Abstract
Recent advances in the understanding of molecular recognition and protein–ligand interactions have facilitated rapid development of potent and selective ligands for therapeutically relevant targets. Over the past two decades, a variety of useful approaches and emerging techniques have been developed to promote the identification and optimization of leads that have high potential for generating new therapeutic agents. Intriguingly, the innovation of a fragment-based drug design (FBDD) approach has enabled rapid and efficient progress in drug discovery. In this critical review, we focus on the construction of fragment libraries and the advantages and disadvantages of various fragment-based screening (FBS) for constructing such libraries. We also highlight the deconstruction–reconstruction strategy by utilizing privileged fragments of reported ligands.
Keywords: fragment-based drug design (FBDD), fragment library, fragment-based screening (FBS), deconstruction-reconstruction, protein-ligand interactions, therapeutic agents
Introduction
Despite significant scientific and technological advances developed to improve the quality and efficiency of drug discovery in the pharmaceutical industry, there is an indisputable fact that the higher investment has not resulted in substantial increase of new chemical entities introduced to the market. More innovative technologies and approaches are needed to address such issues [1,2]. To this end, the efficient use of fragments with weak potency for the targets as starting points for step-wise optimizations has attracted considerable attention recently [3–5]. The concept of FBDD can be traced back to the pioneering work of William Jencks in 1981 [6]. The binding energy of the whole molecule with the target could be considered a summation of individual binding energy between the fragments and the target. Nevertheless, this intriguing viewpoint has not attracted much attention from either the pharmaceutical industry or academia for some time. There are two main obstacles for its practical application: (i) how to identify suitable fragments that bind to the neighboring binding sites and (ii) how to optimize these fragments by merging, linking, or growing to develop drug-like molecules without distortions of their individual binding modes.
The seminal studies and the first successful application of FBDD in drug discovery were done by scientists at Abbott. Together with the traditional high-throughput screening (HTS) approach and combinatorial chemistry [7], FBDD has progressed rapidly and has emerged as one of the most important drug discovery technologies. FBDD has the advantages of comprehensive random screening and structure-based drug design [8]. Conventional HTS approaches after searching huge collections of drug-sized molecules might identify numerous hits or lead compounds, but few of them can reach the market. Limited chemical space, low structural diversity, and unfavorable drug absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties are the major obstacles for further drug development. FBDD enables identification of various active fragments, which can reach into the deep subpockets within the active site. Once the detailed interaction within the cavity is experimentally validated and clearly understood, it could provide a unique opportunity to design potent and efficacious drug-like chemical entities. This strategy offers several attractive features compared with traditional HTS or virtual screening, including higher hit rate, higher binding efficiency, and more effective optimization capacity. From a practical standpoint, the smaller the size of fragment, the more possibilities are available for further structural modifications, making it feasible to search more chemical space. In this critical review, we focus on the construction of fragment libraries and the advantages and disadvantages of various fragment-based screening for fragment mining. We also highlight the deconstruction–reconstruction strategy by utilizing privileged fragments of reported ligands.
Construction of fragment libraries
Construction of fragment libraries is the first step for FBDD. To construct a suitable fragment library, several factors should be considered, including: (i) the distinction between fragments and hits and/or leads. Congreve et al. proposed a rule-of-three (RO3) [9] representing a set of guidelines for the construction of a fragment library (molecular weight is <300, cLogP is ≤3, the number of hydrogen bond donors is ≤3, and the number of hydrogen bond acceptors is ≤3). Recently, RO3 was accredited by most medicinal chemists and could be useful for efficient fragment selection [10]; (ii) the size of the fragment library differs from that in HTS. For instance, screening approaches such as nuclear magnetic resonance (NMR) and X-ray crystallography screening are suitable for a library size in the range of 102–103, whereas approaches such as surface plasmon resonance (SPR) are adaptive for a library size of up to 105 [11]; (iii) structural diversity of the fragment library. The fragment library should cover more chemical space to produce a highly diversified library; (iv) the solubility of fragments. Given that fragments typically bind weakly to the target protein, the measurement of binding interaction is conducted at a higher concentration, which requires a better solubility of fragment to avoid producing false results; and (v) the drug-likeness of fragments [12,13]. Accumulating studies show that most drugs can be divided into two to three fragments according to their scaffolds and side chains. Therefore, the similarity between fragments and the privileged fragments should be considered to improve the druggability of the final drug-like compounds when constructing the fragment library. In addition, the chemical stability and synthetic ease of fragments should also be considered for fragment mining.
