Abstract
Tyrosine sulfation, a well-characterized post-translation modification in eukaryotes, has not previously been reported in prokaryotes. Here we demonstrate that the RaxST protein from the Gram-negative bacterium, Xanthomonas oryzae pv. oryzae, is a tyrosine sulfotransferase. We used a newly developed sulfotransferase assay and ultraviolet photodissociation mass spectrometry (UVPD) to demonstrate that RaxST catalyzes sulfation of tyrosine 22 of the Xoo Ax21 (activator of XA21-mediated immunity). These results demonstrate a previously undescribed post-translational modification in a prokaryotic species with implications extending to host immune response and bacterial cell-cell communication system.
Introduction
Tyrosine sulfation is a common post-translation modification in eukaryotes1. In contrast to phosphorylation, which regulates processes that occur inside the cell2, sulfated proteins/peptides are typically directed outside the cell, where they modulate ligand-receptor interactions3. A notable example is the binding of the glycoprotein 120 (gp120) subunit of the human immunodeficiency virus (HIV), to the human chemokine co-receptors CD4 and CCR54. Sulfation of tyrosine residues in the N-terminal segment of CCR5 appears to be critical for both HIV-1 entry and binding of gp120-CD4 complexes5. In plants, the sulfated peptide, phytosulfokine-α (PSK), is recognized by the carrot phytosulfokine receptor kinase, which is responsible for cellular dedifferentiation and proliferation in plants6.
In prokaryotes, well-known sulfotransferases belong to the carbohydrate, aryl, or glycolipid sulfotransferase families7–10. For example, the Sinorhizobium meliloti Nod factor [a lipo-chitooligosaccharide (LCO)], is sulfated by the NodH sulfotransferase, which transfers the sulfuryl group from 3’-phosphoadenosine 5’-phosphosulfate (PAPS) onto the Nod factor7, 11. NodP and NodQ encode adenosine-5′-triphosphate (ATP) sulfurylase and adenosine-5′-phosphosulfate (APS) kinase, which together catalyze the production of PAPS. S. meliloti strains carrying mutations in the nodP, nodQ, or nodH genes, produce Nod factor that lacks the sulfate group and are severely impaired in their ability to nodulate their normal host alfalfa7. Tyrosine sulfotransferase activity has not previously been reported in prokaryotes1, 11.
We recently reported that the rice XA21 pattern recognition receptor (PRR) binds a sulfated peptide (AENLSYSNFVEGDYVRTP; called axYS22) derived from the Ax21 (Activator of XA21-mediated innate immunity) protein from the Gram-negative bacterium, Xanthomonas oryzae pv. oryzae (Xoo) strain PXO9912, 13. The biological function of Ax21 in Gram-negative bacteria is to mediate quorum sensing (QS), a process where small molecules serve as signals to recognize cell population size, leading to changes in expression of specific genes when the QS factor has accumulated to a certain threshold concentration14, 15. Ax21-mediated QS controls motility, biofilm formation and virulence in Xoo13, 14.
The Xoo genes raxST, raxP and raxQ, are also required for activation of XA21-mediated immunity. Xoo raxP and raxQ are predicted orthologs of S. meliloti nodP and nodQ, respectively. The raxST gene encodes a protein carrying a 3’-phosphoadenosine 5’-phosphosulfate (PAPS) binding motif16. Xoo mutant strains lacking ax21 (PXO99ax21), raxST (PXO99raxST), raxP (PXO99raxP), or raxQ (PXO99raxQ) fail to activate XA21-mediated immunity16–18. The diverse Ax21-mediated biological activities in Xoo also require RaxST14. Based on the presence of the PAPS binding motif in the raxST gene we hypothesized that RaxST utilizes PAPS produced by RaxP and RaxQ to transfer a sulfuryl group (SO3−1) to Ax21.
We show RaxST has tyrosine sulfotransferase activity using a newly developed sulfotransferase assay system. Furthermore, we provide direct evidence confirming Ax21 is a substrate of RaxST through UV photodissociation mass spectrometry. Not only does this represent the first evidence of the post-translational sulfation of a key prokaryotic protein but also the role in host innate immune response and bacterial cell-cell communication is imputed.
