Abstract
Chemokine receptor 1 (CCR1) is a G protein–coupled receptor that binds to members of the C-C chemokine family. Recently, CCL3 (MIP-1α), a high-affinity CCR1 ligand, was identified as part of a model that independently predicts survival in patients with diffuse large B-cell lymphoma (DLBCL). However, the role of chemokine signaling in the pathogenesis of human lymphomas is unclear. In normal human hematopoietic tissues, we found CCR1 expression in intraepithelial B cells of human tonsil and granulocytic/monocytic cells in the bone marrow. Immunohistochemical analysis of 944 cases of hematolymphoid neoplasia identified CCR1 expression in a subset of B- and T-cell lymphomas, plasma cell myeloma, acute myeloid leukemia, and classical Hodgkin lymphoma. CCR1 expression correlated with the non–germinal center subtype of DLBCL but did not predict overall survival in follicular lymphoma. These data suggest that CCR1 may be useful for lymphoma classification and support a role for chemokine signaling in the pathogenesis of hematolymphoid neoplasia.
Keywords: Chemokine, CCR1, Lymphoma, Leukemia
Chemokines are small soluble proteins (8–10 kDa) that function to recruit leukocytes to sites of inflammation. Accumulating evidence suggests that in addition to their role in mediating the immune response, dysregulation of chemokine production and/or signaling may have an important role in carcinogenesis1,2 and hematolymphoid neoplasia.3 Chemokines are classified into 4 groups based on the position of the first 2 cysteine residues in the amino acid sequence at the amino terminus. The C-C family of chemokines includes monocyte chemoattractant protein (CCL2), RANTES (CCL5), and macrophage inhibitory proteins MIP-1α (CCL3) and MIP-1β (CCL4). One member of the C-C chemokine subfamily, CCL3, is inducible in a number of hematopoietic cells, particularly those involved in the adaptive immune response (macrophages, dendritic cells, and B and T lymphocytes).4 CCL3 exerts its biologic effects by binding to 2 related cell surface G protein–coupled receptors, chemokine receptor 1 (CCR1) and chemokine receptor 5 (CCR5). CCR1 is expressed by a diverse group of hematopoietic cells, including monocytes, dendritic cells, T lymphocytes, neutrophils, and CD34+ bone marrow progenitor cells.5 Although the details are not entirely clear, CCR1 activation leads to protein kinase C activation and stimulation of the MAP kinase pathway.6
Recently, the gene (SCYA3) encoding CCL3 was identified as part of a multivariate model based on the expression of 6 genes—LMO2, BCL6, FN1, CCND2, SCYA3, and BCL2—that independently predicts survival in patients with diffuse large B-cell lymphoma (DLBCL).7 In this analysis, SCYA3 expression (along with BCL2 and CCND2) was shown to be a potent predictor of poorer overall patient survival. These results were consistent with prior gene expression experiments that correlated SCYA3 expression with the activated B-cell (non–germinal center) subtype of DLBCL.8,9
In addition to its possible role in the pathogenesis of DLBCL, CCL3 expression has also been implicated in other hematolymphoid neoplasms. For example, increased serum levels of CCL3 have been shown to correlate with lytic bony lesions and overall survival in multiple myeloma.10 Currently, the mechanism by which CCL3 expression contributes to poor outcome in DLBCL and multiple myeloma is unclear. Possible mechanisms include dysregulation of the host immune response through cytokine secretion by tumor or autocrine stimulation of cell proliferation via tumor-associated chemokine receptor molecules such as CCR1. Accumulating evidence suggests that CCR1 signaling may have an important role in the pathogenesis of DLBCL and other human lymphomas. Increased CCR1 messenger RNA (mRNA) expression has been shown in DLBCL,11 small lymphocytic lymphoma/chronic lymphocytic leukemia (SLL/CLL) and hairy cell leukemia,12 MALT lymphoma,13 and the leukemic cells of cutaneous T-cell lymphoma (Sézary syndrome).14 Increased CCR1 mRNA expression has been correlated with shorter survival in patients with follicular lymphoma,15 and cell-surface expression of CCR1 has been associated with disease activity in multiple myeloma.16 Taken together, these data suggest that chemokine signaling mediated through CCR1 may promote neoplastic transformation and perhaps contribute to an aggressive phenotype. Therefore, characterization of CCR1 protein expression may provide important pathogenic and prognostic information for DLBCL and other human hematolymphoid malignancies.
To investigate the expression of CCR1 in hematolymphoid malignancy, we optimized a commercially available anti-CCR1 antibody for use on paraffin-embedded material. We then characterized CCR1 protein expression on normal human hematopoietic tissue and human lymphoma and leukemia samples using tissue microarrays (TMAs) and whole tissue sections. We then correlated CCR1 protein expression with other prognostic markers for DLBCL and investigated the relationship of CCR1 expression to overall survival in follicular lymphoma.
Materials and Methods
Tissue Samples
Formalin fixed, paraffin-embedded tissue samples of normal and neoplastic human hematolymphoid tissue were obtained from the archives of the Department of Pathology, Stanford University Medical Center, Stanford, CA. Tissues were obtained before treatment, and institutional review board approval was obtained for this study. TMAs were constructed as previously described.17 For expression in normal hematopoietic tissue, whole tissue sections of normal human spleen, thymus, bone marrow, and tonsil were used. Hematolymphoid neoplasia was classified according to the 2001 World Health Organization classification.18
Immunohistochemical Studies
TMAs, whole-sections of human lymphoma and leukemia samples, and normal human hematopoietic tissue samples were sectioned at 0.4 µm thickness, deparaffinized in xylene, and hydrated in graduated alcohols. Slides were pretreated with 10 mmol/L citrate buffer, pH 6.0. Slides were then stained with a rabbit polyclonal IgG anti-CCR1 antibody (GenWay Biotech, San Diego, CA) at 1:40 dilution. Slides were developed using the DAKO EnVision method (DAKO, Carpinteria, CA) and coverslipped with aqueous-based mounting medium. TMAs were scored as follows: 0, no staining; 2, weak staining (5%–20% of cells positive); 3, strong staining (>20% of cells positive); and 1, uninterpretable (eg, loss of sample tissue, high background). For the data reported in Table 1, cases were scored as positive if more than 20% of the cells stained positively for CCR1. Each sample was independently scored by 2 pathologists (M.W.A. and Y.N.) with a concordance rate of 97.5%. Discrepant cases were jointly reviewed, and a final score was assigned.