Construction of the fragment library begins with the detection and identification of relatively weak interactions between the fragments and a target macromolecule by using informative biophysical techniques. Currently, there are few available techniques that are sensitive enough for efficient screening of weakly interacting fragments, and each has its advantages and disadvantages (Table 1). Utilizing these various fragment-based screening methods appropriately according to the resource accessibility as well as their pros and cons could facilitate efficient construction of a fragment library. It should be noted that the combination of two or multiple FBS methods could also alleviate the drawbacks of each individual technique and lead to the optimal outcomes for the fragment screening [14].
Table 1.
The pros and cons of various FBS methods
| FBSa | Pros | Cons | Refs |
|---|---|---|---|
| Target-based NMR | High sensitivity Capable of identifying binding sites |
Expensive equipment Need professional knowledge High protein consumption Requirement for isotopically labeled protein with high purity Require suitable solubility and modest molecular weight |
[40–42] |
| Ligand-based NMR | Rapid High sensitivity Moderate protein consumption Without isotopically labeled target and its molecular mass, practically unlimited |
Expensive equipment Need professional knowledge Unable to identify binding sites Prone to false positives |
[40–42] |
| XRC | Provide detailed structural information with high resolution Weak binding fragments can be identified Avoiding false positives Applicable to large proteins |
Requirement for high purity protein Low throughput Need professional knowledge Medium–high protein consumption False negatives can occur |
[43,44] |
| SPR | Rapid and cost effective High throughput Low protein consumption Provide kinetic and thermodynamic information Applicable to most target proteins Ease of automation |
Expensive equipment Prone to artifacts Require immobilization of target or fragment False negatives can occur |
[36,45–47] |
| MS | Highly sensitive and accurate High throughput Rapid and automatable Low protein consumption Significantly reduced constrains on protein purity Label-free |
Expensive equipment Unable to provide fragment binding information |
[48–50] |
| TSA | High throughput, inexpensive, and rapid Low protein consumption Applicable to most target proteins Direct binding assay |
Requirements for high concentrations of fragments Prone to false positives and negatives Difficult to detect very weakly binding fragments |
[51–53] |
| ITC | Direct binding assay Provide information on Kd, H, S and n |
Low throughput and resolution High protein and ligand consumption |
[54–57] |
| HCS | High throughput Various available techniques for detection Small amounts of protein requirement Upfront knowledge of biochemical activity |
Requirements for high concentrations of fragments False negatives and positive Require biochemical function |
[58–60] |
| VS | Fast and cost effective | Low accuracy of predictions Rapid accumulation of errors |
[61,62] |
Abbreviations: HCS, high concentration screening; ITC, isothermal titration calorimetry; MS, mass spectrometry; SPR, surface plasmon resonance; TSA, thermal shift assay; VS, virtual screening; XRC, X-ray crystallography.