Results
PAPS is required for triggering XA21-mediated immunity
To confirm the hypothesis that RaxST uses PAPS generated by RaxP and RaxQ to transfer a sulfuryl group (SO3−1) to Ax21, we tested if the raxQ knockout strain (PXO99ΔraxQ), which cannot make PAPS17, can be complemented by the addition of exogenous PAPS. XA21 rice leaves pretreated with the PXO99ΔraxQ supernatant failed to induce XA21-mediated immunity and were susceptible to subsequent infection by PXO99ΔraxST (Supplementary Fig. S1). By contrast, rice leaves pretreated with supernatant of PXO99ΔraxQ culture incubated with exogenous PAPS induce strong resistance to subsequent infection by PXO99ΔraxST. This result indicates that RaxST utilizes PAPS produced by RaxQ.
RaxST possesses sulfotransferase activity
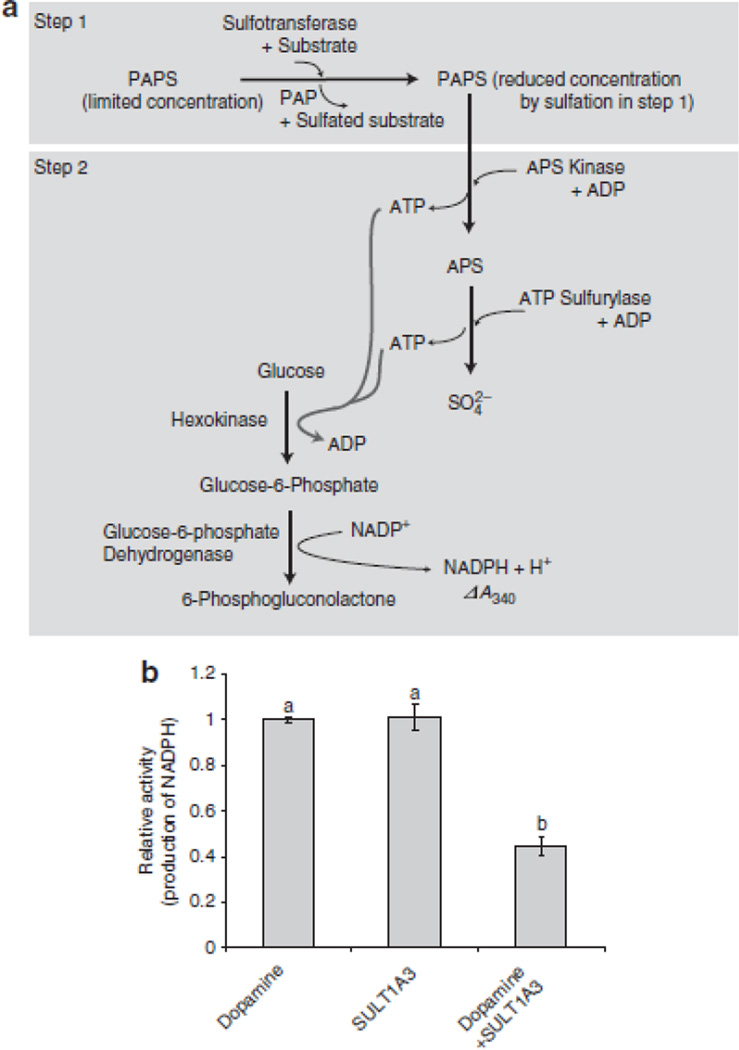
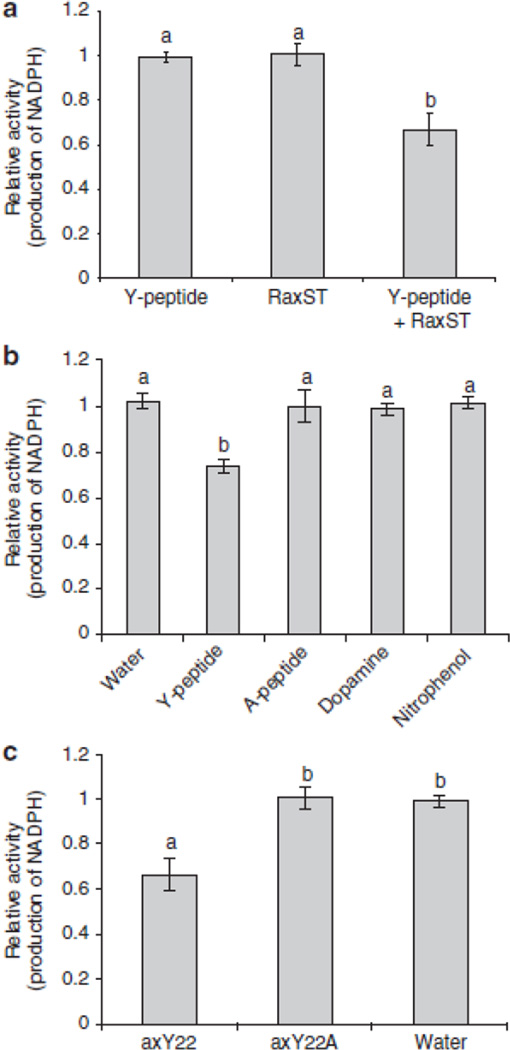
To test if RaxST carries tyrosine sulfotransferase activity, we generated biologically active, soluble recombinant RaxST (Supplementary Fig. S2) and confirmed biological activity (Supplementary Fig. S3). We then developed a system to measure sulfotransferase activity that relies on the measurement of NADPH production (Fig. 1A). This assay system was validated using a well-characterized human dopamine sulfotransferase, SULT1A3, and its substrate, dopamine (Fig. 1B). Next, we examined catalytic activity of RaxST using the N-terminal peptide of the CC-chemokine receptor 5, Y-peptide (DYQVSSPIYDINYYTSE), which is known to be a substrate for two human tyrosine sulfotransferases19. We found that NADPH production decreased when RaxST and the Y-peptide were added in the presence of limiting PAPS concentration (Fig. 2A). The reduction of NADPH was not observed in control reactions lacking either RaxST or the Y-peptide. These results indicate that RaxST has sulfotransferase activity.
Figure 1. Development of a sulfotransferase activity assay.
(A) In the first step, a sulfotransferase, a substrate, and PAPS are incubated under conditions that allow for the production of a sulfated substrate. In the next step, APS kinase and ATP sulfurylase catalyze reverse reactions to produce APS and ATP from the PAPS remaining from the first step. If PAPS is limiting, less APS and ATP will be produced. A subsequent reaction catalyzed