Table 1.
Immunohistologic Analysis of Chemokine Receptor 1 Protein Expression in Human Hematolymphoid Neoplasia
| Lymphoma Subtype | Total Positive/Total No. of Cases (% Positive) |
|---|---|
| B-cell lymphoma (n = 674) | |
| Follicular lymphoma | 122/349 (35.0) |
| Grade 1 | 51/153 (33.3) |
| Grade 2 | 46/120 (38.3) |
| Grade 3 | 25/76 (33) |
| Diffuse large B-cell lymphoma | 77/209 (36.8) |
| Mediastinal large B-cell lymphoma | 2/8 (25) |
| Burkitt lymphoma | 0/4 (0) |
| Extranodal marginal zone lymphoma | 1/21 (5) |
| Splenic marginal zone lymphoma | 0/5 (0) |
| Nodal marginal zone lymphoma | 3/6 (50) |
| Mantle cell lymphoma | 1/17 (6) |
| Small lymphocytic lymphoma/chronic lymphocytic leukemia | 4/35 (11) |
| Lymphoplasmacytic lymphoma | 3/5 (60) |
| Precursor B-lymphoblastic lymphoma | 1/11 (9) |
| Hairy cell leukemia | 0/4 (0) |
| T-cell lymphoma (n = 47) | |
| Precursor T-lymphoblastic lymphoma | 1/14 (7) |
| Peripheral T-cell lymphoma | 10/21 (48) |
| Extranodal NK/T cell lymphoma, nasal type | 0/4 (0) |
| Anaplastic large cell lymphoma | 5/8 (63) |
| Plasma cell neoplasms (n = 106) | |
| Monoclonal gammopathy of undetermined significance | 3/4 (75) |
| Multiple myeloma | 60/83 (72) |
| Plasma cell leukemia | 7/10 (70) |
| Plasmacytoma | 1/9 (11) |
| Hodgkin lymphoma (n = 107) | |
| Lymphocyte predominance Hodgkin lymphoma | 0/18 (0) |
| Classical Hodgkin lymphoma | 26/89 (29) |
| Acute myeloid leukemia (n = 10) | 8/10 (80) |
Double-immunohistochemical labeling was performed by using the DAKO EnVision Doublestain System according to the manufacturer’s specifications. Antibodies to CCR1 (dilution 1:40; GenWay Biotech), CD20 (dilution 1:1,000; DAKO), CD56 (dilution 1:10; Zymed Laboratories, South San Francisco, CA), and CD3 (dilution 1:100; Cell Marque, Rocklin, CA) were used on paraffin-embedded normal human tonsil sections. Heat-induced antigen retrieval was carried out in EDTA/tris(hydroxymethyl) aminomethane (Tris) buffer using a pressure cooker. After endogenous peroxidase blockade with 0.03% hydrogen peroxide, the slides were incubated with the first primary antibody (CD20, CD56, or CD3) at an optimized dilution. The slide was then rinsed in Tris-buffered saline with polysorbate 20, pH 9.0, and incubated with the secondary antibody (labeled polymer–horseradish peroxidase) and liquid diaminobenzidine chromogen (brown stain). Slides were rinsed in distilled water and incubated with the Doublestain Block reagent (DAKO) followed by incubation with the second primary antibody, CCR1, at a dilution of 1:40. Secondary antibody (labeled polymer–alkaline phosphatase) with the Fast red chromogen (red stain) was then applied. The slides were then coverslipped with an aqueous-based mounting medium.
Data Analysis and Visualization
Images of normal human hematolymphoid tissue and TMA immunohistochemical staining results were acquired using a Nikon Eclipse E1000 microscope (Nikon, Tokyo, Japan) equipped with 4×, 10×, 20×, 40×, and 60× objective lenses with numeric apertures ranging from 0.05 to 0.90. Images were captured with a SPOT flex mosaic 15.2 digital camera and software (Diagnostic Instruments, Sterling Heights, MI). Digitized images were processed using Adobe Illustrator software (Adobe Systems, San Jose, CA). The stained lymphoma TMA slides were scanned and stored as high-resolution images using an automated scanner (Bacus Laboratories Slide Scanner, Olympus, Center Valley, PA), and are accessible at http://tma.stanford.edu/tma_portal/CCR1. Cluster analysis19 was used for hierarchical clustering of the immunohistochemical data of 143 cases of DLBCL. Graphic representation of the clustering results was produced using TreeView.20 Positive staining is represented in red, negative staining in green, and uninterpretable staining in white.
Statistical Analysis
The Pearson χ2 test was used to compare the CCR1 expression score (0, 1, 2, 3) for TMAs with the histologic grade and patient Follicular Lymphoma International Prognostic Index (FLIPI) score. The relationship between CCR1 score and overall patient survival was analyzed using the likelihood ratio test. Overall survival was examined by the Kaplan-Meier method (PRISM, GraphPad Software, San Diego, CA).
