (c; PP 1/BS 98) species, respectively, as well as a mixture of L. lubrica and L. viscosa samples (a).
Fig. 4.
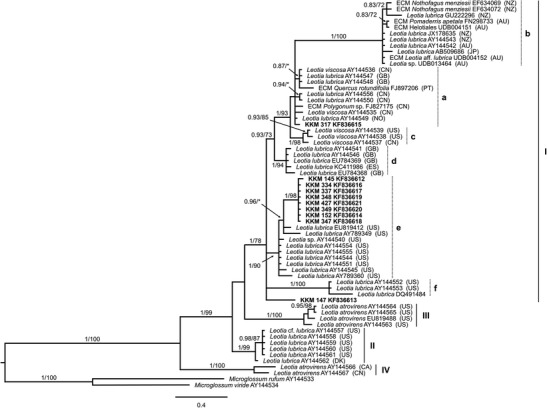
Phylogenetic relationship of ten leotioid mycobionts of Comarostaphylis arbutoides within the genus Leotia. Phylogram was obtained from Bayesian analysis based on ITS sequences. Branch support values were calculated as posterior probability from 2,000,000 generations of Bayesian analysis (first number) and as bootstrap support from RAxML analysis (second number). Values below 70 % are indicated with asterisks or omitted. The phylogram was rooted with Microglossum rufum and M. viride. Sequences were obtained from NCBI and UNITE database complemented by the name of corresponding host plant, if available. Investigated arbutoid mycobionts of C. arbutoides from Costa Rica are marked in bold. Mycorrhizal type: ectomycorrhiza (ECM). Country codes, if applicable: Australia (AU), Canada (CA), China (CN), Denmark (DK), Spain (ES), UK (GB), Japan (JP), Norway (NO), New Zealand (NZ), Portugal (PT), and USA (US). Indicated groups (I–IV) and subgroups (a–f) after Zhong and Pfister (2004)
All investigated leotioid arbutoid mycorrhizas of C. arbutoides can be found in group I, whereby eight group together (PP 1/BS 98) and nest within samples of L. lubrica (subgroup e; PP 1/BS 90). Therefore, these samples were identified as L. cf. lubrica species. The other two mycorrhizas do not group to a specific Leotia species. KKM 317 (KF836615) can be found among various L. lubrica and L. viscosa samples within subgroup a, which is not well supported, whereas the sample KKM 147 (KF836613) does not cluster in one of the above mentioned subgroups.
Discussion
The classification of Ascomycota was traditionally based on the morphology of their fruit bodies, whereas, molecular studies showed that such morphologically defined groups can be phylogenetically misleading (Wang et al. 2006a, b). Currently, the five orders Cyttariales, Erysiphales, Helotiales, Rhytismatales, and Thelebolales are placed in the ascomycetous class Leotiomycetes (Hibbett et al. 2007). The genus Leotia Pers. belongs to the Leotiaceae (Helotiales) whose final composition of genera is not fully resolved. Currently, Lumbsch and Huhndorf (2009) place the genera Geocoryne, Gelatinipulvinella, Leotia, Microglossum, Neobulgaria, and Pezoloma into this family. Sufficient molecular information of currently included genera is rarely or not at all available in NCBI database. Therefore, further reinterpretations within the Leotiaceae family are expected. Nevertheless, the obtained LSU rDNA sequences of the ten arbutoid mycorrhizas of C. arbutoides (KF836622–KF836631) identify them as members of the genus Leotia (Table 1).
Thus far, a detailed phylogenetic investigation within the genus Leotia has been carried out only by Zhong and Pfister (2004). They combined morphological information with phylogenetic analyses and found four groups within the genus Leotia. Group I comprises all L. viscosa and some L. lubrica samples, which are characterized by a yellow stipe in fresh and dry condition, respectively. Another group (II) exclusively formed by L. lubrica samples, however, showed a green stipe when dry. The species L. atrovirens is divided into two groups (III and IV) and differ from each other by the presence or absence of gel in their stipes.
Genetically, eight of ten arbutoid mycorrhizas of C. arbutoides collected in Costa Rica were identified as L. cf. lubrica. The other two leotioid mycorrhizas samples KKM 147 (KF836613) and KKM 317 (KF836615) still remain unidentified, whereby the first one represents a further genotype within the L. lubrica/viscosa complex, because it does not cluster in a specific subgroup within group I. However, those two leotioid mycorrhizas are also assumed to be L. lubrica species, since they were found as mycorrhizal partner of C. arbutoides. This suggests a high genetic variability of L. lubrica species assigned to group I.