by hexokinase and glucose-6-phosphate dehydrogenase utilizes the ATP and glucose to produce the reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) and 6-phosphogluconolactone. NADPH production can be specifically monitored at 340 nm using a spectrophotometer. In this assay system, two moles of NADPH are produced for each mole of PAPS turned over by APS kinase26. Given an excess of PAPS and all other components in each step, the reaction rate will proceed in the absence of a sulfotransferase or its substrate (the sulfuryl-group receiver). If, however, an active sulfotransferase and substrate are incubated together prior to the addition of APS kinase under limited concentration of PAPS, NADPH production is decreased compared to that detected in reactions lacking a sulfotransferase or its substrate. This decrease reflects the presence of sulfotransferase enzymatic activity. Specific conditions are described in Methods. (B) Validation of the enzyme coupling sulfotransferase assay system using dopamine sulfotransferase, SULT1A3 (25 units), and its substrate, dopamine (60 µM). NADPH production was measured at 340 nm using a spectrophotometer. Bars indicate the relative average value obtained for ΔA340/min ± standard deviation in three independent experiments. Different letters on the top of each bar indicate statistically significant differences between treatments [(one way ANOVA analysis (Holm-Sidak test, P<0.05)].
Figure 2. (A) Tyrosine sulfotransferase activity of the RaxST protein.
The reaction was initiated by adding the Y-peptide (DYQVSSPIYDINYYTSE) alone, RaxST protein alone, or both the Y-peptide and 0.5 µg of RaxST protein. (B) RaxST activity is specific to tyrosine peptides. The reaction was incubated with water, Y-peptide, A-peptide (DAQVSSPIADINAATSE), Dopamine, or Nitrophenol. (C) RaxST activity is specific to axY22. The reaction was initiated by adding axY22, axY22A (AENLSANFVEGDYVRTP), or water. NADPH production was measured at 340 nm using a spectrophotometer. Bars indicate the relative average value obtained for ΔA340/min ± standard deviation in three independent experiments. Different letters on the top of each bar indicate statistically significant differences between treatments. [(one way ANOVA analysis (Holm-Sidak test, P<0.05)].
