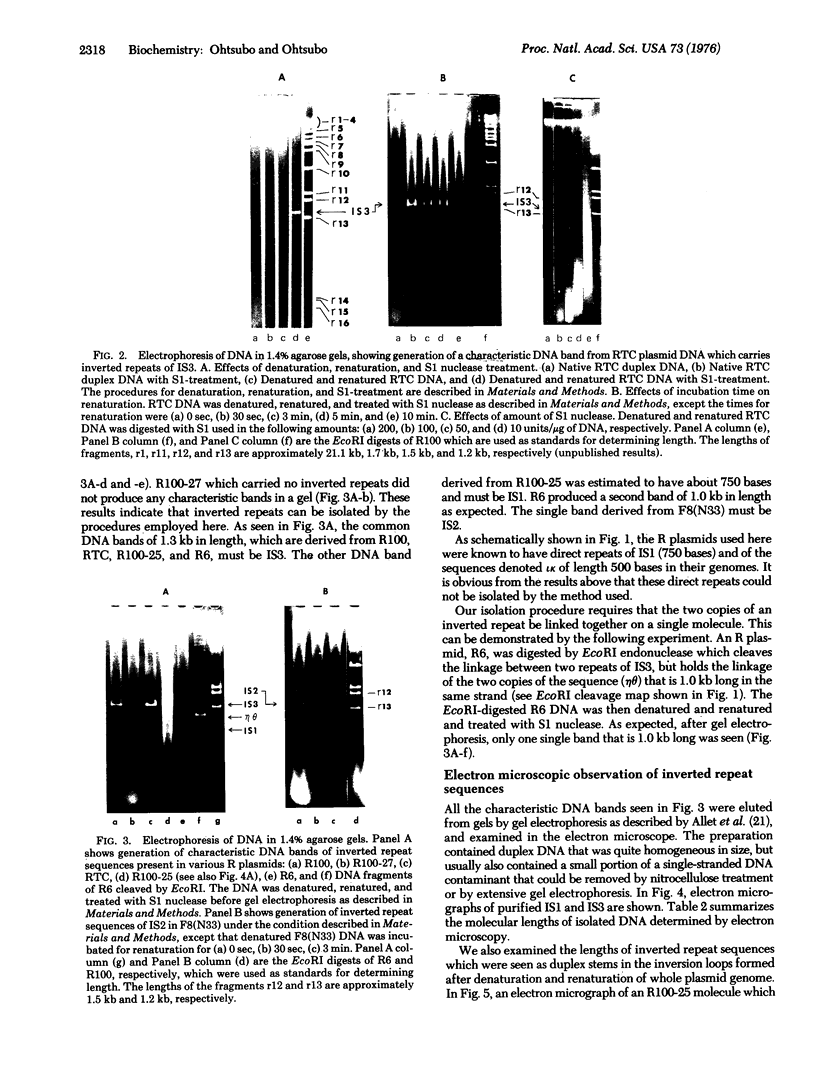
Abstract
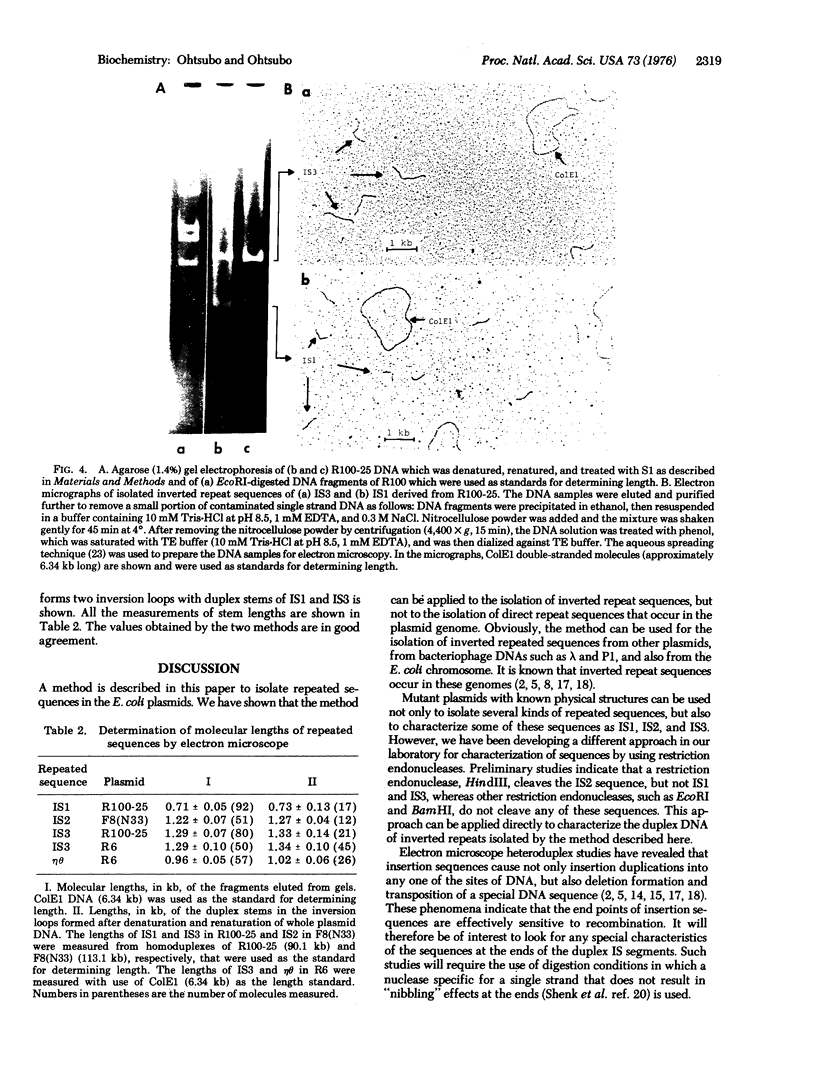
A method is described for isolation of inverted repeat DNA sequences that occur in E. coli plasmids. The procedures of the isolation involved: (a) denaturation of intact plasmid DNA, (b) a rapid, 30 sec, renaturation of inverted-repeat sequences in the genome, (c) digestion of the single-stranded portion by S1 nuclease to recover duplex DNA, and (d) detection and purification of the duplexes using 1.4% agarose gel electrophoresis. If a plasmid DNA carried inverted repeats of either one type or two different types of special DNA sequences, these procedures enabled us to observe either one or two characteristic DNA bands, respectively, in the agarose gels. If a plasmid DNA did not carry any inverted repeats, or if the plasmid DNA only carried direct repeat sequences, no characteristic DNA bands were recovered. Cleavage of the spacer DNA between inverted repeat sequences generated no gel bands. This indicated that the inverted repeat sequences must be in the same strand. Using this method, we isolated and purified several repeated sequences, including IS1, IS2, and IS3, from derivatives of F and R plasmids.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Jeppesen P. G., Katagiri K. J., Delius H. Mapping the DNA fragments produced by cleavage by lambda DNA with endonuclease RI. Nature. 1973 Jan 12;241(5385):120–123. doi: 10.1038/241120a0. [DOI] [PubMed] [Google Scholar]
- Berg D. E., Davies J., Allet B., Rochaix J. D. Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., Rubens C., Falkow S. Translocation of a plasmid DNA sequence which mediates ampicillin resistance: molecular nature and specificity of insertion. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3623–3627. doi: 10.1073/pnas.72.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch H. J., Starlinger P., Brachet P. Two kinds of insertions in bacterial genes. Mol Gen Genet. 1972;119(3):191–206. doi: 10.1007/BF00333858. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Davidson N. Electron microscope heteroduplex study of the heterogeneity of Mu phage and prophage DNA. Virology. 1974 Mar;58(1):229–239. doi: 10.1016/0042-6822(74)90157-3. [DOI] [PubMed] [Google Scholar]