The stipes of the L. lubrica fruit bodies found at site I, were not further investigated regarding a possible color change in dried condition or genetically. As pointed out by Zhong and Pfister (2004), it is difficult to distinguish between L. lubrica species of group I and II, as both have a yellow stipe in fresh conditions. Thus, it is not possible to clearly assign the found fruit bodies to one of both groups. However, thus far mycorrhizal associations are only known from group I L. lubrica samples, not from L. lubrica species belonging to group II. Among C. arbutoides, the species Q. rotundifolia, Polygonum sp., and N. menziesii are reported as host plants of leotioid ECMs (Branco and Ree 2010; Gao and Yang 2010; Orlovich et al. 2013). Orlovich et al. (2013) additionally indicate that Leotia perhaps interact in some way with other ectomycorrhizal fungi, since the fungus was found with either Russula, Clavulina, or Laccaria at the same root tip. Tedersoo et al. (2009) identified L. lubrica from ectomycorrhizal roots formed by Basidiomycota and the Rhamnaceae Pomaderris apetala (FN298733) and suggest a secondary colonization of ECMs by this fungal species (Tedersoo et al. 2010). However, such secondary mycorrhizal association for the described leotioid arbutoid mycorrhizas of C. arbutoides is not assumed. Color of leotioid mycorrhiza is similar to the fruit bodies of L. lubrica and all investigated samples show the same morpho- and anatomotype. Additionally, semi-thin sections reveal a mantle, Hartig net, as well as intracellular hyphae, typical for mycorrhizas of the Arbutoideae (Fig. 3e, f). However, only samples of L. cf. lubrica clustering in subgroup e were investigated, so no conclusion can be made if there are morphological or anatomical differences regarding the leotioid mycorrhizas KKM 147 (KF836613) and KKM 317 (KF836615).
The L. cf. lubrica arbutoid mycorrhiza is morphologically characterized by a hydrophilic, consistently transparent and yellowish colored mantle. According to Agerer and Rambold (2004–2014), these features are in common with the ECM of the Basidiomycete Entoloma nitidum (Montecchio et al. 2006). Given their smooth to moderately hairy surface, the leotioid arbutoid mycorrhiza is assigned to the contact exploration type or short distance exploration type (Agerer 2001). By contrast, the mantle of the E. nitidum ECM features abundant rhizomorphs and is therefore assigned to the medium distance exploration type (Montecchio et al. 2006). Anatomically, all mantle layers lack a matrix and offer hyphae with clamps, which make it also different from the leotioid mycorrhiza.
Anatomically, the L. cf. lubrica arbutoid mycorrhiza is characterized by a continuous plectenchymatous mantle, wherein the middle mantle layer is embedded in a nongelatinous matrix. Anastomoses can be found in all layers, showing various types. Emanating hyphae also show anastomoses, which confirm them as such structures and not as cystidia (Agerer 1999). The yellowish droplets are another characteristic feature, only found in emanating hyphae and in hyphae of the outer mantle layer. The ECM of the hypogeous Ascomycete Gautieria inapire (Palfner and Horak 2001) also show a plectenchymatous mantle in all layers with clampless hyphae and oily droplets, which do not stain in sulpho-vanillin (Agerer and Rambold 2004–2014). However, it differs from L. cf. lubrica arbutoid mycorrhiza in having cystidia, emanating hyphae, and rhizomorphs. A matrix is also present, but shows, by contrast, a gelatinous condition and can be found only in the inner mantle layer.
Color reactions of the leotioid arbutoid mycorrhiza are observed with cotton blue and toluidine blue, which are restricted to the hyphae of the outer mantle layer, emanating hyphae, and the droplets within. Both chemicals cause a metachromatic reaction due to the color change of droplets to red. Additionally, cell walls of the emanating hyphae turn violet in cotton blue and are thus cyanophil. In contrast, other applied chemicals cause no reaction, for instance, dissolution of the droplets.