SULT1A3 is known to utilize both dopamine and tyrosine as substrates20. To determine if the RaxST protein also has dual specificity, we tested nitrophenol and dopamine, which are known substrates of SULT1A2 and SULT1A3, respectively20, 21. In addition, we also tested a variant of the Y peptide in which the tyrosine residues were replaced with alanine residues (A-peptide, DAQVSSPIADINAATSE) to confirm whether the RaxST protein specifically targets tyrosine. The production of NADPH decreased only when the Y-peptide was added but not by the addition of the A-peptide, dopamine or nitrophenol (Fig. 2B), indicating that sulfation by RaxST is specific to a peptide carrying tyrosine, not dopamine or nitrophenol as substrates. These results demonstrate that the RaxST protein specifically catalyzes the sulfation of tyrosine residues. To determine optimal conditions for RaxST activity, we tested RaxST activity using the Y-peptide at different pH and temperatures. RaxST showed maximum activity at weak acidic conditions (approximately pH 6.5 ~ 6.8 and 25 ~ 30°C)(Supplementary Fig. S4A and B). A Michaelis-Menten plot was established to assess the kinetics of the reaction. Under the given conditions, the Km was 1.7 µM and the Vmax was 0.18 units/mg (Supplementary Fig. S4C and D).
A tyrosine residue in axY22 peptide is specifically catalyzed by RaxST
We next tested if RaxST can catalyze sulfation of axY22 and a peptide variant carrying alanine in place of the tyrosine in position 22 (axY22A, AENLSANFVEGDYVRTP). We found that production of NADPH decreased when axY22 was used as a substrate in the presence of RaxST and limiting concentrations of PAPS (Fig. 2C), indicating that axY22 is indeed a substrate for RaxST. By contrast, the peptide variant, axY22A, is not a substrate for RaxST, indicating that the Y residue in position 22 is the only target of RaxST and not the additional tyrosine residue in position 29 (Fig. 2C). Taken together, these studies indicate RaxST catalyzes sulfation of tyrosine 22 on the axY22 peptide to produce the biologically active form.
Confirmation of sulfation of Ax21 by UVPD
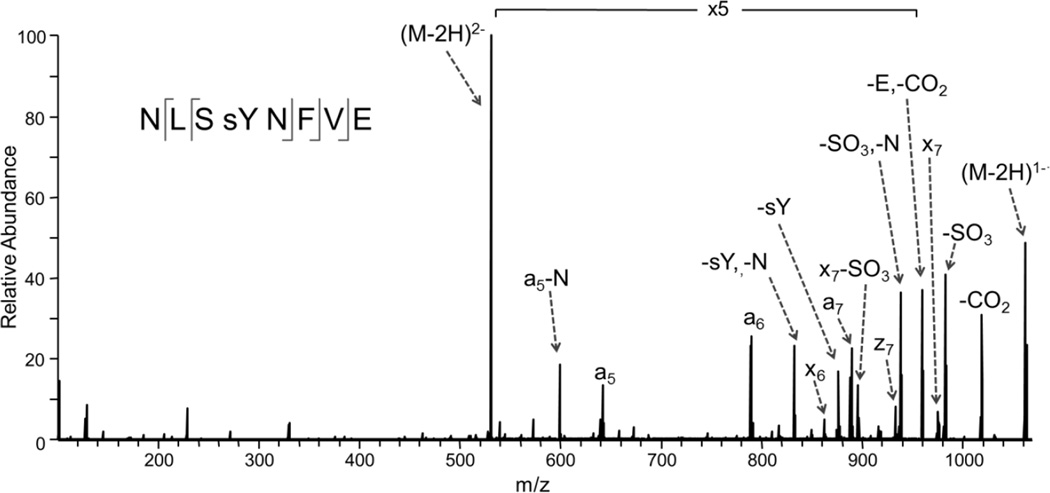
For a more direct determination of the RaxST target and to assess if other tyrosines are sulfated on the Ax21 protein, we expressed and purified the full-length mature Ax21 protein (Supplementary Fig. S5) and subjected it to RaxST sulfation. We then purified the sulfated protein product, subjected it to GluC proteolytic digestion, and then analyzed the resulting peptides via LC-MS/MS in the negative nanoelectrospray mode with ultraviolet photodissociation (UVPD) implemented on a Thermo Scientific Orbitrap Elite mass spectrometer. UVPD has proven to be a versatile MS/MS strategy for proteomics applications22, with particular merits for peptide anions for which conventional collision induced dissociation (CID) and electron-based activation methods (ECD, ETD) are less successful23, 24. Tyrosine sulfation is a particularly labile modification upon CID of protonated or deprotonated peptides25, making UVPD a compelling alternative. Here we demonstrate the first application of the UVPD approach for assessment of tyrosine sulfation. Despite the low abundance of the peptide in the Ax21 digest, the UVPD fragmentation pattern (Fig. 3) confidently confirmed the sequence (NSLYNFVE) and sulfation site (Y22) with diagnostic a5, a6, a7, x7, and x8 ions and retention of the modification on all of the key sequence ions. The ion assignments and mass accuracies corresponding to Figure 3 are summarized in Supplementary Table S1, and the NUVPD mass spectrum of a synthetic sulfated model peptide is displayed in Supplementary Fig. S6. The sulfation modification was not successfully identified using the more conventional CID and ETD methods in the positive ionization mode for tryptic or GluC digests of the Ax21 protein.