Results
Expression of CCR1 Protein in Normal Hematolymphoid Tissue
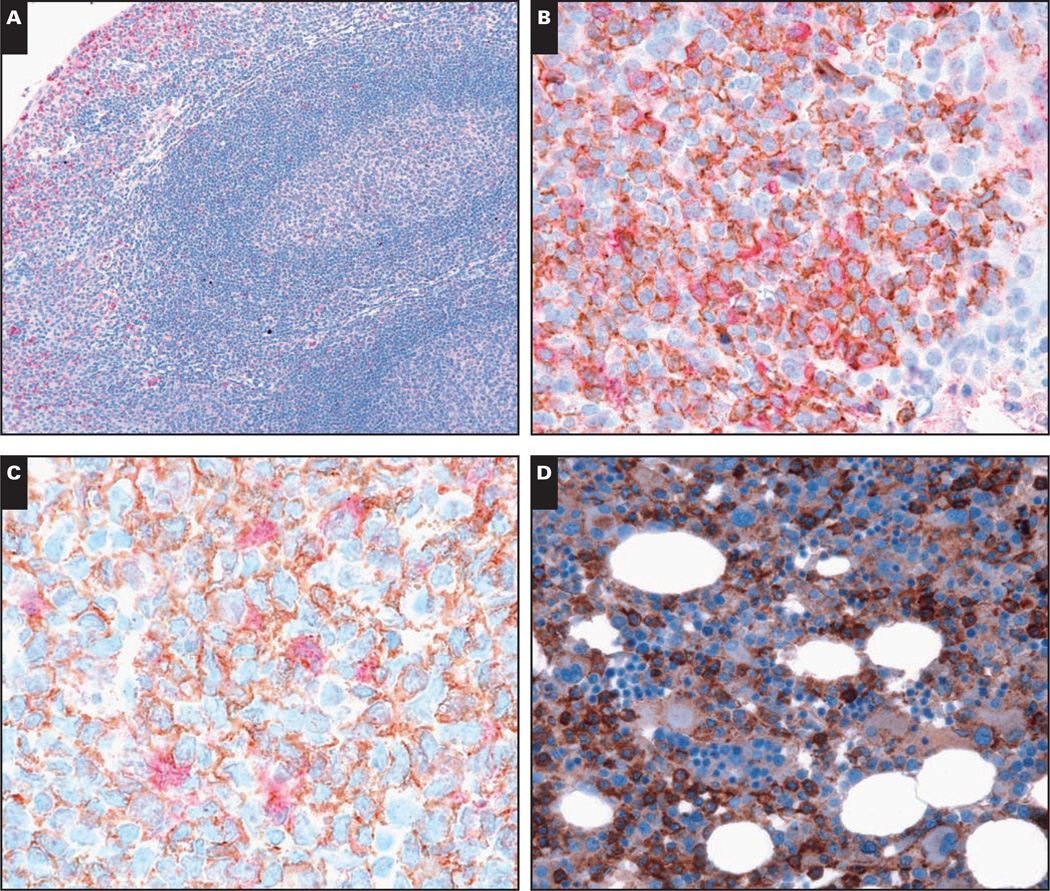
In normal human tonsil, staining for CCR1 protein was preferentially found in intraepithelial lymphocytes within the tonsillar crypts Image 1A and Image 1B. Rare scattered CCR1+ cells were also identified within the germinal center, mantle, parafollicular, and paracortical regions (Image 1A). CCR1 staining was localized to the plasma membrane and cytoplasm. To more precisely define the lineage of CCR1-expressing cells, double immunolabeling experiments were performed. As shown in Image 1B, a significant percentage of the CCR1+ cells within the tonsillar crypt epithelium were of B lineage (CD20+). Only a minority of germinal center B cells and rare mantle zone B lymphocytes showed CCR1 staining. Scattered CCR1-expressing cells in the germinal center were CD20– and showed fine cytoplasmic projections consistent with dendritic cell morphologic features Image 1C. No significant coexpression of CCR1 with CD3 or CD56 was observed (data not shown), suggesting that in normal tonsil, very few T or NK cells express CCR1.
Image 1.
Immunohistologic staining for chemokine receptor 1 (CCR1) protein in normal human hematopoietic tissues. A, CCR1 protein (red, cytoplasm and cell membrane) is predominantly localized to intraepithelial lymphocytes within the tonsillar crypts (×100). B, Double immunolabeling of tonsil sections shows colocalization of CCR1 (red) with CD20 (brown, membrane) on B lymphocytes within the tonsillar crypt epithelium (×400). C, Rare CCR1-expressing cells within the germinal center (red) show dendritic cell morphologic features and do not colocalize with CD20 (brown) (×600). D, In normal bone marrow, CCR1 is expressed by cells of the granulocyte-monocyte lineage but not in cells of the erythroid lineage (×400).
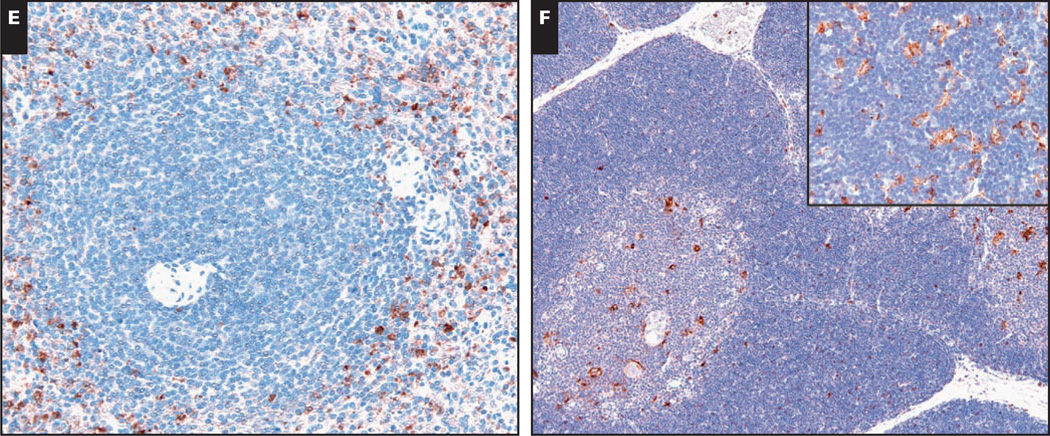
E, In the spleen, CCR1 protein expression is found largely within granulocytes in the red pulp. No significant CCR1 staining is present within the germinal centers of the white pulp (×200). F, In the thymus, CCR1 highlights medullary dendritic cells and the Hassall corpuscles without significant staining of cortical thymocytes (×100). Rare cases show CCR1 expression by cortical epithelial cells (inset, ×400).
In normal human bone marrow, CCR1 was strongly expressed in cells of the monocyte-granulocyte lineage and plasma cells Image 1D. Consistent with previous reports, CCR1 expression was absent from erythroid progenitor cells.21 Megakaryocytes also lacked CCR1 expression.
Sections of normal human spleen Image 1E showed CCR1 expression in histiocytes and granulocytes within the red pulp. Scattered lymphocytes were positive in the mantle and marginal zone of white pulp follicles, but no significant staining was observed within the germinal center.
In normal human thymus Image 1F, CCR1 was strongly expressed by macrophages and Hassall corpuscles within the medulla. In the 11 samples of human thymus stained for CCR1, we found a consistent lack of CCR1 expression by cortical thymocytes. However, in 1 case, CCR1 stained cortical cells with dendritic morphologic features, suggestive of cortical epithelial cells (Image 1F, inset).
