Abstract
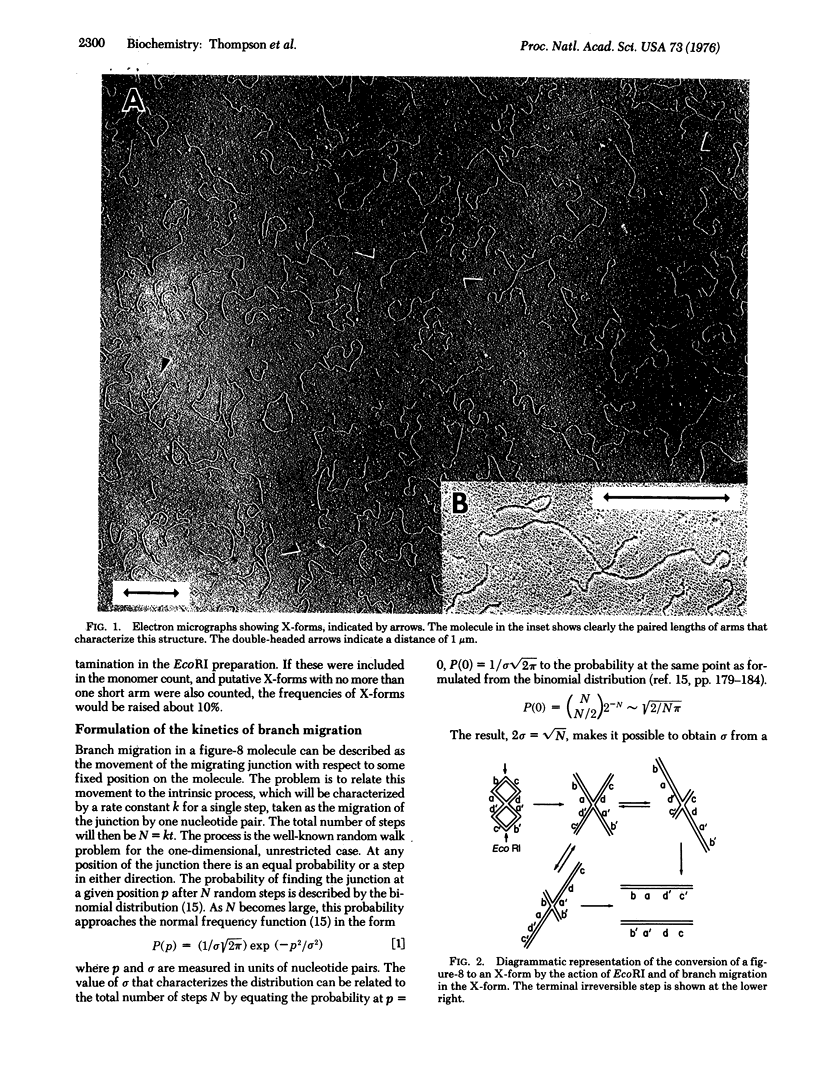
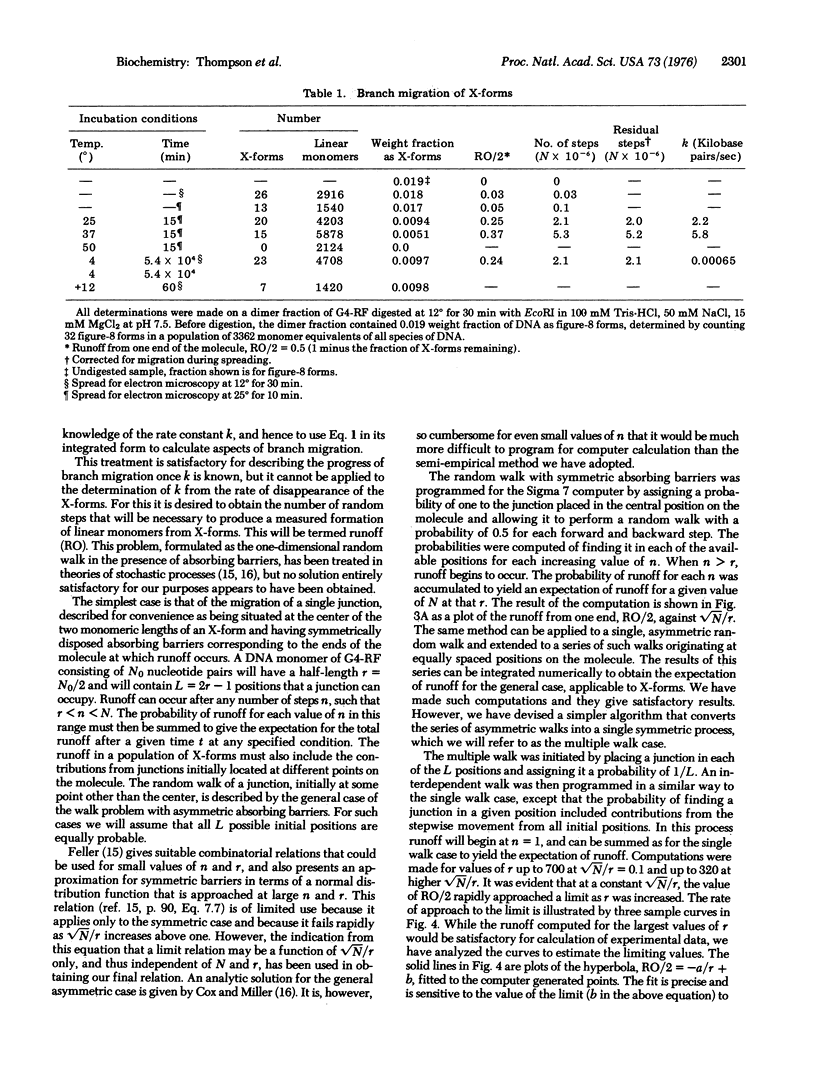
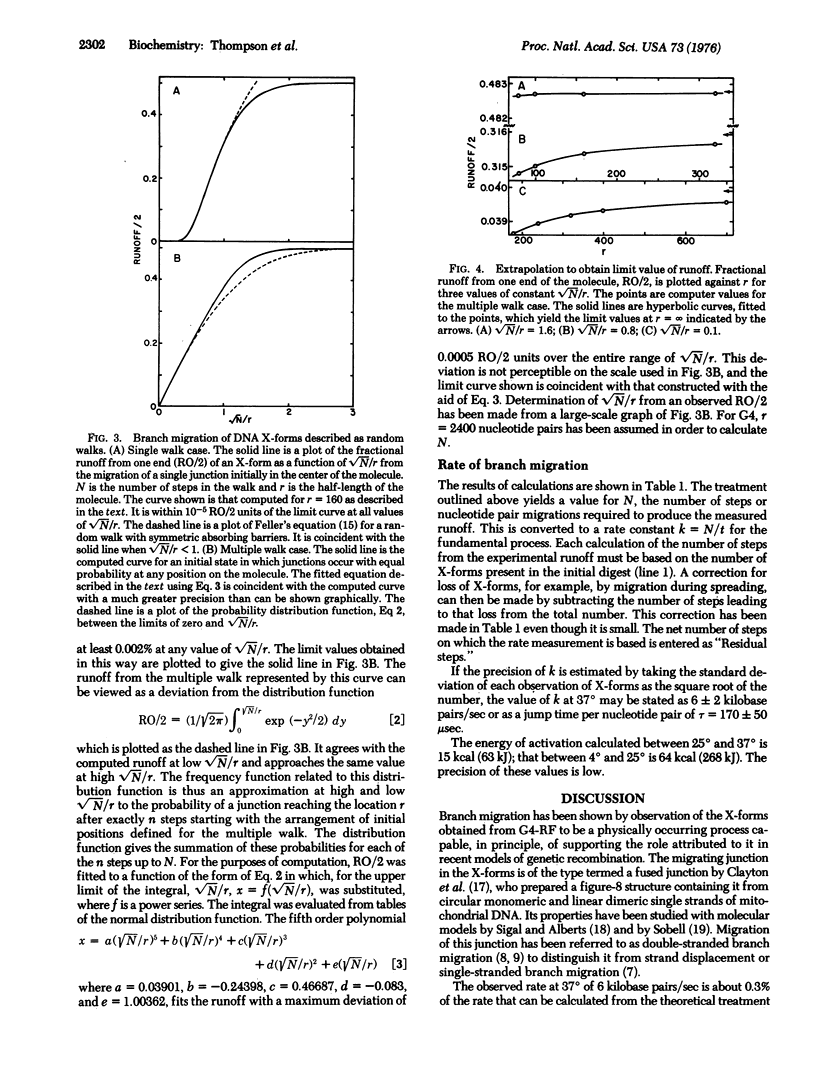
The rate of branch migration in double stranded DNA has been measured by the use of a unique substrate formed by the action of the EcoRI restriction endonuclease on the dimeric figure-8 configuration of the replicative form DNA of phage G4. The figure-8 and the X-form derived from it contain a junction of the kind postulated to occur in the Holliday structure and to be an essential feature of a number of models of recombination. In the X-form this junction can branch migrate to an irreversible terminal configuration consisting of two linear monomers. The disappearance of X-forms was measured by electron microscopy. A treatment of branch migration as a random walk process was developed to permit the determination of the rate of the intrinsic process, a step movement of the junction by a distance of one base pair. A value of about 6 kilobase pairs per sec at 37 degrees was obtained.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blin N., von Gabain A., Bujard H. Isolation of large molecular weight DNA from agarose gels for further digestion by restriction enzymes. FEBS Lett. 1975 Apr 15;53(1):84–86. doi: 10.1016/0014-5793(75)80688-0. [DOI] [PubMed] [Google Scholar]
- Broker T. R. An electron microscopic analysis of pathways for bacteriophage T4 DNA recombination. J Mol Biol. 1973 Nov 25;81(1):1–16. doi: 10.1016/0022-2836(73)90243-x. [DOI] [PubMed] [Google Scholar]