Figure 3. UV photodissociation mass spectrum of Tyr-sulfated peptide NLSYNFVE.
The Tyr-sulfated peptide was identified in the GluC digest of Ax21 after incubation with the sulfotransferase RaxST. The spectrum of the doubly deprotonated (2−) peptide was acquired on an Orbitrap Elite mass spectrometer equipped with an excimer laser (193 nm, 500 Hz, 5 ns pulse, 2 mJ per pulse, two pulses per spectrum). UVPD occurred in the higher-energy collisional dissociation (HCD) cell of the Orbitrap Elite mass spectrometer.
Discussion
Post-translational modifications of prokaryotic proteins are poorly understood compared with those of eukaryotes. The discovery of tyrosine sulfation of Ax21 by RaxST represents a previously unreported post-translational modification in bacteria and provides new insight into the role of such a modification in triggering the host innate immune response and in controlling bacterial cell-cell communication.
Methods
Biological materials
Bacterial strains and plasmids used in this study are listed in Supplementary Table S2.
Sulfotransferase activity assay
Unless otherwise indicated, reactions were initiated by adding a sulfotransferase and its substrate at the desired concentration in 50 µl of sulfotransferase buffer (8 µM PAPS, 10 mM Tris-HCl, pH 7.0, 1 mM adenosine monophosphate, 50 mM NaF, 0.5% Triton-100, and 5 mM MnCl2). Sulfotransferase buffer was freshly prepared for each experiment. After four hours of incubation at 30 °C, the reaction mixture was chilled on ice. The APS cocktail buffer contained 50 mM Tris-HCl, pH 7.0, 70 mM (NH4)2SO4, 5 mM MgCl2, 0.6 mM nicotinamide adenine dinucleotide phosphate (NADP+), 1 mM glucose, 1 mM sodium pyrophosphate, 560 µM adenosine diphosphate, 2.5 units of glucose-6-phosphate dehydrogenase, 5 units of hexokinase, 55 µg of ATP sulfurylase, and 8 µg of APS kinase. The construct containing APS kinase in the pET15b vector was kindly provided by Dr. Andrew Fisher at UC Davis. We purified APS kinase from E. coli expressing the His-tagged APS kinase fusion protein using an Ni-NTA agarose column and tested its activity using a previously established method26. All other co-enzymes and chemicals were purchased from Sigma. For each assay, 50 µl of APS cocktail buffer was added to the ice-chilled sulfotransferase reaction mixture. The coupling assay was finally initiated by adding APS kinase. NADPH production was measured using a SafireII spectrophotometer (Tecan) at 340 nm and 30°C at 30 sec intervals. In the figures, the relative activity of the Y-axis refers to the amount of NADPH produced at a specific time point (7 minutes after the addition of APS kinase). The values are relative to the controls (water or sulfotransferase only), which were set as one.
Enzymatic analysis of RaxTST
To assay the enzymatic activity of RaxTST, 0.15 or 0.45 µg of the partially purified RaxTST and 100 µM synthetic tyrosine peptide (NH2-DYQVSSPIYDINYYTSE, referred to as Y-peptide) were used. To assess the specificity of tyrosine sulfotransferase activity, we generated another peptide in which all the tyrosine residues in the Y-peptide were replaced with alanine (NH2-DAQVSSPIADINAATSE, referred to as A-peptide). In addition, dopamine (1.92 mM) and nitrophenol (640 µM) were tested using the sulfotransferase enzyme-coupling assay described above. To determine the optimum conditions for RaxTST activity, RaxTST and Y-peptide were incubated at 6, 16, 30, or 37 °C and at pH 5, 6, 7, or 8 in 10 mM MES, HEPES, or Tris-HCl buffer at 30 °C. Following the reaction with RaxTST and Y-peptide, the resultant APS kinase activity was monitored at 30 °C, pH 7.0. For kinetic analysis of RaxTST with Y-peptide, we used 0, 0.5, 1, 2.5, 5, 10, 25, or 50 µM Y-peptide. The Km and Vmax values were determined using a double reciprocal plot.