The deconstruction–reconstruction approach
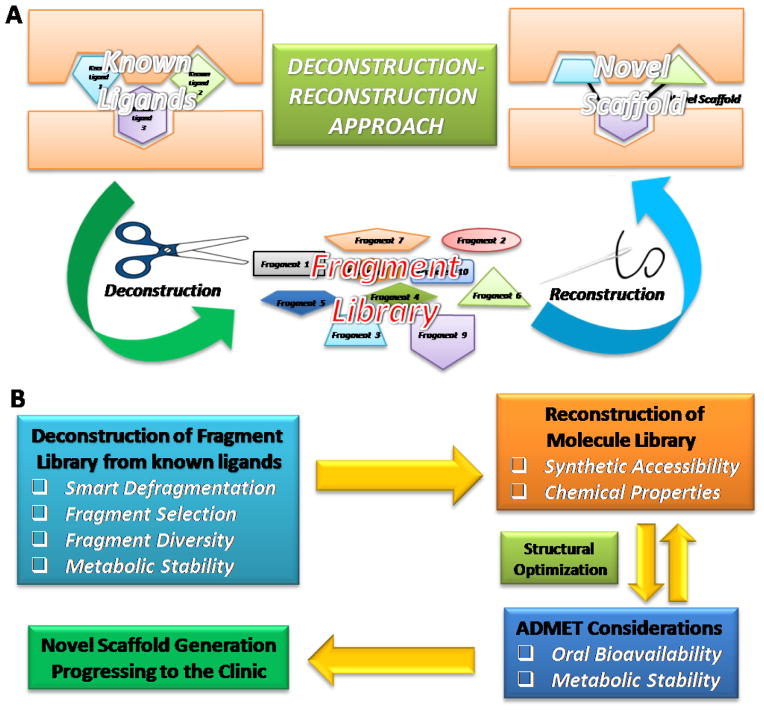
Although different from FBS, deconstruction of known ligands can provide a useful strategy for the construction of a relatively smaller fragment library. The deconstruction–reconstruction approach has gained traction in recent years [15]. As depicted in Figure 1A, the concept for this approach is simple. As already alluded to, traditional FBDD combines fragments into a final molecule [16]. Therefore, it is typically possible to deconstruct a known molecule into several fragments [17,18]. However, some preliminary studies on certain target proteins indicated that the fragments resulting from the deconstruction of known ligands did not recapitulate their positions in a large ligand. For instance, Shoichet et al. reported the deconstructing fragment-based inhibitor discovery from a known β-lactamase inhibitor [19], which was divided into three commercially available fragments. After they grew and compared co-crystals of β-lactamase in complex with these three fragments, the authors found that the binding modes of the three simple fragments differed from their original positions. From these first-hand experimental data, the authors suggested that the converse deconstructive logic need not hold [19]. Krimm and co-workers reported the deconstruction of Bcl-xL inhibitors indicating that these fragments have a preferred binding site of their own [20]. However, most of the derived fragments did not keep the original binding sites that they occupied in the protein–inhibitor complex, indicating that the complexity of the fragment did not guarantee the conservation of the binding mode [20]. More recently, the same group examined fragments from previously developed inhibitors of glycogen phosphorylase by NMR, suggesting that defragmentation not only provides conserved binding pockets, but also uncovers cooperatives between these various binding sites [21]. This study suggests that the deconstruction approach appears to be a valuable tool to probe multiple conserved and nonconserved binding pockets. By contrast, by using a combination of X-ray crystallographic analysis of the peptide–protein complexes, Aalten et al. showed that fragments derived from the natural cyclopentapeptide argifin maintained their binding modes [22]. The authors concluded that these natural product-derived fragments from argifin might represent attractive starting points for further structure-based optimization. Taking into account these representative studies, how to deconstruct rationally the reported ligand into fragments has a crucial role in the process of collecting small functional and efficient fragments.
Figure 1.
(A) Schematic representation of the deconstruction–reconstruction approach in fragment-based drug design (FBDD). (B) Flow diagram of the deconstruction–reconstruction approach in FBDD. Abbreviation: ADMET, absorption, distribution, metabolism, excretion, and toxicity.