- Hu S., Ohtsubo E., Davidson N. Electron microscopic heteroduplex studies of sequence relations among plasmids of Escherichia coli: structure of F13 and related F-primes. J Bacteriol. 1975 May;122(2):749–763. doi: 10.1128/jb.122.2.749-763.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Ptashne K., Cohen S. N., Davidson N. alphabeta sequence of F is IS31. J Bacteriol. 1975 Aug;123(2):687–692. doi: 10.1128/jb.123.2.687-692.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Chan R. K., Tye B. K., Botstein D. Mutagenesis by insertion of a drug-resistance element carrying an inverted repetition. J Mol Biol. 1975 Oct 5;97(4):561–575. doi: 10.1016/s0022-2836(75)80059-3. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Otsubo E., Deonier R. C., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. V. ilv+ Deletion mutants of F14. J Mol Biol. 1974 Nov 15;89(4):585–597. doi: 10.1016/0022-2836(74)90037-0. [DOI] [PubMed] [Google Scholar]
- Malamy M. H., Fiandt M., Szybalski W. Electron microscopy of polar insertions in the lac operon of Escherichia coli. Mol Gen Genet. 1972;119(3):207–222. doi: 10.1007/BF00333859. [DOI] [PubMed] [Google Scholar]
- Otsubo E., Deonier R. C., Lee H. J., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. IV. The F sequences in F14. J Mol Biol. 1974 Nov 15;89(4):565–584. doi: 10.1016/0022-2836(74)90036-9. [DOI] [PubMed] [Google Scholar]
- Ptashne K., Cohen S. N. Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. J Bacteriol. 1975 May;122(2):776–781. doi: 10.1128/jb.122.2.776-781.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif H. J., Saedler H. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol Gen Genet. 1975;137(1):17–28. doi: 10.1007/BF00332538. [DOI] [PubMed] [Google Scholar]
- Saedler H., Reif H. J., Hu S., Davidson N. IS2, a genetic element for turn-off and turn-on of gene activity in E. coli. Mol Gen Genet. 1974;132(4):265–289. doi: 10.1007/BF00268569. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Manning J. E., Davidson N. Inverted repeat sequences in the Drosophila genome. Cell. 1975 Jun;5(2):159–172. doi: 10.1016/0092-8674(75)90024-0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Hsu M. T., Otsubo E., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. I. Structure of F-prime factors. J Mol Biol. 1972 Nov 14;71(2):471–497. doi: 10.1016/0022-2836(72)90363-4. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B. K., Chan R. K., Botstein D. Packaging of an oversize transducing genome by Salmonella phage P22. J Mol Biol. 1974 Jan 5;85(4):485–500. doi: 10.1016/0022-2836(74)90311-8. [DOI] [PubMed] [Google Scholar]