Abstract
Two alleles in cholesteryl ester transfer protein (CETP) gene polymorphisms have been disputably linked to enhanced cognition and decreased risk of Alzheimer’s disease (AD): the V and A alleles of I405V and C-629A. This study investigates whether these polymorphisms affect brain structure in 188 elderly controls and 318 AD or mild cognitive impairment (MCI) subjects from the Alzheimer’s Disease Neuroimaging Initiative cohort. Nominally signficant associations were dependent on APOE ε4 carrier status. In APOE ε4 carriers, the V and A alleles, both of which decrease CETP and increase HDL, associated with greater baseline cortical thickness and less 12-month atrophy in the medial temporal lobe. Conversely, in APOE ε4 non-carriers, the I allele, which increases CETP and decreases HDL, associated with greater baseline thickness, less atrophy and lower risk of dementia. These results suggest CETP may contribute to the genetic variability of brain structure and dementia susceptibility in an APOE-dependent manner.
Keywords: Imaging genetics, Quantitative neuroimaging, CETP, Alzheimer’s disease, Dementia, APOE
Introduction
The role of genetics in brain development and disease susceptibility has recently gained insight from the field of neuroimaging genetics, in which phenotypes are defined by quantitative measures of brain structure or function rather than clinical characteristics such as disease, symptoms or behavior. Association analyses between genetic data and information obtained from structural and functional neuroimaging provide a more direct assessment of genetic influence at the level of neuronal circuitry than other traditional analyses, which only indirectly assess the impact of genes on complex behaviors or cognitive states (Mattay et al. 2008). Thus, the use of imaging measures may provide a closer description of underlying pathology (Saykin et al. 2010) and therefore render association analyses less susceptible to non-genetic variability. Accordingly, neuroimaging genetics may have the potential to examine the biologic impact of genes with greater precision and accuracy than traditional genetic association analyses by examining atrophy as a possible link between genes and behavioral deficits (Stein et al. 2010).
In particular, structural neuroimaging has considerable potential to address whether specific genes may affect brain structure during development or pathological processes. One such gene of interest is the cholesteryl ester transfer protein (CETP) gene, which is located on chromosome 16q21 and contains 14 exons (Arias-Vásquez et al. 2007). CETP is involved in the transfer of insoluble cholesteryl esters between lipoproteins as part of the reverse cholesterol transport system and is known to be synthesized and secreted in the brain (Albers et al. 1992). It has been speculated that CETP may play a role in cholesterol transport within the brain (Albers et al. 1992), perhaps by promoting neuronal uptake of high density lipoprotein (HDL) particles via interactions with receptors for apolipoprotein E (APOE), which is the most common genetic risk factor of Alzheimer’s disease (AD) (Rodríguez et al. 2006). Several studies have shown that cholesterol metabolism is indeed altered in AD (Kivipelto et al. 2001; Pregelj 2008), providing a biologically plausible role for CETP in AD susceptibility (Barzilai et al. 2003, 2006; Sanders et al. 2010).
CETP has been previously implicated in metabolic syndrome, a conglomeration of metabolic risk factors including dyslipidemia, hypertension and altered glucose metabolism, which increases in prevalence with age. Variations in the CETP gene lead to altered CETP serum concentration and activity and subsequent changes in HDL levels and lipoprotein particle sizes (Thompson et al. 2003). Specifically, CETP deficiency is characterized by decreased CETP mass and activity and increased HDL and lipoprotein size (Qureischie et al. 2008). Two functional CETP polymorphisms associated with CETP deficiency have been controversially implicated as protecting against cognitive decline and neurodegenerative disease susceptibility: the A allele of promoter polymorphism C-629A (rs1800775) and the V allele of the amino acid exchange polymorphism I405V in exon 14 (rs5882). Three studies have reported that I405V VV genotype was associated with increased cognitive function and decreased risk of dementia (Barzilai et al. 2003, 2006; Sanders et al. 2010), although these effects have not been replicated in other studies (Arias-Vásquez et al. 2007; Johnson et al. 2007; Qureischie et al. 2009; Rodríguez et al. 2006). Likewise, one study reported a decreased risk of AD associated with C-629A AA genotype (Rodríguez et al. 2006), but this has been contradicted by others (Qureischie et al. 2008, 2009).
Inconsistent findings involving these two polymorphisms may be due to a variety of factors, including discrepancies in the age and ethnic composition of study samples, or the possibility that these polymorphisms are merely surrogates for another polymorphism that acts as the true causal variant for modulating disease susceptibility. Further, it is possible that the associations found were due to chance alone. However, the inability to replicate findings may also be due to imprecision of the phenotype when defined by neuropsychological measures or clinical disease status. To address this concern, we examined the effect of the CETP C-629A and I405V polymorphisms on structural brain measures in 188 healthy elderly controls and 318 subjects with AD or mild cognitive impairment (MCI) from the Alzheimer’s Disease Neuroimaging Initiative (ADNI). To reduce the number of statistical comparisons, the analysis was restricted to the hippocampus, parahippocampal gyrus and entorhinal cortex, three brain regions in which structural change is considered to be a sensitive indicator of memory impairment and early pathological processes occurring in AD (Braak and Braak 1991; Dickerson et al. 2009; Jack et al. 1997; McEvoy et al. 2009).
Methods
Alzheimer’s Disease Neuroimaging Initiative
Raw data used in this paper were obtained from the ADNI public database (http://www.loni.ucla.edu/ADNI/). ADNI is a multi-site, five-year observational study of elderly individuals started in 2004 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies and nonprofit organizations. Elderly controls, subjects with MCI and AD subjects underwent longitudinal MRI scans and neuropsychological assessment at specified intervals for 2 to 3 years.
The Principal Investigator of this initiative is Michael W. Weiner, M.D. at the VA Medical Center and University of California, San Francisco. ADNI is the result of the efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. For more information, see www.adni-info.org.
Standard protocol approvals, enrollment and patient consents
ADNI was conducted according to Good Clinical Practice guidelines, the Declaration of Helsinki, US 21CFR Part 50—Protection of Human Subjects, and Part 56—Institutional Review Boards, and pursuant to state and federal HIPAA regulations. Written informed consent for the study was obtained from all subjects and/or authorized representatives and study partners (http://www.ADNI-info.org).
Subjects
ADNI eligibility criteria are described at http://adni-info.org. In general, all enrolled subjects were between 55 and 90 (inclusive) years of age, had a study partner available to provide an independent evaluation of functioning, and spoke either English or Spanish. A total of 188 healthy controls (HC), 327 subjects with MCI, and 149 AD subjects were included in this study based on their availability of genetic data and baseline MRI data that passed local quality control. One-hundred and nine ADNI subjects were excluded on account of missing data or failure of imaging data to pass local quality control.