Evidence of amyloidity or dextrinoidity in fungal structures is an important characteristic feature in taxonomy (Baral 1987; Agerer and Rambold 2004–2014). These blue (amyloid) or red to red-brown (dextrinoid) iodine-based reactions are induced by Melzer’s reagent as well as Lugol’s solution. According to Baral (1987) there exists a special case of amyloidity, called hemiamyloidity. Here, Lugol’s solution provokes a red reaction, whereas Melzer’s reagent yields no reaction at all due to chloral hydrate contained within. However, a pretreatment with KOH causes a blue reaction in both, Lugol’s and Melzer’s. This hemiamyloid color reaction is so far only known in Ascomycetes, including many Helotiales (Baral 2009). Among mycologists, Melzer’s reagent is prefered over Lugol’s solution. This makes it more difficult to observe clearly a hemiamyloid reaction as Melzer’s reagent solely used may falsely indicate inamyloidity (Baral 2009). For that reason, a possible hemiamyloid reaction of Ascomycete mycorrhizas could be overlooked. Regardless which treatment was applied, the arbutoid mycorrhiza of L. cf. lubrica reacts inamyloid. Zhong and Pfister (2004) confirm this reaction also for the cap of the ascoma for all three species of the genus Leotia.
The present study is the first morpho-anatomical proof that the species L. cf. lubrica is actually mycorrhizal. Besides C. arbutoides (Ericaceae) as mycorrhizal partner, an association likewise with Q. costaricensis has not been found as it was shown for the Basidiomycete Sebacina sp. (Kühdorf et al. 2014). However, an ectomycorrhizal association with Q. costaricensis is likewise assumed, since Branco and Ree (2010) reported a leotioid ECM with Q. rotundifolia. Nevertheless, it is interesting that mycorrhizas of L. lubrica were overlooked so far, although the fungus occurs worldwide. One possibility could be a low competitive ability, e.g., due to slow colonization of root tips (Kennedy 2010). In addition, a dual lifestyle (mycorrhizal as well as saprotrophic) may be also possible, as is known for the ectomycorrhizal Basidiomycete Laccaria bicolor (Vincent et al. 2012). However, these assumptions need further investigation. Further sampling could reveal if group II L. lubrica species also have the potential to be mycorrhizal or if this ability is limited to members of group I.
Acknowledgments
The authors thank Prof. R. Agerer for his help with studying mycorrhizas. The authors are grateful to Monika Roth for excellent technical assistance. The authors also thank Federico Valverde and Silvia Lobo Cabezas for their assistance in the cloud forests of Costa Rica. We are indebted to Roy E. Halling and the National Geographic Society (grant no. 7341-02) for supporting the photo of Comarostaphylis arbutoides. We would also like to thank Mary Lavin-Zimmer from the German Research Centre (GFZ) for English corrections. The authors are indebted to the German Research Foundation (DFG) for funding this project (Mu 1035/15-1).
Footnotes
J. Gómez-Laurito is deceased.
References
- Acsai J, Largent DL. Fungi associated with Arbutus menziesii, Arctostaphylos uva-ursi in central and northern California. Mycologia. 1983;75:544–547. doi: 10.2307/3792696. [DOI] [Google Scholar]
- Acsai J, Largent DL. Mycorrhizae of Arbutus menziesii Pursh. and Arctostaphylos manzanita Parry in northern California. Mycotaxon. 1983;16:519–536. [Google Scholar]
- Agerer R (1987–2012) Colour atlas of ectomycorrhizae. 1st–15th delivery, Einhorn, Schwäbisch Gmünd
- Agerer R. Characterization of ectomycorrhiza. In: Norris JR, Read DJ, Varma AK, editors. Methods in microbiology. London: Academic Press; 1991. pp. 25–74. [Google Scholar]
- Agerer R. Elaphomyces aculeatus Tul. + Quercus robur L. In: Agerer L, Danielson RM, Egli S, Ingleby K, Luoma D, Treu R, editors. Descriptions of ectomycorrhizae. Schwäbisch Gmünd: Einhorn; 1999. pp. 37–41. [Google Scholar]