Ax21 is a substrate of RaxST
To test if peptide axY22 is a substrate of RaxST, we generated two peptides based on the amino acid sequence of the Ax21 protein (AENLSYNFVEGDYVRTP, axY22, and AENLSANFVEGDYVRTP, axY22A). The reaction was initiated by adding 0.45 µg of partially purified RaxTST protein and 100 µM axY22 or axY22A in 50 µl of sulfotransferase buffer. After four hours of incubation at 30°C, the reaction mixture was chilled on ice. Fifty µl of APS cocktail buffer was added to the ice-chilled sulfotransferase reaction mixture. The coupling assay was finally initiated by adding APS kinase. NADPH production was measured using a SafireII spectrophotometer (Tecan) at 340 nm and 30 °C. To determine if the mature Ax21 protein also serves as substrate for RaxST and if other tyrosine residues could serve as sites for RaxST-mediated sulfation we used E. coli-purified Ax21 and RaxTST. We incubated 20 µg of RaxTST with 200 µg highly purified MBP-Ax21 fusion in freshly made sulfation buffer (25 mM Tris pH 7.5, 150 mM NaCl, 20 mM MgCl2, 480 µM PAPS) for 4 h at 30 °C. Samples were then kept on dry ice, or −80 °C until analyzed by LC-MS/MS.
Sample Preparation for LC-MS/MS analysis
20 ug of the RaxTST/MBP-Ax21 fusion protein mixture was prepared for digestion by performing a buffer exchange using a 10 KDa molecular weight cut-off filter. The filtered protein was diluted in 1% Protease Max (Promega) in 1× PBS. The protein was reduced with dithiothreitol (DTT) for 30 minutes (55 °C) then incubated with iodoacetamide at room temperature for 30 minutes in the dark. For GluC digestion, 1 uL of 1 ug/uL GluC protease in water was added, giving a protein to enzyme ratio of 20:1. The digestion solution was incubated for four hours at 37 °C. For trypsin digestion, 1µL of trypsin (1 µg/µL) was added to 20 µg of buffer exchanged, reduced and alkylated protein solution and incubated overnight at 37 °C. Following digestion, the solutions was desalted using C18 spin columns (Thermo/Pierce) and reconstituted in 98% water, 2% acetonitrile for LCMS/MS analysis.
LC-MS/MS and UVPD Analysis
LCMS analysis was undertaken using a Dionex Ultimate 3000 NSLC system operated in a reversed phase nano-liquid chromatography mode at a flow rate of 300 nL/min. 3 µL of the GluC digest was injected onto a New Objective IntegraFrit 100 µm inner diameter trap column (Woburn, MA) packed with Michrom Magic C18 AQ (Auburn, CA) to 3.6 cm. Samples were preconcentrated for 3 minutes using 2% acetonitrile/0.05% acetic acid at a flow rate of 5 µL/min. The preconcentration column was then switched in-line with a New Objective PicoFrit analytical column (Woburn, MA) (75 µm × 20 cm) packed with 5 µm Michrom Magic C18 AQ. Separation was performed with eluent A consisting of 0.05% acetic acid in water and eluent B consisting of 0.05% acetic acid in acetonitrile with a 140 min linear gradient from 3% to 30% eluent B at a flow rate of 300 nL/min.
Mass spectrometric analysis was performed on a Thermo Scientific Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a Coherent (Santa Clara, CA) ExciStar XS excimer laser (Coherent Inc., Santa Clara, CA) operated at 193 nm. The coupling of the laser to an orbitrap mass spectrometer was described in detail previously27. Data-dependent nanoLC-MS/UVPD was performed as follows: the first event was the survey negative mass scan (m/z range of 400 – 2000) at resolution 120,000 (1 µscan) followed by ten UVPD events on the ten most abundant ions from the survey scan at resolution 15,000. The isolation width was set to 2.00 and the minimum required signal was 4000.00. Dynamic exclusion settings included a repeat count of 1 during a 30.00 sec timeframe. The exclusion duration was 60.00 sec. Exclusion mass width relative to low and high (ppm) was set to 10. A series of two 2 mJ UV pulses (applied during a 4 ms activation period) was used per MS/MS scan with normalized collision energy set to 1. One µscan was performed per MS/MS scan. After an initial run in which the entire range was monitored in a data dependent manner and the elution of the sulfated peptide (m/z 531) was identified, the mass spectrometer was then set to monitor m/z 531 and to perform UVPD for increased sensitivity during the elution range of 40 and 105 minute. All spectra were processed using the MassMatrix database search engine.