Expression of CCR1 Protein in Hematolymphoid Neoplasia
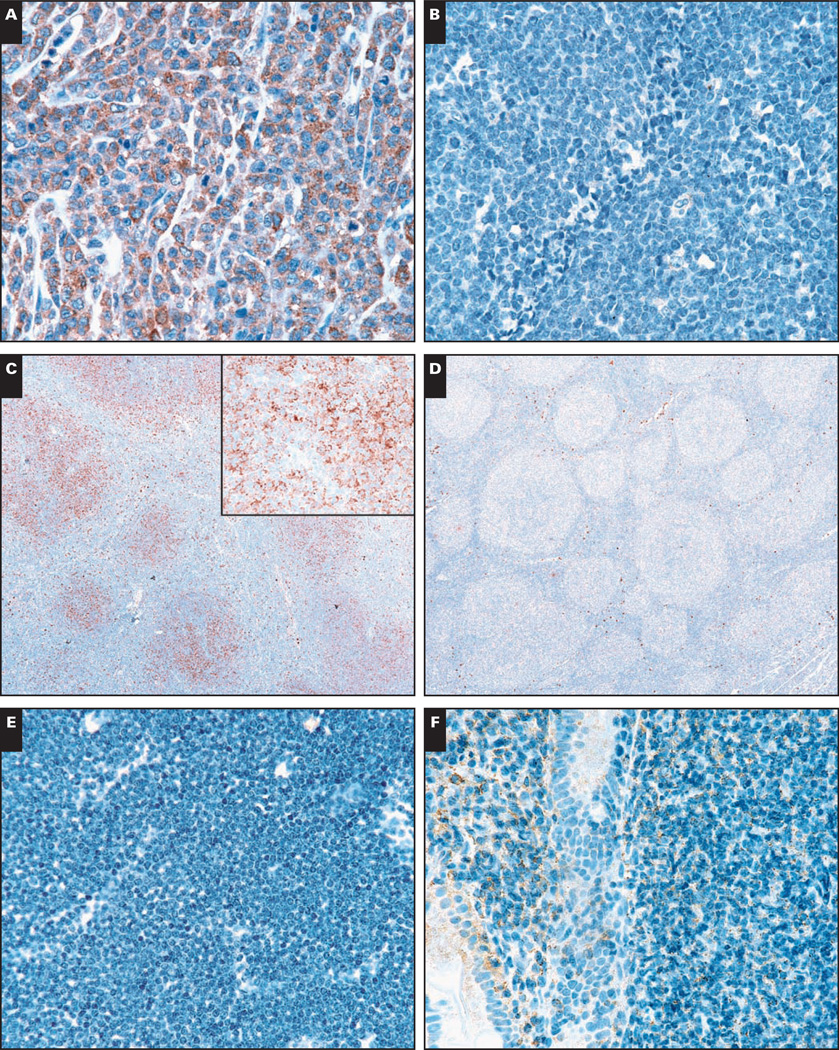
TMAs and whole tissue sections encompassing 944 samples of human lymphomas and leukemias were used to investigate the expression of CCR1 protein in human hematopoietic neoplasms. As shown in Table 1 and Image 2, immunoreactivity for CCR1 protein was observed in lymphomas of T- and B-cell derivation. Examples included follicular lymphoma (all histologic grades) (122/349) (Image 2C), DLBCL (77/209) (Image 2A), mediastinal large B-cell lymphoma (2/8), peripheral T-cell lymphoma (10/21) (Image 2I), and anaplastic large cell lymphoma (5/8) (Image 2J). Negative examples of DLBCL (Image 2B) and follicular lymphoma (Image 2D) showed minimal background staining.
Image 2.
Immunohistologic staining for chemokine receptor 1 (CCR1) protein expression in human hematolymphoid neoplasia. Representative examples of CCR1 immunostaining in human lymphomas and leukemia are shown. CCR1 is expressed in cases of diffuse large B-cell lymphoma (A, ×400) and follicular lymphoma (C, ×40), with negative examples provided for comparison (B, ×400; D, ×40). In follicular lymphoma, CCR1 expression is present on the neoplastic cells (C, inset, ×400).
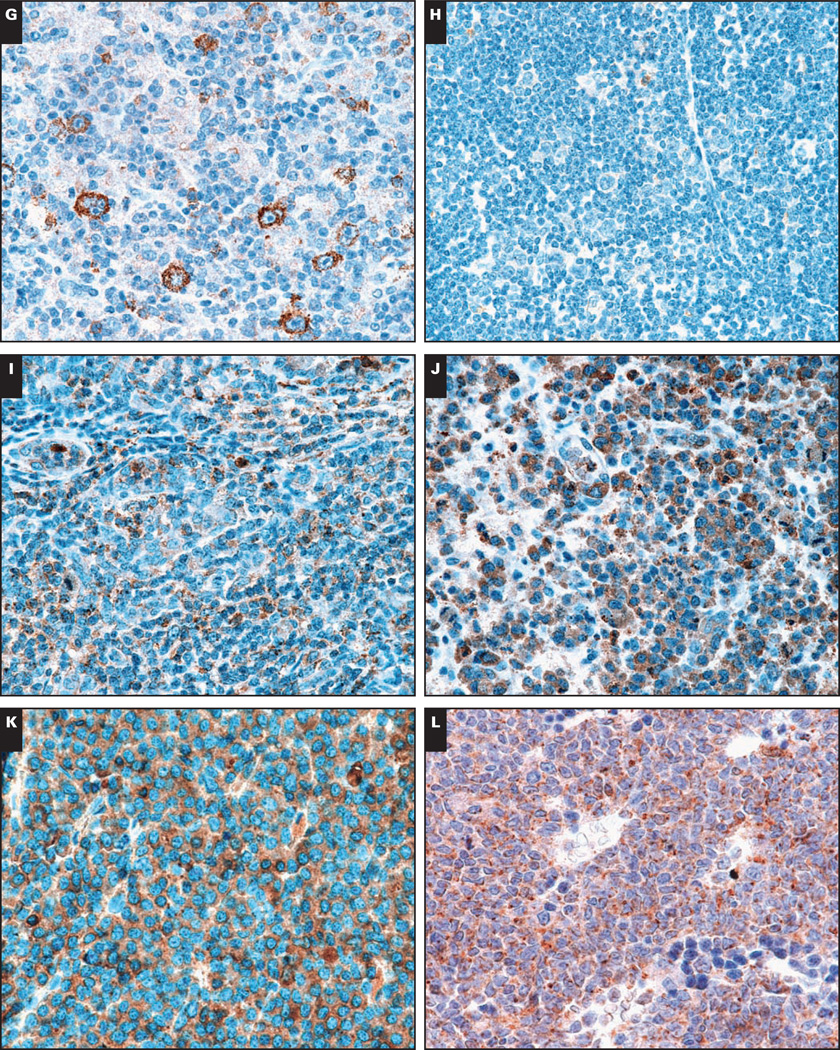
While CCR1 expression was present in cases of classical Hodgkin lymphoma (G, ×400), expression was largely absent in cases of extranodal marginal zone lymphoma (F, ×400), small lymphocytic lymphoma/chronic lymphocytic leukemia (E, ×400), and nodular lymphocyte predominant Hodgkin lymphoma (H, ×400). CCR1 was expressed by T-cell lymphomas, including peripheral T-cell lymphoma, not otherwise specified (I, ×400) and anaplastic large cell lymphoma (J, ×400). CCR1 expression was present in a majority of cases of multiple myeloma (K, ×400) and acute myeloid leukemia (L, ×400).