To increase the power of analysis, subjects with MCI and AD were treated as a single diagnostic group after selecting only the 169 MCI subjects who were classified as most at risk for imminent conversion to AD based on their AD-like pattern of regional atrophy, as described in McEvoy et al. (2009). Briefly, stepwise linear discriminant analysis was used to identify eight cortical and subcortical regions to best aid discrimination of HC and AD subjects. A classifier trained on data from HC and AD subjects was applied to data from all 327 MCI subjects to determine whether they had patterns of regional atrophy characteristic of mild AD. The discrimination analysis correctly classified 92% of HC subjects and 86% of AD subjects, which is comparable to the clinical diagnostic accuracy of AD (Gearing et al. 1995; Mok et al. 2004; Schneider et al. 2007). The discriminant model was predictive of the rate of clinical decline and progression of MCI patients to an AD diagnosis. Further, the rates of cognitive decline in these prodromal AD patients were in line with those of AD patients in ADNI (McEvoy et al. 2009).
Demographics of the 188 HC, and 318 MCI and AD subjects are described in Table 1. Of the 506 total subjects, 420 had 12-month follow-up imaging data that passed local quality control and were thus included in the longitudinal analysis.
Table 1.
Subject characteristics
| Healthy controls (n = 188) |
MCI & AD subjects (n = 318) |
|
|---|---|---|
| Age (years) | 76.12±5.13 | 74.91±7.52 |
| Percent female subjects | 46% | 46% |
| Education (years) | 16.10±2.81* | 15.34±3.01* |
| Percent carriers of APOE ε4 | 28%* | 64%* |
| MMSE | 29.13±.97* | 25.16±2.49* |
| ADAS-cog | 6.15±2.85* | 15.38±5.86* |
Data are given as mean±SD unless otherwise specified as a percent.
The MCI and AD subjects had a significantly lower level of education (p = .005), higher frequency of the APOE ε4 allele (p < .001) and significantly more impaired MMSE (p < .001) and ADAS-cog (p < .001) scores at baseline than the healthy controls
Genotyping
DNA was extracted from blood and genotyped with the Illumina Human610-Quad BeadChip. Since the Illumina chip does not include the APOE alleles, the Alzheimer’s Disease Neuroimaging Initiative (ADNI) did a separate DNA extraction from blood samples, and APOE genotyping was done via PCR amplification and HhaI restriction enzyme digestion (Potkin et al. 2009). Smartpca was used to identify ADNI subjects who were outliers relative to the genetic cluster of ADNI subjects who self-reported their race as exclusively “White,” and one subject who reported “More than one race” (Joyner et al. 2009). Fifty-three genetic outliers were identified. These and 30 subjects with missing genetic data were excluded from the study and, therefore, are not included in the total subjects enumerated above.
MR acquisition and analysis
Two three-dimensional, T1-weighted volumes per subject per visit were downloaded from the public ADNI database (http://www.loni.ucla.edu/ADNI/). All image processing and analyses occurred at the Multimodal Imaging Laboratory at the University of California, San Diego. Images were corrected for gradient nonlinearities (Jovicich et al. 2006) and B1 field inhomogeneity (Sled et al. 1998). The two baseline images were rigid-body aligned, averaged to improve signal-to-noise ratio and resampled to isotropic 1-mm voxels. An automated segmentation procedure based on FeeSurfer software (Fischl et al. 2002) and customized Matlab code was used to obtain volumetric segmentation. Cortical surface reconstruction yielded a measure of thickness at each vertex, and the surface was parceled into distinct regions of interest (ROIs) (Dale and Sereno 1993; Dale et al. 1999; Fischl et al. 1999, 2004).
The methods to obtain 12-month change are described in detail in Holland et al. (2009). Briefly, follow-up images were fully affine-registered to the baseline images and corrected for intensity nonuniformity. Nonlinear registration was then used to align voxel centers in the baseline with the appropriate location in the follow-up scans, and a volume-change field was calculated. This field was averaged over each ROI to compute the percentage change from baseline. This method has been validated using models where amount of change was known and where noise was added to approximate that seen in human brain imaging of demented patients. In these studies, technical measurement error for the computed change in volume was within 0.2% of the structure volume. That is, for a 6,000 mm3 hippocampal volume undergoing a change of 1%, the technical measurement error in estimating the 60 mm3 volumetric change would be ±12 mm3 in an individual subject if voxels were each 1 mm isotropic (Murphy et al. 2010).
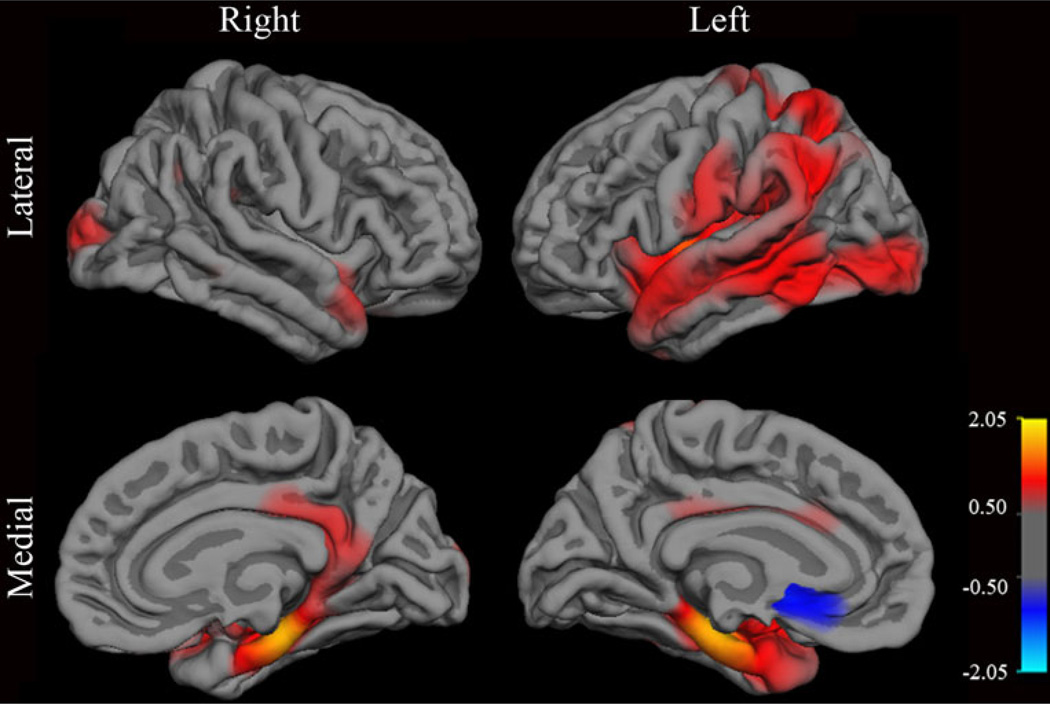
The methods to visualize CETP by APOE interactions at each vertex on the cortical surface are described in detail in Joyner et al. (2009). Briefly, the cortical surface was reconstructed to measure thickness at each vertex. Continuous maps of cortical surface area were smoothed with a Gaussian kernel and mapped onto a standardized spherical atlas space. Association of cortical thickness and CETP by APOE interactions was assessed at each surface vertex after controlling for the effects of age, sex and diagnosis, and the strength of association was mapped as negative log p-values.