The LCMS CID method incorporated the same survey scan and dynamic exclusion parameters described above followed by a series of top ten data dependent scans for which an isolation width of 2 was used and the minimum required signal was 8000.0. Normalized collision energy for CID was 35.0 with an activation q of 0.250. The number of microscans was set to 1.
In the negative mode the FTMS full AGC target was 1000000.00 and the FTMS MSn AGC target was 50000.00. The maximum ion time was 500.00 ms for FTMS full and MSn scans. The source voltage was 1.60 kV and the capillary temperature was 275 °C. In the positive mode the FTMS full AGC target was 1000000.00 and the ion trap MSn AGC target was 10000.00. The maximum ion time was 100.00 ms and 200 ms for FTMS full and ion trap MSn scans. The source voltage was 1.70 kV and the capillary temperature was 275 °C.
Supplementary Material
Acknowledgements
We thank Sharon Long, Carolyn Bertozzi and Edward Marcotte for helpful discussions as well as Joseph Mougous for critical reading of the manuscript. This work was funded by NIH 2-R01-GM055962-09 (to PCR) and NIH R21 GM099028 (to JSB) and the Welch Foundation (F155 to JSB). This work, in part, was also supported by grants from the Next-Generation BioGreen 21 Program (PJ009006 to S.-W.H and PJ008098 to S.-W. L.), Rural Development Administration, Republic of Korea and the United States - Israel Binational Agricultural Research and Development Fund (OB). We thank Thermo Fisher Scientific with helping on the modifications to the Orbitrap Elite mass spectrometer to allow UVPD.
Footnotes
Author contributions
S.W.H., S.W.L., and P.C.R. designed the project. S.W. H., O. B., B. S., M.R.R., J.B.S., and J.A.M. conducted the experimental work. S.W.H., J.S.B. and P.C.R. wrote the paper.
Competing interests
The authors declare no competing financial interests.
Accession codes:
The mass spectrometry data has been deposited in Peptide Atlas under accession codes:
Ax21 Trypsin positive CID orbitrap: PASS00077
http://www.peptideatlas.org/PASS/PASS00077
Ax21 GluC negative UVPD orbitrap: PASS00078
http://www.peptideatlas.org/PASS/PASS00078
Ax21 GluC positive CID orbitrap: PASS00079
References
- 1.Moore KL. Protein tyrosine sulfation: a critical posttranslation modification in plants and animals. Proc Natl Acad Sci U S A. 2009;106:14741–14742. doi: 10.1073/pnas.0908376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 3.Stone MJ, Chuang S, Hou X, Shoham M, Zhu JZ. Tyrosine sulfation: an increasingly recognised post-translational modification of secreted proteins. N Biotechnol. 2009;25:299–317. doi: 10.1016/j.nbt.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Rizzuto CD, et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 5.Farzan M, et al. Tyrosine Sulfation of the Amino Terminus of CCR5 Facilitates HIV-1 Entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 6.Matsubayashi Y, Ogawa M, Morita A, Sakagami Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science. 2002;296:1470–1472. doi: 10.1126/science.1069607. [DOI] [PubMed] [Google Scholar]
- 7.Roche P, et al. Molecular basis of symbiotic host specificity in Rhizobium meliloti:nodH and nodPQ genes encode the sulfation of lipochito-oligosaccharide signals. Cell. 1991;67:1131–1142. doi: 10.1016/0092-8674(91)90290-f. [DOI] [PubMed] [Google Scholar]
- 8.Mougous JD, et al. Discovery of sulfated metabolites in mycobacteria with a genetic and mass spectrometric approach. Proc Natl Acad Sci U S A. 2002;99:17037–17042. doi: 10.1073/pnas.252514899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keating DH. Sinorhizobium meliloti SyrA mediates the transcriptional regulation of genes involved in lipopolysaccharide sulfation and exopolysaccharide biosynthesis. J Bacteriol. 2007;189:2510–2520. doi: 10.1128/JB.01803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimshaw JP, et al. DsbL and DsbI form a specific dithiol oxidase system for periplasmic arylsulfate sulfotransferase in uropathogenic Escherichia coli. J Mol Biol. 2008;380:667–680. doi: 10.1016/j.jmb.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 11.Ehrhardt DW, et al. In vitro sulfotransferase activity of NodH, a nodulation protein of Rhizobium meliloti required for host-specific nodulation. J Bacteriol. 1995;177:6237–6245. doi: 10.1128/jb.177.21.6237-6245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SW, et al. A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science. 2009;326:850–853. doi: 10.1126/science.1173438. [DOI] [PubMed] [Google Scholar]