Consistent with previous studies,22,23 the majority of multiple myeloma cases (60/83) showed CCR1 positivity (Image 2K). Although cases of monoclonal gammopathy of undetermined significance and plasma cell leukemia showed similar rates of positive staining (~70%), we observed significantly less CCR1 staining in cases of extramedullary plasmacytoma.
A significant percentage of classical Hodgkin lymphoma showed strong membrane CCR1 immunoreactivity (Image 2G), but nodular lymphocyte predominant Hodgkin lymphoma did not (Image 2H). Significantly lower expression was observed in marginal zone lymphoma (with the exception of the nodal subtype), mantle cell lymphoma, and SLL/CLL (Images 2E and 2F). The relative lack of staining in SLL/CLL in our series may be due to a preponderance of SLL cases.12 No significant staining was observed in examples of Burkitt lymphoma, hairy cell leukemia, or NK cell lymphomas. Rare cases of precursor B- and T-cell lymphomas (1/11 and 1/14, respectively) showed positive CCR1 staining. Consistent with the strong CCR1 expression by cells of the myeloid lineage (Image 1D), a majority of the cases of acute myeloid leukemia (8/10) showed strong CCR1 expression (Image 2L). Within this small number of cases, the expression of CCR1 appeared to be independent of FAB subtype, although cases of the M6 subtype were not included in this analysis.
CCR1 Expression and DLBCL Subtype
Previous gene expression profiling experiments have defined 2 biologically distinct subtypes of DLBCL, the germinal center B-cell type (GCB) and the activated B-cell type (ABC).8,24 In addition, expression of genes associated with the GCB subtype (LMO2 and BCL6) or the ABC subtype (BCL2) has also been shown to strongly predict patient survival.7 In that study, the expression of the gene (SCYA3) encoding the CCR1 ligand CCL3 was correlated with poorer overall patient survival. Although the relationship between SCYA3 gene expression and the ABC subtype is unknown, we asked whether CCR1 protein expression would correlate with the GCB or ABC subtype of DLBCL.
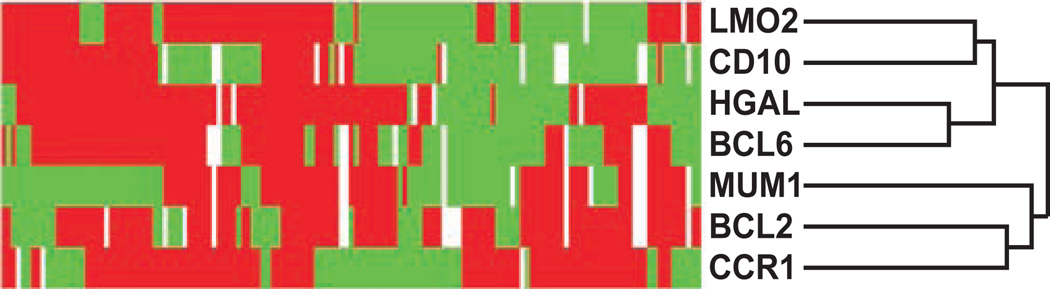
To address this question, we analyzed CCR1 protein expression in 143 cases of DLBCL that had previously been stained with antibodies to proteins associated with germinal center or non–germinal center B-cell derivation (human germinal center–associated lymphoma [HGAL], BCL6, CD10, BCL2, LMO2, and MUM1).25–27 Through the use of hierarchical cluster analysis, we found that CCR1 protein expression correlated with the expression of the ABC markers BCL2 and MUM1 but not with the GCB markers LMO2, BCL6, CD10, and HGAL Image 3. These results indicated that CCR1 expression is associated with the ABC subtype of DLBCL and provide a possible biologic mechanism for poorer patient outcome in association with high SCYA3 mRNA expression levels.
Image 3.
Hierarchical cluster analysis of immunohistologic results for diffuse large B-cell lymphoma (DLBCL). The expression patterns of LMO2, human germinal center–associated lymphoma (HGAL), CD10, BCL6, BCL2, MUM1, and chemokine receptor 1 (CCR1) in 143 cases of DLBCL are shown. Positive staining is represented in red, negative staining in green, and uninformative data (loss of core tissue) in white. CCR1 protein expression clusters on the dendrogram branch, which includes the non–germinal center (GC) markers BCL2 and MUM1, and away from the GC-associated markers LMO2, BCL6, CD10, and HGAL.