Generally, the first step of the deconstruction–reconstruction approach is to deconstruct known ligands into several fragments that are likely to act as key pharmacophores for FBDD (Figure 1B). This step could be utilized to construct a general landscape of binding sites for fragments, defining the direction for further structural elaboration and optimization. After the construction of a fragment library derived from the known ligands, it is expected that structural analysis will be beneficial for assessing the suitability of fragments for rational decoration. The second step is to reconstruct these fragments selected from the relevant fragment library into a new scaffold. Although it is relatively straightforward to deconstruct biologically active fragments of drugs, there are usually more challenges in the reconstruction procedure, such as to optimize the fragments by merging, linking, or growing them to develop drug-like molecules without distortions of their individual binding modes. To this end, the overall reconstruction approach should be governed by classic guidelines, such as Lipinski’s Rule of Five [23] and Veber’s rules [24], together with public algorithms, to ensure suitable drug-like physicochemical properties, including LogP, topological polar surface area (tPSA), molecular weight, and volume, are maintained. Computer-assisted molecular modeling and docking with the target protein, as well as the analysis of bioinformatics and chemoinformatics, could facilitate a better binding mode prediction during reconstruction. Meanwhile, the aforementioned RO3 fragment selection rules might also be useful guidelines for getting rid of undrug-like motifs to improve metabolic stability and oral bioavailability for a more efficient reconstruction. In addition, the synthetic accessibility should be considered for converting fragments into a final compound. This approach is useful for target proteins that have been explored for many years because accumulating studies could provide abundant information to support a more rational reconstruction design. To date, a good number of reported ligands generated in drug discovery efforts by industry and academic laboratories can provide powerful and efficient data to create novel and promising scaffolds for further development. Therefore, the deconstruction–reconstruction approach based on the available chemical and biological data has proven highly effective in FBDD, and several representative examples are discussed below to illustrate this approach.
Signal transducers and activators of transcription 3 inhibitors
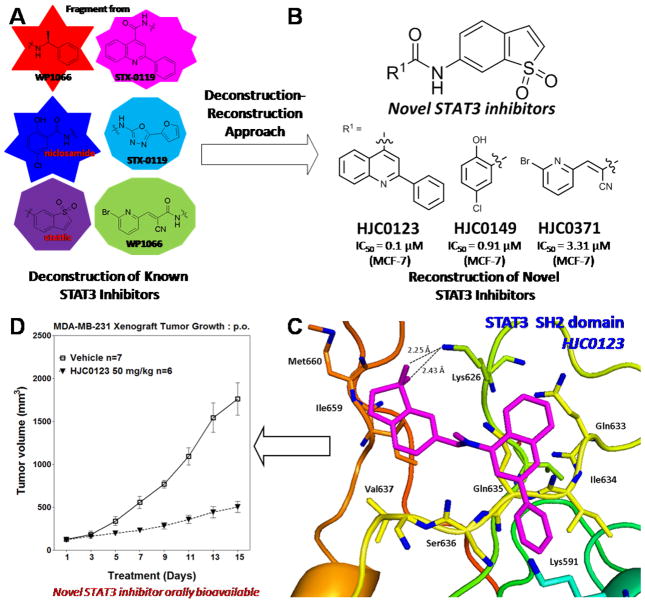
As an attractive therapeutic or preventive target for various human cancers, signal transducers and activators of transcription 3 (STAT3) has attracted increasing attention in recent years. Over the past decade, dozens of STAT3 inhibitors with diversified scaffolds have been developed. Despite these significant advances, no US Food and Drug Administration (FDA)-approved STAT3 inhibitor drugs are currently available in the clinic because of their limited potency, efficacy and drug-like properties. Figure 2 illustrates the discovery of HJC0123, an orally bioavailable drug candidate that is now in preclinical studies for the treatment of breast cancer and other malignancies [25]. After deconstruction of privileged fragments from known STAT3 inhibitors to generate a small fragment library (Figure 2A), several novel scaffolds were reconstructed and chemically synthesized (Figure 2B). Further pharmacological evaluation led to the identification of several compounds, including HJC0123, HJC0149, and HJC0371, as advanced chemical leads for further optimization [26]. The results from the predicted binding mode of HJC0123 with the SH2 domain of STAT3 (Figure 2C) are in full agreement with previous established structure–activity relation (SAR) data [25]. Further biological evaluations resulted in the discovery of HJC0123 as an orally bioavailable anticancer drug candidate targeting STAT3 (Figure 2D).
Figure 2.
The discovery of orally bioavailable signal transducers and activators of transcription 3 (STAT3) inhibitor HJC0123 by utilizing a deconstruction–reconstruction approach. (A) Deconstruction of known STAT3 inhibitors to identify privileged fragments. (B) Reconstruction of novel STAT3 inhibitors. (C) The predicted binding mode of HJC0123 to the STAT3 SH2 domain. (D) In vivo efficacy of HJC0123 in inhibiting growth of xenograft tumors in mice (p.o.). Reproduced, with permission, from [49].