- Agerer R. Exploration types of ectomycorrhizae—a proposal to classify ectomycorrhizal mycelial systems according to their patterns of differentiation and putative ecological importance. Mycorrhiza. 2001;11:107–114. doi: 10.1007/s005720100108. [DOI] [Google Scholar]
- Agerer R, Rambold G (2004–2014) DEEMY—an information system for characterization and determination of ectomycorrhizae. München, Germany. http://www.deemy.de. Accessed 05 Mar 2014
- Arora D. Mushrooms demystified. A comprehensive guide to the fleshy fungi. 2. Berkeley: Ten Speed Press; 1986. p. 959. [Google Scholar]
- Baral H-O. Lugol's solution/IKI versus Melzer's reagent: hemiamyloidity, a universal feature of the ascus wall. Mycotaxon. 1987;29:399–450. [Google Scholar]
- Baral H-O (2009) Iodine reaction in Ascomycetes: why is Lugol’s solution superior to Melzer’s reagent? http://www.gbif-mycology.de/HostedSites/Baral/IodineReaction.htm. Accessed 14 Jan 2014
- Branco S, Ree RH. Serpentine soils do not limit mycorrhizal fungal diversity. Plos One. 2010;5:e11757. doi: 10.1371/journal.pone.0011757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg A. Effect of soil humus cover on the establishment and development of mycorrhiza on containerised Pinus sylvestris L. and Pinus contorta spp. latifolia Engelm. after outplanting. Scand J For Res. 1990;5:103–112. doi: 10.1080/02827589009382597. [DOI] [Google Scholar]
- Durand EJ. The Geoglossaceae of North America. Ann Mycol. 1908;6:387–477. [Google Scholar]
- Gao Q, Yang ZL. Ectomycorrhizal fungi associated with two species of Kobresia in an alpine meadow in the eastern Himalaya. Mycorrhiza. 2010;20:281–287. doi: 10.1007/s00572-009-0287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. ITS primers with enhanced specificity for Basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Grund DW, Harrison KA. Nova Scotian fungi: geoglossaceae. Can J Bot. 1967;45:1625–1641. doi: 10.1139/b67-169. [DOI] [Google Scholar]
- Hagerman SM, Sakakibara SM, Durall DM. The potential for woody understory plants to provide refuge for ectomycorrhizal inoculum at an interior Douglas-fir forest after clear-cut logging. Can J For Res. 2001;31:711–721. doi: 10.1139/x00-199. [DOI] [Google Scholar]
- Halling RE, Mueller GM. Leccinum (Boletaceae) in Costa Rica. Mycologia. 2003;95:488–499. doi: 10.2307/3761891. [DOI] [PubMed] [Google Scholar]
- Halling RE, Mueller GM (2004) Common mushrooms of the Talamanca mountains, Costa Rica. New York Botanical Garden Pr Dept., p 195
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Lumbsch HT, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai Y-C, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson K-H, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo J-M, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schüßler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao Y-J, Zhang N. A higher-level phylogenetic classification of the fungi. Mycol Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Hileman LC, Vasey MC, Parker VT. Phylogeny and biogeography of the Arbutoideae (Ericaceae): implications for the Madrean-Tethyan hypothesis. Syst Bot. 2001;26:131–143. [Google Scholar]
- Horton TR, Bruns TD, Parker VT. Ectomycorrhizal fungi associated with Arctostaphylos contribute to Pseudotsuga menziesii establishment. Can J Bot. 1999;77:93–102. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy P. Ectomycorrhizal fungi and interspecific competition: species interactions, community structure, coexistence mechanisms, and future research directions. New Phytol. 2010;187:895–910. doi: 10.1111/j.1469-8137.2010.03399.x. [DOI] [PubMed] [Google Scholar]
- Kennedy PG, Smith DP, Horton TR, Molina RJ. Arbutus menziesii (Ericaceae) facilitates regeneration dynamics in mixed evergreen forests by promoting mycorrhizal fungal diversity and host connectivity. Am J Bot. 2012;99:1691–1701. doi: 10.3732/ajb.1200277. [DOI] [PubMed] [Google Scholar]