Statistical analysis
All statistical analysis was performed using PASW Statistics 18. Genotype distributions and allele frequencies were compared using χ2-statistics. Genotype was modeled as minor allele SNP-dosage (homozygote of minor allele = 2, heterozygote = 1, homozygote of major allele = 0) and separated into two regressor variables to account for sex-specific effects. Thus, genotype was modeled as dosage of the A allele at C-629A and the V allele at I405V. Sex was additionally included as a covariate in regression analyses to ensure that males and females homozygous for major alleles would not be equivalent in the overall model.
To examine the relationship between CETP genotype and diagnosis, binary logistic regression analysis was used with age, sex and APOE ε4 carrier status as covariates. Independent-sample t-tests were used to compare the effects of specific genotypes on diagnosis. To study the association between genotype and brain structure, standardized morphometric measures were entered as dependent variables into linear regression models. Morphometric measures were standardized by converting raw values to standardized residuals in a linear regression model built upon data from only the HC subjects. Cortical thickness measures were standardized with respect to age and sex. Subcortical volumes were standardized with respect to age, sex and intracranial volume. Likewise, longitudinal morphometric measures were entered as dependent variables into linear regression models to determine whether genotype influenced atrophy. These analyses were also performed with stratification for APOE ε4 carrier status to determine whether the effects were dependent on genotype at this locus. Given the imaging and genetic data currently available, we conducted a restricted exploratory analysis without correction for multiple comparisons. The two-sided significance level was set at α = .05 for all regression models, each of which was repeated for two CETP polymorphisms and nine subject groups to demonstrate the effect of these polymorphisms on every possible combination of subject diagnosis and APOE ε4 carrier status.
To determine whether the aging process had different effects on brain structure and atrophy depending upon genotype at the two CETP polymorphisms, a univariate General Linear Model factor by covariate analysis of variance was used. Genotype was a grouping factor and genotype*age, age, sex, diagnosis and APOE ε4 carrier status were covariates. In this analysis, cortical thickness measures were standardized with respect to sex, and subcortical volumes with respect to sex and intracranial volume, but no measures were adjusted for age. Likewise, to determine whether APOE ε4 carrier status affected the influence of CETP genotype on brain structure, genotype*APOE ε4 carrier status was entered along with the main effects in the following interaction model: y = b0 + b1*CETP + b2*APOE + b3*CETP*APOE + b4*age + b5*sex + b6*diagnosis.
Results
Influence of CETP polymorphisms on risk of MCI and AD
Genotype distributions and allele frequencies of CETP I405V and C-629A for the 188 HCs and 318 subjects with AD or MCI are presented in Table 2. Both polymorphisms were in Hardy-Weinberg equilibrium in the study sample (I405V: χ2 = .043, p = .979; C-629A: χ2 = 2.289, p = .318).
Table 2.
Genotype distribution and allele frequencies of CETP polymorphisms
| Diagnosis (n) | Genotypes | Allele frequencies | χ2-test | |||||
|---|---|---|---|---|---|---|---|---|
| C-629A | CC(%) | AC(%) | AA(%) | C | A | χ2 | df | p |
| HC (188) | 50 (26.7) | 97 (51.9) | 40 (21.4) | 52.7 | 47.3 | 2.289 | 2 | .318* |
| MCI, AD (318) | 76 (23.9) | 172 (54.1) | 70 (22.0) | 50.9 | 49.1 | |||
| I405V | II(%) | IV(%) | VV(%) | I | V | χ2 | df | p |
| HC (188) | 99 (52.7) | 73 (38.8) | 16 (8.5) | 72.1 | 27.9 | 0.043 | 2 | .979* |
| MCI, AD (318) | 139 (43.7) | 147 (46.2) | 32 (10.1) | 66.8 | 33.2 | |||
Distribution of the CETP C-629A and I405V polymorphisms in healthy controls and subjects with MCI or AD.
Both polymorphisms were in Hardy-Weinberg equilibrium in the study population
Logistic regression analysis revealed no association of either CETP polymorphism with the risk of MCI and AD. Likewise, age and sex were not significantly associated with the risk of MCI and AD although APOE ε4 carrier status was significantly associated with the risk of MCI and AD (χ2 = 61.60, df = 1, p = 4.21E-15).
When the study sample was stratified by gender and APOE ε4 carrier status, I405V genotype was associated with the risk of MCI and AD in both male (n = 135, χ2 = 9.89, df = 2, p = .007) and female (n = 115, χ2 = 9.89, df = 2, p = .019) non-carriers. Males with genotype IV were more likely to have MCI or AD than males with genotype II (t(122) = −3.151, p = .002), and likewise, females with genotype IV showed a trend of being more likely to have MCI or AD than females with genotype II (t(96) = −1.973, p = .051). In APOE ε4 carriers, I405V genotype showed no influence on diagnosis. Thus, the I405V polymorphism appeared to influence the risk of MCI and AD only in the absence of the APOE ε4 allele. In contrast, C-629A genotype was not significantly associated with the risk of MCI and AD in either carriers or non-carriers of APOE ε4.
Influence of CETP polymorphisms on brain structure
In the female population, C-629A and I405V genotype were associated with morphometric measures in two of the three brain structures tested, dependent upon diagnosis and APOE ε4 carrier status (Table 3). In particular, the I allele of the I405V polymorphism was associated with greater entorhinal (β = .222; SE = .232; p = .007) and parahippocampal thickness (β = .197; SE = .120; p = .016) in APOE ε4 non-carriers, and this effect appeared primarily driven by healthy individuals (β = .233; SE = .191; p = .032 for entorhinal; β = .256; SE = .139; p = .017 for parahippocampal), as it was not seen in the MCI and AD group. In contrast, the protective allele relationship was reversed in APOE ε4 carriers, where the V allele was associated with greater parahippocampal thickness (β = .194; SE = .130; p = .018), and this effect appeared primarily driven by individuals with MCI or AD (β = .230; SE = .139; p = .011). For the C-629A polymorphism, the A allele was associated with greater entorhinal (β = .547; SE = .376; p = .016) and parahippocampal thickness (β = .577; SE = .212; p = .020) in healthy controls. No significant associations were detected with hippocampal volume in females.