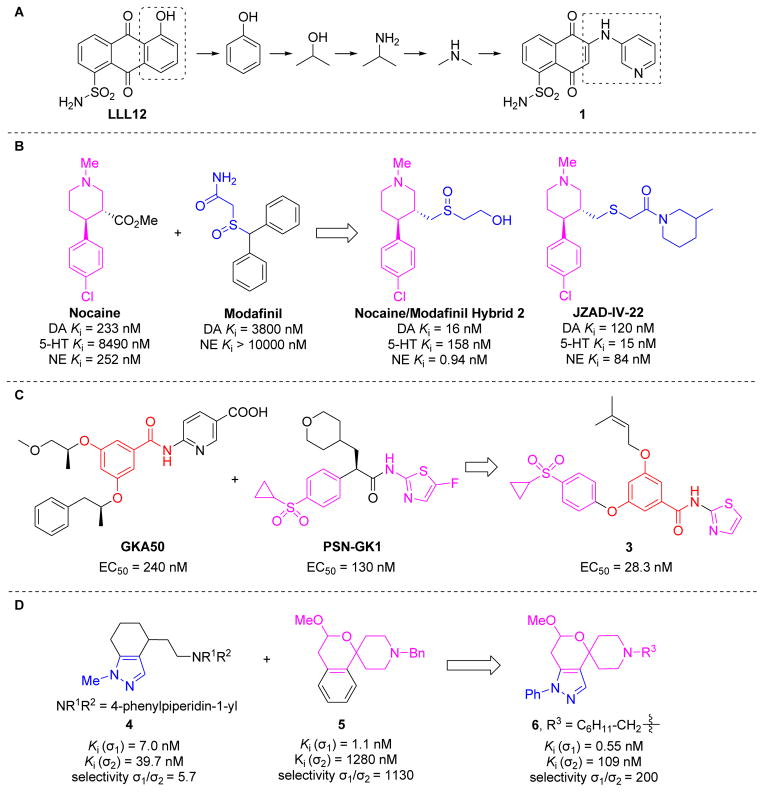
Li and co-workers also reconstructed fragment libraries from known STAT3 inhibitors [27]. They divided representative STAT3 dimerization inhibitors into specific sublibraries according to docking poses. They built a new molecule library by linking selected fragments from different fragment sublibraries. Taking inspiration from the in silico site-directed FBDD approach, compound 1 based on the chemical scaffold of previous lead compound LLL12 was synthesized and identified as a novel STAT3 inhibitor with enhanced potency and drug-like properties (Figure 3A).
Figure 3.
(A) The evolution of linker design and discovery of novel signal transducers and activators of transcription 3 (STAT3) inhibitor 1 via in silico site-directed fragment-based drug design (FBDD). (B) Rational design of nocaine/modafinil hybrid ligand 2 and novel triple reuptake inhibitor JZAD-IV-22 by utilizing a deconstruction–reconstruction approach. (C) A benzamide derivative 3 as the glucokinase activator was designed and assembled by using a deconstruction–reconstruction approach through a privileged fragment-merging strategy. (D) Discovery of the spirocyclic pyranopyrazole 6 as a novel sigma 1 (σ1) receptor ligand by the combination of the privileged fragments from known σ1 receptor ligands 4 and 5.