- Kõljalg U, Larsson K-H, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Vrålstad T, Ursing BM. UNITE: a database providing Web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005;166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- Kühdorf K, Münzenberger B, Begerow D, Karasch-Wittmann C, Gómez-Laurito J, Hüttl RF. Sebacina sp. is a mycorrhizal partner of Comarostaphylis arbutoides (Ericaceae) Mycol Prog. 2014 [Google Scholar]
- Lumbsch HT, Huhndorf SM (2009) Myconet Volume 14. Part One. Outline of Ascomycota—2009. Part Two. Notes on Ascomycete Systematics. Nos. 4751–5113. 10.3158/1557.1 [DOI]
- Mains EB. North American species of the Geoglossaceae. Tribe Cudonieae. Mycologia. 1956;48:694–710. doi: 10.2307/3755379. [DOI] [Google Scholar]
- Massicotte HB, Melville LH, Molina R, Peterson L. Structure and histochemistry of mycorrhizae synthesized between Arbutus menziesii (Ericaceae) and two Basidiomycetes, Pisolithus tinctorius (Pisolithaceae) and Piloderma bicolor (Corticiaceae) Mycorrhiza. 1993;3:1–11. doi: 10.1007/BF00213461. [DOI] [Google Scholar]
- Massicotte HB, Molina R, Luoma DL, Smith JE. Biology of the ectomycorrhizal genus Rhizopogon. II. Patterns of host-fungus specificity following spore inoculation of diverse hosts grown in mono- and dual-cultures. New Phytol. 1994;126:677–690. doi: 10.1111/j.1469-8137.1994.tb02962.x. [DOI] [Google Scholar]
- Molina R, Trappe JM. Lack of mycorrhizal specificity by the ericaceous hosts Arbutus menziesii and Arctostaphylos uva-ursi. New Phytol. 1982;90:495–509. doi: 10.1111/j.1469-8137.1982.tb04482.x. [DOI] [Google Scholar]
- Molina R, Trappe JM. Patterns of ectomycorrhizal host specificity and potential among Pacific Northwest conifers and fungi. For Sci. 1982;28:423–458. [Google Scholar]
- Molina R, Massicotte H, Trappe JM. Specificity phenomena in mycorrhizal symbioses: community-ecological consequences and practical implications. In: Allen MF, editor. Mycorrhizal functioning: an integrative plant-fungal process. New York: Routledge, Chapman and Hall; 1992. pp. 357–423. [Google Scholar]
- Molina R, Smith JE, McKay D, Melville LH. Biology of the ectomycorrhizal genus Rhizopogon. III. Influence of co-cultured conifer species on mycorrhizal specificity with the arbutoid hosts Arctostaphylos uva-ursi and Arbutus menziesii. New Phytol. 1997;137:519–528. doi: 10.1046/j.1469-8137.1997.00836.x. [DOI] [PubMed] [Google Scholar]
- Moncalvo J-M, Lutzoni FM, Rehner SA, Johnson J, Vilgalys R. Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Syst Biol. 2000;49:278–305. doi: 10.1093/sysbio/49.2.278. [DOI] [PubMed] [Google Scholar]
- Montecchio L, Rossi S, Courty P-E, Garbaye JG. Entoloma nitidum Quél. + Carpinus betulus L. In: Agerer L, Danielson RM, Egli S, Ingleby K, Luoma D, Treu R, editors. Descriptions of ectomycorrhizae, vol 9/10. Schwäbisch Gmünd: Einhorn; 2006. pp. 33–38. [Google Scholar]
- Mühlmann O, Göbl F. Mycorrhiza of the host-specific Lactarius deterrimus on the roots of Picea abies and Arctostaphylos uva-ursi. Mycorrhiza. 2006;16:245–250. doi: 10.1007/s00572-006-0038-9. [DOI] [PubMed] [Google Scholar]
- Münzenberger B, Kottke I, Oberwinkler F. Ultrastructural investigations of Arbutus unedo-Laccaria amethystea mycorrhiza synthezised in vitro. Trees. 1992;7:40–47. doi: 10.1007/BF00225230. [DOI] [Google Scholar]
- Münzenberger B, Bubner B, Wöllecke J, Sieber TN, Bauer R, Fladung M, Hüttl RF. The ectomycorrhizal morphotype Pinirhiza sclerotia is formed by Acephala macrosclerotiorum sp. nov., a close relative of Phialocephala fortinii. Mycorrhiza. 2009;19:481–492. doi: 10.1007/s00572-009-0239-0. [DOI] [PubMed] [Google Scholar]
- Orlovich DA, Draffin SJ, Daly RA, Stephenson SL. Piracy in the high trees: ectomycorrhizal fungi from an aerial ‘canopy soil’ microhabitat. Mycologia. 2013;105:52–60. doi: 10.3852/11-307. [DOI] [PubMed] [Google Scholar]