Table 3.
Regression coefficients and significance levels (p-values) for associations between CETP polymorphisms and brain structure in a priori selected ROIs in females
| SNP | Diagnosis | APOE ε4 carrier status |
N | Entorhinal cortex thickness β (p-value) |
Parahippocampal gyrus thickness β (p-value) |
|---|---|---|---|---|---|
| C-629A | All | All | 506 | 0.047 (0.507) | 0.003 (0.966) |
| HC | All | 188 | 0.267 (0.023*) (A) | 0.250 (0.031*) (A) | |
| MCI, AD | All | 318 | −0.017 (0.850) | −0.118 (0.178) | |
| All | Non-carriers | 250 | 0.005 (0.956) | 0.040 (0.684) | |
| HC | Non-carriers | 135 | 0.098 (0.479) | 0.164 (0.224) | |
| MCI, AD | Non-carriers | 115 | −0.115 (0.439) | −0.128 (0.387) | |
| All | Carriers | 256 | 0.052 (0.606) | −0.096 (0.321) | |
| HC | Carriers | 53 | 0.547 (0.016*) (A) | 0.577 (0.020*) (A) | |
| MCI, AD | Carriers | 203 | 0.042 (0.703) | −0.135 (0.212) | |
| I405V | All | All | 506 | 0.088 (0.134) | 0.203 (0.686) |
| HC | All | 188 | 0.151 (0.115) | 0.242 (0.011*) (I) | |
| MCI, AD | All | 318 | 0.033 (0.658) | −0.125 (0.090) | |
| All | Non-carriers | 250 | 0.222 (0.007*) (I) | .0197 (0.016*) (I) | |
| HC | Non-carriers | 135 | 0.233 (0.032*) (I) | 0.256 (0.017*) (I) | |
| MCI, AD | Non-carriers | 115 | 0.100 (0.442) | 0.023 (0.861) | |
| All | Carriers | 256 | −0.076 (0.370) | −0.194 (0.018*) (V) | |
| HC | Carriers | 53 | −0.066 (0.741) | 0.236 (0.282) | |
| MCI, AD | Carriers | 203 | −0.061 (0.506) | −0.230 (0.011*) (V) |
Regression coefficients and significance levels were obtained from linear regression models in which CETP genotype was the independent variable and morphometric measures were the dependent variables. The valence assigned to β coefficients corresponds to the directionality of association. For CETP C-629A, positive values mean that the A allele is associated with greater volumes, and negative values mean that the C allele is associated with greater volumes. For CETP I405V, positive values mean that the I allele is associated with greater volumes, and negative values mean that the V allele is associated with greater volumes. CETP polymorphisms are associated with two morphometric measures in females, dependent upon diagnosis and APOE ε4 carrier status. No significant associations were detected with hippocampal volume in females. The only significant associations detected in males were between C-629A and hippocampal volume in MCI and AD carriers of APOE ε4 (β = .230; p = .017, protective allele = A); I405V and entorhinal cortex thickness in non-carriers of APOE ε4 (β = .165; p = .048, protective allele = I); and I405V and hippocampal volume in non-carriers of APOE ε4 (β = .176; p = .036, protective allele = I).
Statistically significant p-values (p < .05) are represented in bold, and the allele corresponding to greater thickness (i.e. the “protective” allele) is denoted in parentheses
Fewer significant associations among these polymorphisms and brain structures were detected in males, but those that were found matched well with the findings in females. Namely, the I allele of the I405V polymorphism was associated with greater entorhinal cortex thickness (β = .165; SE = .249; p = .048) and hippocampal volume (β = .176; SE = .215; p = .036) in APOE ε4 non-carriers; the A allele of the C-629A polymorphism was associated with greater hippocampal volume in APOE ε4 carriers with MCI or AD (β = .230; SE = .127; p = .017).
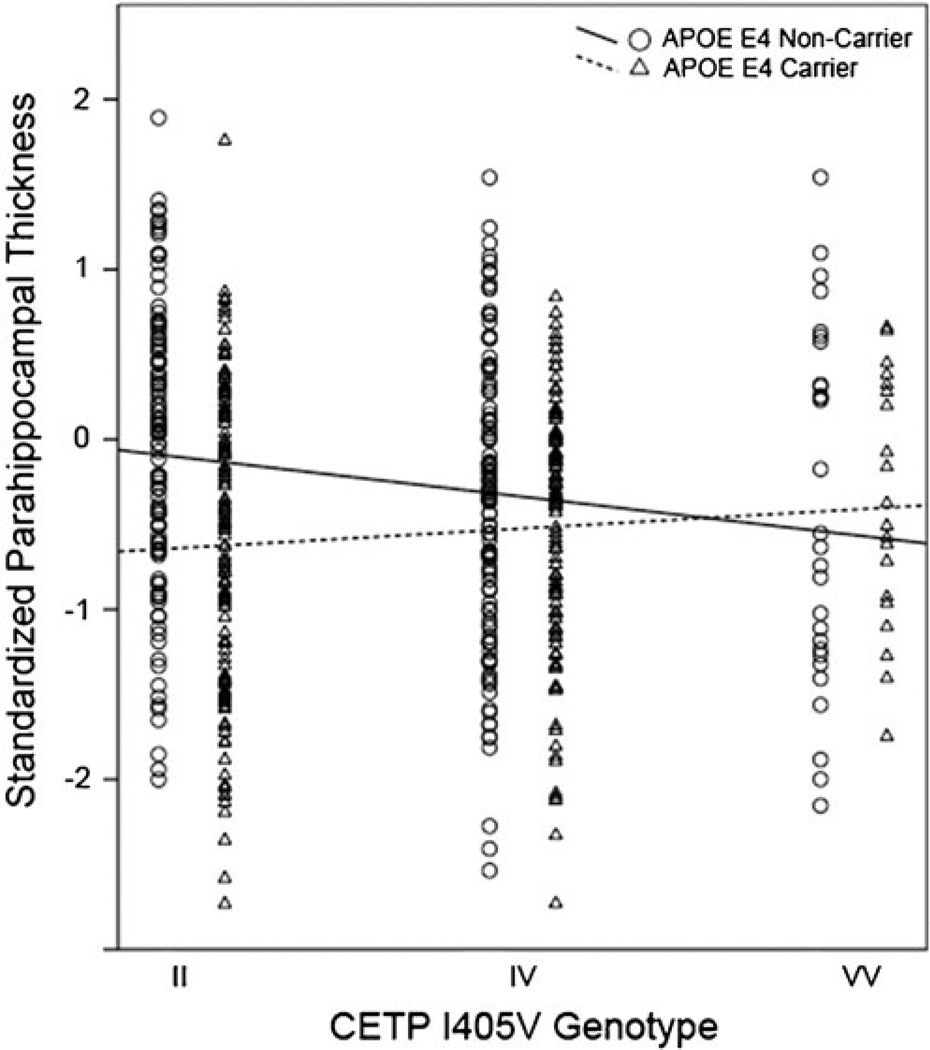
An interaction between CETP I405V and APOE ε4 carrier status was found with parahippocampal gyrus thickness after controlling for age, sex and diagnosis (F = 2.459; p = .007). Namely, the I allele was found to be associated with thicker parahippocampal cortex in APOE ε4 non-carriers but with thinner parahippocampal cortex in APOE ε4 carriers (Figs. 1 and 2). Likewise, an interaction between CETP C-629A and APOE ε4 carrier status was found with parahippocampal gyrus thickness after controlling for age, sex and diagnosis (F = 1.983; p = .031). The A allele was found to be associated with thicker parahippocampal cortex in APOE ε4 carriers but with thinner parahippocampal cortex in APOE ε4 non-carriers.