Expression of CCR1 in Follicular Lymphoma
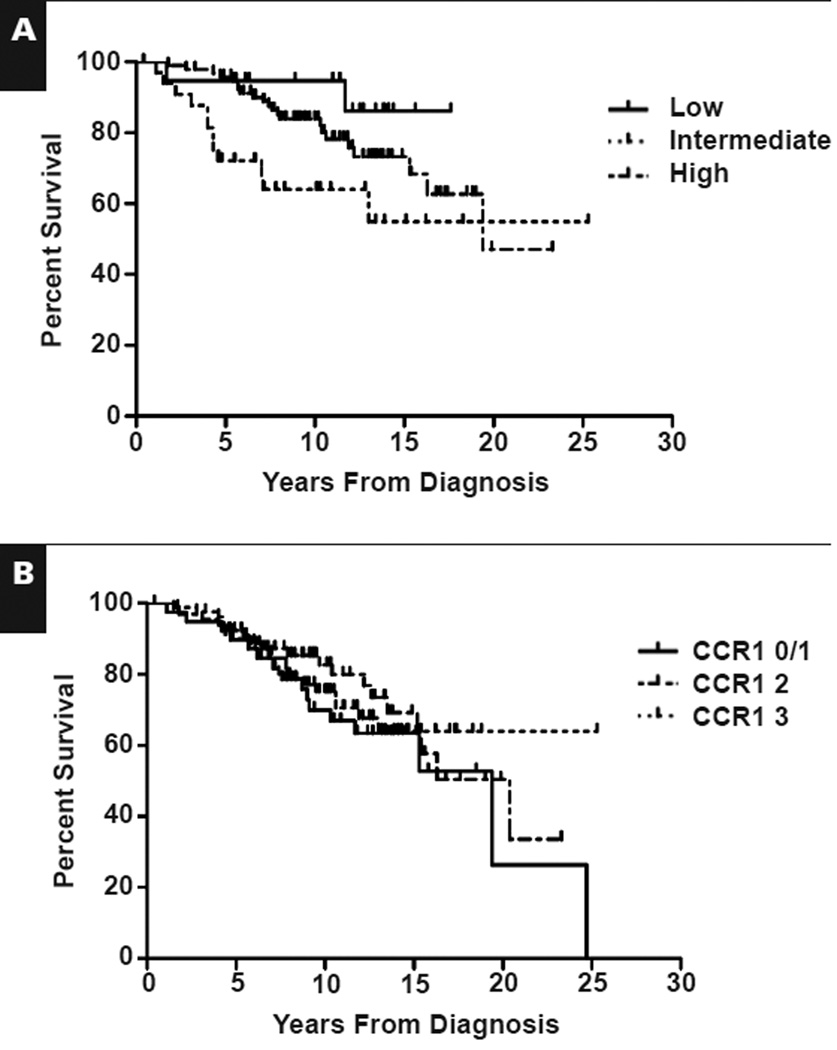
To evaluate CCR1 protein expression in follicular lymphoma, we analyzed 187 cases of untreated follicular lymphoma of all grades and correlated CCR1 protein expression with histologic grade and patient outcome. The characteristics of the patients are described in Table 2. FLIPI28 scores were available for 147 of 187 patients, with a distribution of 12.9% low, 64.6% intermediate, and 22.4% high. As expected, FLIPI groups correlated with overall survival Figure 1A, but we found no correlation between CCR1 expression and overall survival Figure 1B. In addition, we found no significant correlation with CCR1 expression and histologic grade (P = .88; Pearson χ2) or the FLIPI score (P = .842; Pearson χ2) (data not shown). These results suggested that in follicular lymphoma, CCR1 protein expression is not correlated with patient outcome and does not correlate with FLIPI score or histologic grade.
Table 2.
Characteristics of 187 Patients With Follicular Lymphoma*
| Characteristic | Result |
|---|---|
| Median (range) age (y) | 45 (15–17) |
| ≤60 | 164 (87.7) |
| >60 | 23 (12.3) |
| Median follow-up (y) | 10.0 |
| Stage | |
| I | 1 (0.5) |
| II | 2 (1.1) |
| III | 51 (27.3) |
| IV | 130 (69.5) |
| Unknown | 3 (1.6) |
| B symptoms (n = 184) | |
| Absent | 166 (88.8) |
| Present | 18 (9.6) |
| Unknown | 3 (1.6) |
| Histologic grade | |
| 1 | 112 (59.9) |
| 2 | 65 (34.8) |
| 3 | 10 (5.3) |
| Lactate dehydrogenase level | |
| Abnormal | 21 (11.2) |
| Normal | 130 (69.5) |
| Unknown | 36 (19.3) |
| FLIPI group | |
| Low | 19 (10.2) |
| Intermediate | 95 (50.8) |
| High | 33 (17.6) |
| Unknown | 40 (21.4) |
FLIPI, Follicular Lymphoma International Prognostic Index.
Data are given as number (percentage) unless otherwise indicated.
Figure 1.
Kaplan-Meier estimate of overall survival in patients with follicular lymphoma. A, Follicular Lymphoma International Prognostic Index scores predict overall survival. P = .027. B, Chemokine receptor 1 (CCR1) expression level is not correlated with overall survival. CCR1 levels were scored as follows: 0, negative; 1, uninterpretable; 2, 5%–20% of cells positive; and 3, >20% of cells positive. P = .60.
Discussion
In a previous flow cytometric analysis of normal human peripheral blood hematopoietic cells, CCR1 protein expression was restricted to T lymphocytes, monocytes, and granulocytes, with a relative lack of expression by CD19+ B lymphocytes.5 In this work, we also found a relative absence of CCR1 protein expression in cells of B lineage. However, a notable exception was the strong expression of CCR1 in CD20+ crypt intraepithelial B lymphocytes of normal human tonsil. Intraepithelial tonsillar B lymphocytes have been proposed to be related to marginal zone B cells in other organs such as the gastrointestinal tract.29 Therefore, CCR1 may have a role in trafficking of these specialized B lymphocytes to mucosal sites. CCR1 did not seem to be significantly expressed by other B-cell populations in the tonsil or spleen. It is interesting that we found a relative lack of CCR1 expression in cases of extranodal marginal zone lymphoma, suggesting that CCR1 down-regulation may have a role in the pathogenesis of this disease. In support of this hypothesis, previous studies have shown a relative decrease in CCR1 mRNA expression levels in extragastric marginal zone lymphomas as compared with extranodal DLBCL.13
Although we found CCR1 protein expression in cells of the monocytic and granulocytic lineage in bone marrow and other tissues, we did not detect significant CCR1 protein expression in immature T cells in the thymus or mature T cells in normal tonsil. These results are consistent with the phenotype of CCR1-deficient mice that showed disordered trafficking and proliferation of myeloid progenitors and mature neutrophils without significant abnormalities in the distribution of mature lymphocytes.30
In this work, we show CCR1 protein expression in a number of human lymphomas of B-, T-, and plasma cell differentiation, supporting a role for CCR1 expression in the pathogenesis of lymphomas derived from multiple lymphoid lineages. In B cells, CCR1 expression was not specific for germinal center derivation, as a significant number of follicular lymphoma and DLBCL cases showed CCR1 expression, but nodular lymphocyte predominant Hodgkin and Burkitt lymphomas did not. It is interesting that despite significant CCR1 expression in B-cell lymphomas, CCR1 was not present at significant levels in normal germinal center or marginal zone B cells in tonsil or spleen. Therefore, up-regulation of CCR1 expression by B cells may occur during neoplastic transformation.