- Osmundson TW, Halling RE, den Bakker H. Morphological and evidence supporting an arbutoid mycorrhizal relationsphip in the Costa Rican páramo. Mycorrhiza. 2007;17:217–222. doi: 10.1007/s00572-006-0098-x. [DOI] [PubMed] [Google Scholar]
- Palfner G, Horak E. Gautieria inapire sp. nov., a new hypogeous species from Nothofagus forest in southern Central Chile. Sydowia. 2001;53:140–151. [Google Scholar]
- Richard F, Millot S, Gardes M, Selosse M-A. Diversity and specificity of ectomycorrhizal fungi retrieved from an old-growth Mediterranean forest dominated by Quercus ilex. New Phytol. 2005;166:1011–1023. doi: 10.1111/j.1469-8137.2005.01382.x. [DOI] [PubMed] [Google Scholar]
- Rinaldi AC, Comandini O, Kuyper TW (2008) Ectomycorrhizal fungal diversity: separating the wheat from the chaff. Fungal Divers 33:1–45
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitzman BH, Ouimette A, Mixon RL, Hobbie EA, Hibbett DS. Conservation of biotrophy in Hygrophoraceae inferred from combined stable isotope and phylogenetic analyses. Mycologia. 2011;103:280–290. doi: 10.3852/10-195. [DOI] [PubMed] [Google Scholar]
- Selosse M-A, Setaro S, Glatard F, Richard F, Urcelay C, Weiß M. Sebacinales are common mycorrhizal associates of Ericaceae. New Phytol. 2007;174:864–878. doi: 10.1111/j.1469-8137.2007.02064.x. [DOI] [PubMed] [Google Scholar]
- Spurr AR. A low viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/S0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Pärtel K, Jairus T, Gates G, Põldmaa K, Tamm H. Ascomycetes associated with ectomycorrhizas: molecular diversity and ecology with particular reference to the Helotiales. Environ Microbiol. 2009;11:3166–3178. doi: 10.1111/j.1462-2920.2009.02020.x. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, May TW, Smith ME. Ectomycorrhizal lifestyle in fungi: global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza. 2010;20:217–263. doi: 10.1007/s00572-009-0274-x. [DOI] [PubMed] [Google Scholar]
- Vincent D, Kohler A, Claverol S, Solier E, Joets J, Gibon J, Lebrun MH, Plomion C, Martin F. Secretome of the free-living mycelium from the ectomycorrhizal Basidiomycete Laccaria bicolor. J Proteome Res. 2012;11:157–171. doi: 10.1021/pr200895f. [DOI] [PubMed] [Google Scholar]
- Visser S. Ectomycorrhizal fungal succession in jack pine stands following wildfire. New Phytol. 1995;129:389–401. doi: 10.1111/j.1469-8137.1995.tb04309.x. [DOI] [Google Scholar]
- Wang Z, Binder M, Schoch CL, Johnston PR, Spatafora JW, Hibbett DS. Evolution of helotialean fungi (Leotiomycetes, Pezizomycotina): a nuclear rDNA phylogeny. Mol Phylogenet Evol. 2006;41:295–312. doi: 10.1016/j.ympev.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Wang Z, Johnston PR, Takamatsu S, Spatafora JW, Hibbett DS. Toward a phylogenetic classification of the Leotiomycetes based on rDNA data. Mycologia. 2006;98:1065–1075. doi: 10.3852/mycologia.98.6.1065. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JN, White TJ, editors. PCR Protocols: a guide to method and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Zak B. Pure culture synthesis of Pacific Madrone ectendomycorrhizae. Mycologia. 1976;68:362–369. doi: 10.2307/3759006. [DOI] [Google Scholar]
- Zak B. Pure culture synthesis of bearberry mycorrhizae. Can J Bot. 1976;54:1297–1305. doi: 10.1139/b76-142. [DOI] [Google Scholar]
- Zeller B, Brechet C, Maurice J-P, Le Tacon F. 13C and 15N isotopic fractionation in trees, soils and fungi in a natural forest stand and a Norway spruce plantation. Ann For Sci. 2007;64:419–429. doi: 10.1051/forest:2007019. [DOI] [Google Scholar]
- Zhong Z, Pfister DH. Phylogenetic relationships among species of Leotia (Leotiales) based on ITS and RPB2 sequences. Mycol Prog. 2004;3:237–246. doi: 10.1007/s11557-006-0094-8. [DOI] [Google Scholar]