- 13.Han SW, Lee SW, Ronald PC. Secretion, modification, and regulation of Ax21. Curr Opin Microbiol. 2011;14:62–67. doi: 10.1016/j.mib.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Han SW, et al. Small protein-mediated quorum sensing in a Gram-negative bacterium. PloS one. 2011;6:e29192. doi: 10.1371/journal.pone.0029192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.McCarthy Y, Dow JM, Ryan RP. The Ax21 protein is a cell-cell signal that regulates virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J Bacteriol. 2011;193:6375–6378. doi: 10.1128/JB.05949-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.da Silva FG, et al. Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol. Plant Microbe Interact. 2004;17:593–601. doi: 10.1094/MPMI.2004.17.6.593. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y, Sharma P, Silva FG, Ronald P. The Xanthomonas oryzae pv. oryzae raxP and raxQ genes encode an ATP sulfurylase and APS kinase that are required for AvrXa21 avirulence activity. Mol. Microbiol. 2002;44:37–48. doi: 10.1046/j.1365-2958.2002.02862.x. [DOI] [PubMed] [Google Scholar]
- 18.Lee SW, Han SW, Bartley LE, Ronald PC. Unique characteristics of Xanthomonas oryzae pv. oryzae AvrXa21 and implications for plant innate immunity. Proc Natl Acad Sci U S A. 2006;103:18395–18400. doi: 10.1073/pnas.0605508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seibert C, Cadene M, Sanfiz A, Chait BT, Sakmar TP. Tyrosine sulfation of CCR5 N-terminal peptide by tyrosylprotein sulfotransferases 1 and 2 follows a discrete pattern and temporal sequence. Proc Natl Acad Sci U S A. 2002;99:11031–11036. doi: 10.1073/pnas.172380899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu JH, et al. Crystal structure of human sulfotransferase SULT1A3 in complex with dopamine and 3'-phosphoadenosine 5'-phosphate. Biochem Biophys Res Commun. 2005;335:417–423. doi: 10.1016/j.bbrc.2005.07.091. [DOI] [PubMed] [Google Scholar]
- 21.Meinl W, Meerman JH, Glatt H. Differential activation of promutagens by alloenzymes of human sulfotransferase 1A2 expressed in Salmonella typhimurium. Pharmacogenetics. 2002;12:677–689. doi: 10.1097/00008571-200212000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Madsen JA, Boutz DR, Brodbelt JS. Ultrafast ultraviolet photodissociation at 193 nm and its applicability to proteomic workflows. Journal of proteome research. 2010;9:4205–4214. doi: 10.1021/pr100515x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madsen JA, Kaoud TS, Dalby KN, Brodbelt JS. 193-nm photodissociation of singly and multiply charged peptide anions for acidic proteome characterization. Proteomics. 2011;11:1329–1334. doi: 10.1002/pmic.201000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brodbelt JS. Shedding light on the frontier of photodissociation. Journal of the American Society for Mass Spectrometry. 2011;22:197–206. doi: 10.1007/s13361-010-0023-6. [DOI] [PubMed] [Google Scholar]
- 25.Cook SL, Jackson GP. Metastable atom-activated dissociation mass spectrometry of phosphorylated and sulfonated peptides in negative ion mode. Journal of the American Society for Mass Spectrometry. 2011;22:1088–1099. doi: 10.1007/s13361-011-0123-y. [DOI] [PubMed] [Google Scholar]
- 26.Renosto F, Seubert PA, Segel IH. Adenosine 5'-phosphosulfate kinase from Penicillium chrysogenum. Purification and kinetic characterization. J. Biol. Chem. 1984;259:2113–2123. [PubMed] [Google Scholar]
- 27.Vasicek LA, et al. Implementing photodissociation in an Orbitrap mass spectrometer. Journal of the American Society for Mass Spectrometry. 2011;22:1105–1108. doi: 10.1007/s13361-011-0119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.