Fig. 1.
Association of baseline cortical thickness at each vertex with the interaction of CETP SNP I405V by APOE ε4 carrier status in all subjects after controlling for the effects of age, gender and diagnosis. The map shows the distribution of nominal −log p-values across the reconstructed cortical surface. The sign represents the direction of the effect. Red values correspond to the I allele associating with greater cortical thickness in APOE ε4 non-carriers and the V allele associating with greater cortical thickness in APOE ε4 carriers. Blue values represent the reverse relationship. The most significant effects of this genotype interaction are seen in the parahippocampal gyrus
Fig. 2.
Standardized values for parahippocampal gyrus thickness plotted against CETP I405V genotype for APOE ε4 non-carriers (circles; solid line) and carriers (triangles; dashed line). In APOE ε4 non-carriers, the II genotype seems to be protective with respect to parahippocampal thickness. In contrast, in APOE ε4 carriers, VV seems protective. Regression lines computed using the least squares method to best fit the data points. For APOE ε4 non-carriers, R2 = .029. For APOE ε4 carriers, R2 = .007
Influence of CETP polymorphisms on brain atrophy
Not only did genotype at these two CETP polymorphisms affect baseline brain structure, but also it influenced 12-month brain atrophy in a consistent manner (Table 4). In females, the A allele of the C-629A polymorphism was protective against thinning of the entorhinal cortex (β = .188; SE = .142; p = .011). This protective effect remained significant when considering only those with a diagnosis of MCI or AD (β = .233; SE = .164; p = .013), as well as only those with a diagnosis of MCI or AD who were APOE ε4 carriers (β = .240; SE = .189; p = .043). In males, the I allele of the I405V genotype was protective against volume reduction in the hippocampus in APOE ε4 non-carriers (β = .227; SE = .235; p = .011), an effect still seen when the sample was limited to non-carriers with MCI or AD (β = .324; SE = .443; p = .031).
Table 4.
Proposed model of CETP effects on MCI/AD risk, baseline volumes and one-year atrophy in medial temporal lobe structures
| Protective for baseline volumes | Protective for one-year atrophy | Protective for risk of MCI/AD | |
|---|---|---|---|
| APOE ε4 carriers | V allele (low CETP) | – | – |
| HC | A allele (low CETP) | – | – |
| MCI or AD | V allele (low CETP) | – | |
| A allele (low CETP) | A allele (low CETP) | ||
| APOE ε4 non-carriers | I allele (high CETP) | I allele (high CETP) | I allele (high CETP) |
| HC | I allele (high CETP) | – | – |
| MCI or AD | – | I allele (high CETP) | – |
Alleles conferring low CETP and high HDL (V at I405V and A at C-629A) appear to be protective in APOE ε4 carriers, whereas alleles conferring high CETP and low HDL (I at I405V and C at C-629A) appear protective in non-carriers
Effect of age on the interaction between CETP polymorphism and brain structure
In the female population, CETP I405V genotype by age interactions were found with parahippocampal gyrus thickness after controlling for diagnosis and APOE ε4 carrier status (Table 5). Females with the I405V II genotype showed less decline in parahippocampal gyrus thickness with age than those with VV (F = 4.374; p = .013). No significant CETP C-629A genotype by age interactions were found in the three brain structures tested, nor were any significant effects found in males.
Table 5.
Effect of age on CETP I405V influence on parahippocampal gyrus thickness
| Variable | df | F | Sig. |
|---|---|---|---|
| Male I405V | 2 | 2.309 | .100 |
| Female I405V | 2 | 4.504 | .012* |
| Male I405V*Age | 2 | 2.343 | .097 |
| Female I405V*Age | 2 | 4.374 | .013* |
| Age | 1 | 15.208 | 1.10E-4* |
| Diagnosis | 1 | 125.971 | 3.45E-26* |
| Sex | 1 | .166 | .684 |
| APOE ε4 status | 1 | .251 | .617 |
Univariate general linear model factor by covariate analysis of variance was used for parahippocampal gyrus thickness, with I4045V genotype as a grouping factor and age, sex, diagnosis and APOE ε4 carrier status as covariates. There were no significant I405V genotype by age interactions with entorhinal cortex thickness or hippocampal volume. There were no significant C-629A genotype by age interactions in any of the three brain structures tested.
Statistically significant p-values (p < .05) are represented in bold
Discussion
The present findings suggest that genotype at CETP polymorphisms I405V and C-629A may contribute to the genetic variability of brain structure and neurodegenerative disease susceptibility. This study is the first to detect APOE-specific associations between the I405V and C-629A polymorphisms and brain structure, and to relate these findings to longitudinal change in brain structure with aging. In APOE ε4 non-carriers, the I allele of the I405V polymorphism, which leads to increased CETP and decreased HDL, was associated with larger medial temporal lobe structures at baseline, less 12-month atrophy in these structures, and lower risk of MCI and AD. This relationship was reversed in APOE ε4 carriers, in whom the V allele of I405V and the A allele of C-629A, both of which lower CETP and raise HDL, were found to be protective. Although the primary ROI analysis was restricted to three medial temporal structures selected a priori, a cortical surface analysis revealed the greatest effects of these polymorphisms were indeed found in the medial temporal lobe, and specifically in the parahippocampal gyrus.