- Broker T. R., Lehman I. R. Branched DNA molecules: intermediates in T4 recombination. J Mol Biol. 1971 Aug 28;60(1):131–149. doi: 10.1016/0022-2836(71)90453-0. [DOI] [PubMed] [Google Scholar]
- Clayton D. A., Davis R. W., Vinograd J. Homology and structural relationships between the dimeric and monomeric circular forms of mitochondrial DNA from human leukemic leukocytes. J Mol Biol. 1970 Jan 28;47(2):137–153. doi: 10.1016/0022-2836(70)90335-9. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T., Sinsheimer R. L. The process of infection with bacteriophage phi-X174. 3. Phage maturation and lysis after synchronized infection. J Mol Biol. 1965 Jul;12(3):641–646. doi: 10.1016/s0022-2836(65)80318-7. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T., Sinsheimer R. L. The process of infection with bacteriophage phi-X174. IV. Replication of the viral DNA in a synchronized infection. J Mol Biol. 1965 Jul;12(3):647–662. doi: 10.1016/s0022-2836(65)80319-9. [DOI] [PubMed] [Google Scholar]
- Doniger J., Warner R. C., Tessma I. Role of circular dimer DNA in the primary recombination mechanism of bacteriophage S13. Nat New Biol. 1973 Mar 7;242(114):9–12. doi: 10.1038/newbio242009a0. [DOI] [PubMed] [Google Scholar]
- Hotchkiss R. D. Models of genetic recombination. Annu Rev Microbiol. 1974;28(0):445–468. doi: 10.1146/annurev.mi.28.100174.002305. [DOI] [PubMed] [Google Scholar]