To our knowledge, this is also the first report of CCR1 expression in classical Hodgkin lymphoma. Recently, members of our group reported that expression of the germinal center–associated marker HGAL in classical Hodgkin lymphoma was correlated with improved overall survival in univariate analysis.31 As CCR1 clustered with non–germinal center markers BCL2 and MUM1 in DLBCL, it will be interesting to determine if CCR1 expression correlates with poorer overall survival in classical Hodgkin lymphoma. CCR1 was also highly expressed by plasma cells in cases of multiple myeloma. It is interesting that although the majority of cases of multiple myeloma showed strong CCR1 expression, there was a relative lack of CCR1 staining in plasmacytomas located in extramedullary sites. As CCL3 and CCR1 promote the association of multiple myeloma cells to bone marrow stromal cells,23 it is tempting to speculate that loss of CCR1 expression by multiple myeloma cells may contribute to the formation of plasmacytomas at extramedullary sites. However, additional cases of extramedullary plasmacytoma are required to address this possibility in a statistically significant manner. Although CCR1 is strongly expressed by plasma cells of multiple myeloma, the strong staining of granulocytic-monocytic precursors in the marrow likely limits the diagnostic usefulness of CCR1 immunohistochemical analysis to detect small numbers of myeloma cells.
The demonstration of CCR1 protein expression in DLBCL also provides a possible biologic mechanism for the observation that CCL3 expression by DLBCL is associated with poorer overall patient survival. An attractive hypothesis would be that coordinate up-regulation of CCL3 and CCR1 expression would lead to an autocrine stimulation of cell proliferation. However, CCR1 binds to multiple chemokines of the C-C subfamily.32 Therefore, an alternative hypothesis might be that CCL3 expression by DLBCL recruits inflammatory cells that release other C-C chemokines that act to stimulate cell proliferation through CCR1. To resolve this issue, immunohistochemical analysis of CCL3 expression by human lymphomas and its relationship to CCR1 protein expression is currently underway.
What might be the prognostic significance of CCR1 expression in DLBCL and other lymphomas? Although CCR1 was not included in the 36-gene panel used to identify prognostically relevant genes in a univariate analysis,7 CCR1 mRNA has been previously found to be up-regulated (82-fold) in 4 cases of DLBCL.11 In this report, each DLBCL case was classified as the ABC subtype based on expression of other genes. Our data provide additional support for CCR1 as a marker of the ABC subtype of DLBCL, as we found that CCR1 clustered with the non–germinal center markers BCL2 and MUM1. Taken together, these data suggest that chemokine signaling though the CCL3/CCR1 pathway may be associated with the ABC subtype of DLBCL.
In a series of 187 cases of follicular lymphoma with clinical follow-up, we observed no statistically significant relationship between CCR1 protein expression and overall patient survival. CCR1 protein expression also did not correlate with histologic grade or FLIPI score.
The lack of a relationship between CCR1 protein expression and overall survival in our patient cohort is in contrast with findings reported by Byers et al,15 who found an association with increased CCR1 mRNA levels and a shorter patient survival interval. These results were correlated with increased numbers of tumor-infiltrating macrophages based on immunohistochemical staining for CD68.
By using immunohistochemical stains for CD163, members of our group have identified very few macrophages per neoplastic follicle in our patient cohort (Gratzinger et al, unpublished data, 2008). Indeed, 87% of cases with strong CCR1 staining (3+) had fewer than 5 macrophages per neoplastic follicle (data not shown), suggesting that in our series, macrophages were not significant contributors to the overall CCR1 staining intensity.
Furthermore, in the study by Byers et al,15 immunohistochemical analysis of CCR1 protein expression was not reported. As we detected CCR1 immunoreactivity on the follicular lymphoma cells, it is possible that CCR1 expression by the tumor cells may have significantly contributed to the observed increase in CCR1 mRNA expression. Differences in the 2 patient groups may have also contributed to the discrepant results. Our patient series is significantly larger, with a longer median follow-up period. Many of the patients in our study were also treated with anti-idiotype vaccine strategies,33 an immune-modulating therapy that may alter the expression patterns of chemokine receptors such as CCR1.
Finally, the demonstration of CCR1 protein expression in a variety of human lymphoma types has important clinical implications. With the development of small molecule antagonists of CCR1,34 the expression of CCR1 by lymphomas and acute myeloid leukemia becomes an attractive potential target for drug therapy.
We used CCR1 immunohistochemical analysis to define CCR1 protein expression in normal and neoplastic human hematolymphoid tissues. These results provide further support for the importance of chemokine signaling pathways in promoting and maintaining a neoplastic phenotype. Furthermore, signaling through the CCR1/CCL3 pathway may lead to a more aggressive phenotype in DLBCL and contribute to poorer patient outcome.
Acknowledgments
Supported by grant CA34233 from the National Institutes of Health, Bethesda, MD.