The results suggest that these CETP polymorphisms may modify brain structure and neurodegenerative disease susceptibility in a manner dependent upon APOE. I405V genotype influenced the risk of MCI and AD in this study sample only in non-carriers of APOE ε4, and the protective allele relationships reversed in carriers and non-carriers of APOE ε4. Two prior studies found the effects of these polymorphisms on the risk of AD to be similarly dependent upon APOE. One study of 286 AD subjects found that risk of AD was lower in carriers of APOE ε4 with the C-629A AA genotype (Rodríguez et al. 2006), and another study of 544 AD subjects reported that risk of AD was higher in non-carriers of APOE ε4 with the I405V VV genotype (Arias-Vásquez et al. 2007), analogous to the present finding that the I allele was protective in non-carriers. The finding of alleles that lower CETP levels being protective in APOE ε4 carriers is further supported by a study of 107 AD subjects in which another CETP polymorphism, D442G, was associated with lower levels of CETP and decreased risk of AD in APOE ε4 carriers (Chen et al. 2008).
It has been demonstrated that protein-protein interactions occur between CETP and APOE due to their joint participation in cholesterol metabolism (Sorlí et al. 2006). CETP-immunoreactivity has been demonstrated in astrocytes from AD brain tissue, a cell known to produce APOE (Yamada et al. 1995), and a physiological cooperation between CETP and APOE has been established in cholesterol ester transport in plasma (Marcel et al. 1990). Thus, it is reasonable that the influence of CETP on brain cholesterol metabolism and structure might be modulated by APOE activity within the brain, and that a dysfunction in cholesterol metabolism involving both proteins may play a central role in the AD pathology (Yamada et al. 1995).
The APOE-dependent reversal of the protective allele relationship observed in this study might be explained by such interactions between CETP and APOE. For example, it has been proposed that CETP may modify the risk of AD by altering HDL concentration in the brain, whereas the amount of HDL generated varies with APOE isoform (Rodríguez et al. 2006). CETP is believed to promote neuronal uptake of HDL particles via interactions with APOE receptors (Rodríguez et al. 2006). Indeed, gene-gene interactions between APOE and CETP have been shown to influence HDL concentrations (Sorlí et al. 2006), and it is known that HDL particles interact with amyloid beta to inhibit its aggregation into fibrils (Olesen and Dago 2000).
The results further suggest that these two polymorphisms have a differential degree of influence on brain structure in males and females. Although both males and females were affected by these polymorphisms in a similar manner, the majority of associations between genotype and brain structure were found in females. This may be due to sex-related differences in cholesterol metabolism (De Gennes et al. 1983) or due to underlying structural differences in the male and female brains imposed by distinct hormonal or genetic expression environments during the time of development (Negri-Cesi et al. 2004).
In APOE ε4 non-carriers, risk of MCI and AD increased with carriage of Vat I405V. Since the Vallele corresponds to lower levels of CETP and subsequently higher levels of HDL (Arias-Vásquez et al. 2007), this suggests that low CETP and high HDL may actually increase the risk of AD in non-carriers. One mechanism for this increased risk proposed by Arias-Vásquez et al. (2007) is that low CETP levels lead to a reduction in neuronal repair. Other studies have found that variations in I405V had no effect on cognitive function in 525 subjects who participated in the Scottish Mental Survey of 1932 (Johnson et al. 2007) and did not influence the risk of vascular dementia in 163 subjects (Qureischie et al. 2009). Two additional studies reported no difference in distribution of allele and genotype frequencies for I405V between AD subjects and elderly controls (Arias-Vásquez et al. 2007; Rodríguez et al. 2006); however, after segregating by APOE ε4 status, VV homozygosity was more frequent in AD non-carriers than in elderly control non-carriers (Arias-Vásquez et al. 2007), similar to the present findings.
A recent prospective cohort study by Sanders et al. 2010, followed 523 mixed-ethnicity subjects aged 70 years or older over a time period during which 40 subjects developed dementia. This group reported that the I405V VV genotype was associated with slower age-associated memory decline and lower risk of incident dementia and AD. Further, the hazard ratio of dementia for IV heterozygotes was between that for II and VV homozygotes, suggesting a possible gene-dose relationship (Sanders et al. 2010). This finding supported two previous studies in the Ashkenazi Jewish population that found the VV genotype to be more prevalent in 158 subjects with exceptional longevity (aged≥95 years) and to be associated with better cognitive function in these subjects (Barzilai et al. 2003, 2006), although the specific ethnic composition of these studies limit their generalizability. Further, the fact that these studies adjusted for APOE ε4 status rather than stratifying by this allele makes their findings difficult to compare with those of the present study.
The effect of variations in C-629A on cognition and disease susceptibility is similarly unclear. One group found that AA homozygotes had a significantly decreased risk of AD associated with carriage of the APOE ε4 allele (Rodríguez et al. 2006), consistent with the present findings. Another group studying 388 AD subjects found no influence of C-629A on risk of AD in either carriers or non-carriers of APOE ε4 (Qureischie et al. 2008) but later reported increased risk of vascular dementia in AA homozygotes who were APOE ε4 non-carriers (Qureischie et al. 2009).
The discrepancies between prior research and the present study may be due to myriad factors, including variation in the age and ethnic compositions of study samples. One characteristic to note in this study sample is the high rate of APOE ε4 carriers among MCI and AD subjects as compared to rates reported among other study samples (Crean et al. 2011), which is a difference that may limit the generalizability of these findings. Additionally, the discrepancies between prior research and the present study may reflect the limitation of examining polymorphisms in the CETP gene whose functional consequences are uncertain (Qureischie et al. 2008). For example, it is possible that the I405V polymorphism is a surrogate for another polymorphism that acts as the true causal variant for modulating brain structure and disease susceptibility; I405V may be a better surrogate for this true variant in the Ashkenazi Jewish population, which would be expected to have wider linkage disequilibrium blocks than in the ADNI study sample. Variable findings in prior research may also reflect the difficulty in trying to evaluate the genetic contribution of a single polymorphism to a heterogenous disorder such as AD, which is likely impacted by the interaction of multiple genes, proteins and non-genetic factors. That the apparent heritability of late-onset AD is seemingly unexplained by the known contributions of APOE and other well-replicated genes (e.g., presenilin-1, presenilin-2, APP) (Seshadri et al. 2010) suggests the likelihood that many genes contribute to AD susceptibility to a lesser degree. Additionally, this study suggests the importance of accounting for interactions with APOE, as failure to do so may produce inconsistent findings.