- Kim J., Sharp P. A., Davidson N. Electron microscope studies of heteroduplex DNA from a deletion mutant of bacteriophage phiX-174. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1948–1952. doi: 10.1073/pnas.69.7.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. Formation of hybrid DNA by rotary diffusion during genetic recombination. J Mol Biol. 1972 Nov 28;71(3):795–798. doi: 10.1016/s0022-2836(72)80040-8. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Molecular mechanisms in genetic recombination. Annu Rev Genet. 1973;7:87–111. doi: 10.1146/annurev.ge.07.120173.000511. [DOI] [PubMed] [Google Scholar]
- Rush M. G., Warner R. C. Alkali denaturation of covalently closed circular duplex deoxyribonucleic acid. J Biol Chem. 1970 May 25;245(10):2704–2708. [PubMed] [Google Scholar]
- Sigal N., Alberts B. Genetic recombination: the nature of a crossed strand-exchange between two homologous DNA molecules. J Mol Biol. 1972 Nov 28;71(3):789–793. doi: 10.1016/s0022-2836(72)80039-1. [DOI] [PubMed] [Google Scholar]
- Sobell H. M. Symmetry in protein-nucleic acid interaction and its genetic implications. Adv Genet. 1973;17:411–490. doi: 10.1016/s0065-2660(08)60175-3. [DOI] [PubMed] [Google Scholar]
- Thompson B. J., Escarmis C., Parker B., Slater W. C., Doniger J., Tessman I., Warner R. C. Figure-8 configuration of dimers of S13 and phiX174 replicative form DNA. J Mol Biol. 1975 Feb 5;91(4):409–419. doi: 10.1016/0022-2836(75)90269-7. [DOI] [PubMed] [Google Scholar]