References
- 1.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 2.Rollins BJ. Inflammatory chemokines in cancer growth and progression. Eur J Cancer. 2006;42:760–767. doi: 10.1016/j.ejca.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Laurence AD. Location, movement and survival: the role of chemokines in haematopoiesis and malignancy. Br J Haematol. 2006;132:255–267. doi: 10.1111/j.1365-2141.2005.05841.x. [DOI] [PubMed] [Google Scholar]
- 4.Maurer M, von Stebut E. Macrophage inflammatory protein-1. Int J Biochem Cell Biol. 2004;36:1882–1886. doi: 10.1016/j.biocel.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Su SB, Mukaida N, Wang J, et al. Preparation of specific polyclonal antibodies to a C-C chemokine receptor, CCR1, and determination of CCR1 expression on various types of leukocytes. J Leukoc Biol. 1996;60:658–666. doi: 10.1002/jlb.60.5.658. [DOI] [PubMed] [Google Scholar]
- 6.Kim IS, Jang SW, Sung HJ, et al. Differential CCR1-mediated chemotaxis signaling induced by human CC chemokine HCC-4/CCL16 in HOS cells. FEBS Lett. 2005;579:6044–6048. doi: 10.1016/j.febslet.2005.09.064. [DOI] [PubMed] [Google Scholar]
- 7.Lossos IS, Czerwinski DK, Alizadeh AA, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 8.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 9.Shaffer AL, Yu X, He Y, et al. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 10.Uneda S, Hata H, Matsuno F, et al. Macrophage inflammatory protein-1 alpha is produced by human multiple myeloma (MM) cells and its expression correlates with bone lesions in patients with MM. Br J Haematol. 2003;120:53–55. doi: 10.1046/j.1365-2141.2003.04040.x. [DOI] [PubMed] [Google Scholar]
- 11.Mahadevan D, Spier C, Della Croce K, et al. Transcript profiling in peripheral T-cell lymphoma, not otherwise specified, and diffuse large B-cell lymphoma identifies distinct tumor profile signatures. Mol Cancer Ther. 2005;4:1867–1879. doi: 10.1158/1535-7163.MCT-05-0146. [DOI] [PubMed] [Google Scholar]
- 12.Trentin L, Cabrelle A, Facco M, et al. Homeostatic chemokines drive migration of malignant B cells in patients with non-Hodgkin lymphomas. Blood. 2004;104:502–508. doi: 10.1182/blood-2003-09-3103. [DOI] [PubMed] [Google Scholar]
- 13.Deutsch A, Aigelsreiter A, Steinbauer E, et al. Distinct signatures of B-cell homeostatic and activation-dependent chemokine receptors in the development and progression of extragastric MALT lymphomas. J Pathol. 2008;215:431–444. doi: 10.1002/path.2372. [DOI] [PubMed] [Google Scholar]
- 14.Capriotti E, Vonderheid EC, Thoburn CJ, et al. Chemokine receptor expression by leukemic T cells of cutaneous T-cell lymphoma: clinical and histopathological correlations. J Invest Dermatol. 2007;127:2882–2892. doi: 10.1038/sj.jid.5700916. [DOI] [PubMed] [Google Scholar]
- 15.Byers RJ, Sakhinia E, Joseph P, et al. Clinical quantitation of immune signature in follicular lymphoma by RT-PCR-based gene expression profiling. Blood. 2008;111:4764–4770. doi: 10.1182/blood-2007-10-115915. [DOI] [PubMed] [Google Scholar]
- 16.Vande Broek I, Leleu X, Schots R, et al. Clinical significance of chemokine receptor (CCR1, CCR2 and CXCR4) expression in human myeloma cells: the association with disease activity and survival. Haematologica. 2006;91:200–206. [PubMed] [Google Scholar]
- 17.Natkunam Y, Warnke RA, Montgomery K, et al. Analysis of MUM1/IRF4 protein expression using tissue microarrays and immunohistochemistry. Mod Pathol. 2001;14:686–694. doi: 10.1038/modpathol.3880373. [DOI] [PubMed] [Google Scholar]
- 18.Jaffe E, Harris NL, Stein H, et al. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. World Health Organization Classification of Tumours. Lyon, France: IARC Press; 2001. [Google Scholar]
- 19.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 21.de Wynter EA, Heyworth CM, Mukaida N, et al. CCR1 chemokine receptor expression isolates erythroid from granulocyte-macrophage progenitors. J Leukoc Biol. 2001;70:455–460. [PubMed] [Google Scholar]
- 22.Moller C, Stromberg T, Juremalm M, et al. Expression and function of chemokine receptors in human multiple myeloma. Leukemia. 2003;17:203–210. doi: 10.1038/sj.leu.2402717. [DOI] [PubMed] [Google Scholar]
- 23.Oba Y, Lee JW, Ehrlich LA, et al. MIP-1alpha utilizes both CCR1 and CCR5 to induce osteoclast formation and increase adhesion of myeloma cells to marrow stromal cells. Exp Hematol. 2005;33:272–278. doi: 10.1016/j.exphem.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 25.Natkunam Y, Zhao S, Mason DY, et al. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood. 2007;109:1636–1642. doi: 10.1182/blood-2006-08-039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Natkunam Y, Lossos IS, Taidi B, et al. Expression of the human germinal center–associated lymphoma (HGAL) protein, a new marker of germinal center B-cell derivation. Blood. 2005;105:3979–3986. doi: 10.1182/blood-2004-08-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 28.Solal-Celigny P, Roy P, Colombat P, et al. Follicular Lymphoma International Prognostic Index. Blood. 2004;104:1258–1265. doi: 10.1182/blood-2003-12-4434. [DOI] [PubMed] [Google Scholar]
- 29.Morente M, Piris MA, Orradre JL, et al. Human tonsil intraepithelial B cells: a marginal zone–related subpopulation. J Clin Pathol. 1992;45:668–672. doi: 10.1136/jcp.45.8.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao JL, Wynn TA, Chang Y, et al. Impaired host defense, hematopoiesis, granulomatous inflammation and type 1–type 2 cytokine balance in mice lacking CC chemokine receptor. J Exp Med. 1997;1185:1959–1968. doi: 10.1084/jem.185.11.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natkunam Y, Hsi ED, Aoun P, et al. Expression of the human germinal center–associated lymphoma (HGAL) protein identifies a subset of classic Hodgkin lymphoma of germinal center derivation and improved survival. Blood. 2007;109:298–305. doi: 10.1182/blood-2006-04-014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen SJ, Crown SE, Handel TM. Chemokine: receptor structure, interactions, and antagonism. Annu Rev Immunol. 2007;25:787–820. doi: 10.1146/annurev.immunol.24.021605.090529. [DOI] [PubMed] [Google Scholar]
- 33.Weng WK, Czerwinski D, Timmerman J, et al. Clinical outcome of lymphoma patients after idiotype vaccination is correlated with humoral immune response and immunoglobulin G Fc receptor genotype. J Clin Oncol. 2004;22:4717–4724. doi: 10.1200/JCO.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Cheng JF, Jack R. CCR1 antagonists. Mol Divers. 2008;12:17–23. doi: 10.1007/s11030-008-9076-x. [DOI] [PubMed] [Google Scholar]