A limitation of this study is its inability to elucidate whether genetic variation in CETP modulates brain structure at the time of development, during aging, or solely in the context of disease. However, the regional specificity of findings suggests that CETP may influence susceptibility to AD given that these polymorphisms are associated with cortical thickness in the parahippocampal gyrus and entorhinal cortex, regions particularly damaged in AD (Braak and Braak 1991). Further, the significant I405V genotype by age interaction in females, independent of disease status, was also seen in the parahippocampal gyrus. This could suggest that these age-related effects might still be related to AD pathology, including pathology present in healthy controls.
Although a sample size of 188 elderly controls and 318 AD or MCI subjects may seem inadequate to identify a genetic risk factor for atrophy and disease susceptibility, the ADNI dataset is currently the largest dataset with complete genetic and imaging information available for such an analysis. Prior studies found similar effects using comparably sized, and even smaller (Chen et al. 2008), samples. Given the data currently available, this study was limited to a restricted exploratory analysis for subtle effects using relaxed correction for multiple comparisons. To reduce the need to correct for multiple comparisons across all brain regions and genetic variants, the study examined an a priori selection of targeted brain regions involved in Alzheimer’s disease (Braak and Braak 1991; Dickerson et al. 2009; Jack et al. 1997; McEvoy et al. 2009) and targeted genetic variants based on extant evidence of their involvement in aging and neurodegeneration (Barzilai et al. 2003, 2006; Sanders et al. 2010; Rodríguez et al. 2006). No doubt, the future will bring wider access to even larger datasets with complete genetic and imaging information in order to independently replicate our results. These results build on evidence of an interaction between APOE, cholesterol metabolism, and neurodegeneration with aging and AD and indicate the need for future studies examining this interplay.
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., and Wyeth, as well as non-profit partners the Alzheimer’s Association and Alzheimer’s Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation.
J.B.B. is supported by NINDS K02 NS067427, NIA U01 AG10483, NIA P50 AG005131, NIA RC2AG036535 and General Electric Medical Foundation and is an investigator for, and receives research funds from Janssen Alzheimer Immunotherapy. He has served on advisory boards for Elan and Avanir Pharmaceuticals, holds stock options in CorTechs Labs, Inc., and serves as an editor for the International Journal of Alzheimer’s Disease; E.A.M. is supported in part by NIGMS Training Grant GM007198.
A.M.D. receives funding to his laboratory from General Electric Medical Systems as part of a Master Research Agreement with UCSD; and is a founder of, holds equity in, and serves on the scientific advisory board for CorTechs Labs, Inc.
Footnotes
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu/ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators is available at http://www.loni.ucla.edu/ADNI/Collaboration/ADNI_Manuscript_Citations.pdf.
The terms of this arrangement have been reviewed and approved by UCSD in accordance with its conflict of interest policy.
Contributor Information
Elizabeth A. Murphy, Department of Neurosciences, University of California, San Diego, CA, USA
John Cooper Roddey, Multimodal Imaging Laboratory, University of California, San Diego, CA, USA.
Linda K. McEvoy, Department of Radiology, University of California, San Diego, CA, USA Multimodal Imaging Laboratory, University of California, San Diego, CA, USA.
Dominic Holland, Department of Neurosciences, University of California, San Diego, CA, USA; Multimodal Imaging Laboratory, University of California, San Diego, CA, USA.
D. J. Hagler, Jr, Department of Radiology, University of California, San Diego, CA, USA; Multimodal Imaging Laboratory, University of California, San Diego, CA, USA.
Anders M. Dale, Department of Neurosciences, University of California, San Diego, CA, USA Department of Radiology, University of California, San Diego, CA, USA; Multimodal Imaging Laboratory, University of California, San Diego, CA, USA.
James B. Brewer, Email: jbrewer@ucsd.edu, Department of Neurosciences, University of California, San Diego, CA, USA; Department of Radiology, University of California, San Diego, CA, USA; Human Memory Laboratory, 8950 Villa La Jolla Drive C212, La Jolla, CA 92037, USA.
References
- Albers JJ, Tollefson JH, Wolfbauer G, Albright RE., Jr Cholesteryl ester transfer protein in human brain. International Journal of Clinical & Laboratory Research. 1992;21(3):264–266. doi: 10.1007/BF02591657. [DOI] [PubMed] [Google Scholar]
- Arias-Vásquez A, Isaacs A, Aulchenko YS, Hofman A, Oostra BA, Breteler M, et al. The cholesteryl ester transfer protein (CETP) gene and the risk of Alzheimer’s disease. Neurogenetics. 2007;8(3):189–193. doi: 10.1007/s10048-007-0089-x. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Atzmon G, Schechter C, Schaefer EJ, Cupples AL, Lipton R, et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290(15):2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Atzmon G, Derby CA, Bauman JM, Lipton RB. A genotype of exceptional longevity is associated with preservation of cognitive function. Neurology. 2006;67(12):2170–2175. doi: 10.1212/01.wnl.0000249116.50854.65. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathologica. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Chen DW, Yang JF, Tang Z, Dong XM, Feng XL, Yu S, et al. Cholesteryl ester transfer protein polymorphism D442G associated with a potential decreased risk for Alzheimer’s disease as a modifier for APOE epislon4 in Chinese. Brain Research. 2008;1187:52–57. doi: 10.1016/j.brainres.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Crean S, Ward A, Mercaldi CJ, Collins JM, Cook MN, Baker NL, Arrighi HM. Apolipoprotein E ε4 prevalence in Alzheimer’s disease patients varies across global populations: a systematic literature review and meta-analysis. 2011 doi: 10.1159/000321984. [DOI] [PubMed] [Google Scholar]
- Dale AM, Sereno MI. Improved localization of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction: a linear approach. Journal of Cognitive Neuroscience. 1993;5:162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. NeuroImage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- De Gennes JL, Dairou F, Gardette J, Truffert J. Sex hormones and metabolism of lipoproteins. Annales de Endocrinologie. 1983;44(1):59–65. [PubMed] [Google Scholar]
- Dickerson BC, Feczko E, Augustinack JC, Pacheco J, Morris JC, Fischl B, et al. Differential effects of aging and Alzheimer’s disease on medial temporal lobe cortical thickness and surface area. Neurobiology of Aging. 2009;30(3):432–440. doi: 10.1016/j.neurobiolaging.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, et al. Automatically parcellating the human cerebral cortex. Cerebral Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Gearing M, Mirra SS, Hedreen JC, Sumi SM, Hansen LA, Heyman A. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). X. Neuropathology confirmation of the clinical diagnosis of Alzheimer’s disease. Neurology. 1995;45:461–466. doi: 10.1212/wnl.45.3.461. [DOI] [PubMed] [Google Scholar]
- Holland D, Brewer JB, Hagler DJ, Fennema-Notestine C, Dale AM the Alzheimer’s Disease Neuroimaging Initiative. Subregional neuroanatomical change as a biomarker for Alzheimer’s disease. Proc Natl Acad Sci USA. 2009;106(49):20954–20959. doi: 10.1073/pnas.0906053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu YC, Waring SC, O’Brien PC, Tangalos EG, et al. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer’s disease. Neurology. 1997;49(3):786–794. doi: 10.1212/wnl.49.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W, Harris SE, Collins P, Starr JM, Whalley LJ, Deary IJ. No association of CETP genotype with cognitive function or age-related cognitive change. Neuroscience Letters. 2007;420(2):189–192. doi: 10.1016/j.neulet.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, et al. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. NeuroImage. 2006;30(2):436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Joyner AH, Roddey JC, Bloss CS, Bakken TE, Rimol LM, Melle I, et al. A common MECP2 haplotype associates with reduced cortical surface area in humans in two independent populations. Proc Natl Acad Sci USA. 2009;106(36):15483–15488. doi: 10.1073/pnas.0901866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivipelto M, Helkala EL, Laaksom MP, Hanninen T, Hallikainen M, Alhainen K, et al. Midlife vascular risk factors and Alzheimer’s disease in later life: longitudinal, population based study. BMJ. 2001;322:1447–1451. doi: 10.1136/bmj.322.7300.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel YL, McPherson R, Hogue M, Czarnecka H, Zawadzki Z, Weech PK, et al. Distribution and concentration of cholesteryl ester transfer protein in plasma of normolipemic subjects. The Journal of Clinical Investigation. 1990;85:10–17. doi: 10.1172/JCI114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattay VS, Goldberg TE, Sambataro F, Weinberger DR. Neurobiology of cognitive aging: insights from imaging genetics. Biological Psychology. 2008;79(1):9–22. doi: 10.1016/j.biopsycho.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy LK, Fennema-Notestine C, Roddey JC, Hagler DJ, Jr, Holland D, Karow DS, et al. Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009;251(1):195–205. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok W, Chow TW, Zheng L, Mack WJ, Miller C. Clinicopathological concordance of dementia diagnoses by community versus tertiary care clinicians. American Journal of Alzheimer's Disease and Other Dementias. 2004;19:161–165. doi: 10.1177/153331750401900309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy EA, Holland D, Donohue M, McEvoy LK, Hagler DJ, Jr, Dale AM, et al. Six-month atrophy in MTL structures is associated with subsequent memory decline in elderly controls. NeuroImage. 2010;53(4):1310–1317. doi: 10.1016/j.neuroimage.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri-Cesi P, Colciago A, Celotti F, Motta M. Sexual differentiation of the brain: role of testosterone and its active metabolites. Journal of Endocrinological Investigation. 2004;27(6):120–127. [PubMed] [Google Scholar]
- Olesen OF, Dago L. High density lipoprotein inhibits assembly of amyloid beta-peptides into fibrils. Biochemical and Biophysical Research Communications. 2000;270(1):62–66. doi: 10.1006/bbrc.2000.2372. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as quantitative trait in genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PloS One. 2009;4(8):e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregelj P. Involvement of cholesterol in the pathogenesis of Alzheimer’s disease: role of statins. Psychiatria Danubina. 2008;20(2):162–167. [PubMed] [Google Scholar]
- Qureischie H, Heun R, Lütjohann D, Popp J, Jessen F, Ledschbor-Frahnert C, et al. CETP polymorphisms influence cholesterol metabolism but not Alzheimer’s disease risk. Brain Research. 2008;1232:1–6. doi: 10.1016/j.brainres.2008.07.047. [DOI] [PubMed] [Google Scholar]
- Qureischie H, Heun R, Popp J, Jessen F, Maier W, Schmitz S, et al. Association of CETP polymorphisms with the risk of vascular dementia and white matter lesions. Journal of Neural Transmission. 2009;116(4):467–472. doi: 10.1007/s00702-008-0180-y. [DOI] [PubMed] [Google Scholar]
- Rodríguez E, Mateo I, Infante J, Llorca J, Berciano J, Combarros O. Cholesteryl ester transfer protein (CETP) polymorphism modifies the Alzheimer’s disease risk associated with APOE ε4 allele. Journal of Neurology. 2006;253(2):181–185. doi: 10.1007/s00415-005-0945-2. [DOI] [PubMed] [Google Scholar]
- Sanders AE, Wang C, Katz M, Derby CA, Barzilai N, Ozelius L, et al. Association of a functional polymorphism in the cholesteryl ester transfer protein (CETP) gene with memory decline and incidence of dementia. JAMA. 2010;303(2):150–158. doi: 10.1001/jama.2009.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer’s Disease neuroimaging initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimer’s & Dementia. 2010;6(3):265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JA, Arvanitakis Z, Bang W, Bennett DA. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology. 2007;69:2197–2204. doi: 10.1212/01.wnl.0000271090.28148.24. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1864–1865. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging. 1998;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Sorlí JV, Corella D, Francés F, Ramírez JB, González JI, Guillén M, et al. The effect of the APOE polymorphism on HDL-C concentrations depends on the cholesterol ester transfer protein gene variation in a Southern European population. Clinica Chimica Acta. 2006;366(1–2):196–203. doi: 10.1016/j.cca.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Stein JL, Hua X, Morra JH, Lee S, Hibar DP, Ho AJ, et al. Genome-wide analysis reveals novel genes influencing temporal lobe structure with relevance to neurodegeneration in Alzheimer’s disease. NeuroImage. 2010;51(2):542–554. doi: 10.1016/j.neuroimage.2010.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JF, Lira ME, Durham LK, Clark RW, Bamberger MJ, Milos PM. Polymorphisms in the CETP gene and association with CETP mass and HDL levels. Atherosclerosis. 2003;167:195–204. doi: 10.1016/s0021-9150(03)00005-4. [DOI] [PubMed] [Google Scholar]
- Yamada T, Kawata M, Arai H, Fukasawa M, Inoue K, Sato T. Astroglial localization of cholesteryl ester transfer protein in normal and Alzheimer’s disease brain tissues. Acta Neuropathologica. 1995;90(6):633–636. doi: 10.1007/BF00318577. [DOI] [PubMed] [Google Scholar]