Monoamine transporter inhibitors
Monoamine transporters of dopamine (DA), serotonin (5-HT), and norepinephrine (NE) have been empirically used as classic medication targets for several psychological and neurological disorders in the central nervous system (CNS) [28,29]. Over the past two decades, a growing number of monoamine transporter inhibitors were reported in the literature exhibiting varying levels of transporter selectivity, and some of them are widely used in the clinic. Such abundantly available information provides a unique opportunity to develop highly transporter-specific agents with diverse pharmacological profiles using the deconstruction–reconstruction approach. Zhou et al. designed and synthesized a series of piperidine-based hybrid inhibitors by deconstruction of the parent structures of nocaine and modafinil, followed by a reconstruction–elaboration approach (Figure 3B) [30]. After replacement of the hydrolyzable ester of nocaine with a sulfur-containing side chain of modafinil, some highly active monoamine transporter inhibitors with low nanomolar to subnanomolar potency and diversified selectivity profiles were identified. Ligand 2 displayed excellent reuptake inhibitory effects for both DA and NE transporters and exhibited higher efficacy compared with cocaine in an open-field locomotor activity study in vivo. JZAD-IV-22 has been identified as a novel triple reuptake inhibitor (TRI) by targeting DA/5-HT/NE transporters with antidepressant-like activity similar to that of DOV 216,303 [31]. The striking feature that distinguishes these two TRIs is that locomotor sensitization, a common underlying feature of drug abuse liability, is seen with DOV 216,303 but is completely lacking in JZAD-IV-22, a promising antidepressant drug candidate in preclinical development [31].
Glucokinase activators
Glucokinase (GK) was identified as a promising drug target for type 2 diabetes (T2D) over the past four decades. To date, GK activators (GKAs) have been designed, synthesized, and pharmacologically evaluated for the treatment of patients with T2D patients. Despite the limited success of effective and safe drug candidates, the reported GKAs provide a good arsenal of fragments for the rational design of new scaffolds. Zhang et al. recently designed and synthesized a series of benzamide derivatives by utilizing a deconstruction–reconstruction approach (Figure 3C) [32]. Analysis of currently available GKAs suggests that benzamide is the key fragment in phenylacetamide GKA50 and the clinical candidate PSN-GK1 contains cyclopropylsulfonyl and aminothiazolyl moieties as two privileged fragments. The authors reconstructed these fragments followed by elaboration and optimization and successfully identified a promising GKA (3) with a favorable potency and activation profile. Further structural optimization is needed to improve the pharmacokinetic properties of this class of compounds; however, this work represents the first rational design of GKAs using the deconstruction–reconstruction approach.
Sigma 1 receptor ligands
The sigma 1 (σ1) receptor represents an interesting target for the development of novel CNS drugs because of its important role associated with various neurological processes [33]. A range of structurally diverse compounds can bind to the σ1 receptor with high affinity. Taking advantage of the privileged fragments through the deconstruction of reported σ1 receptor ligands, Wünsch et al. designed and synthesized a series of new spirocyclic pyranopyrazole derivatives with high affinity based upon their excellent σ1 affinity and extraordinarily high σ1/σ2 selectivity. Using the benzofuran derivative 5 and the high analgesic activity of the indazole derivative 4 (Figure 3D) [34], ligand 6 displayed an outstanding affinity and a good selectivity for σ1 receptor (Ki (σ1) = 0.55 nM, selectivity σ1/σ2 = 200). The combination of the privileged fragments from the reported ligands could provide an additional strategy for developing novel σ1 receptor ligands.
AMPA receptor positive allosteric modulators
Despite significant advances in the development of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor-positive allosteric modulators (PAMs) over the past few years, no drug candidate has reached the market. However, numerous co-crystal ligand–protein structures with diverse ligands of the GluA2 isoform of the AMPA receptors are now available following two decades of research on this druggable target. After detailed overlay analysis of these crystal structures, a deconstruction–reconstruction approach was applied to design and synthesize two AMPA receptor PAMs with enhanced potency in vitro and remarkable efficacy in vivo for preventing neuroapoptosis (Figure 4) [35]. The first step of this drug discovery campaign was to deconstruct these known ligands into a small fragment library. The structural analysis led to a general landscape of binding sites for these fragments (Figure 4A). It was found that fragment 3, N-(2-phenylpropyl)methanesulfonamide, accommodated the deep cavity and covered more space in pocket S3. Reconstruction and design a series of asymmetric bivalent ligands were conducted by merging fragment 3 with various privileged fragments selected from the fragment library (Figure 4B). After physicochemical calculation and molecular docking studies (Figure 4C) of this focused new compound library, two promising compounds were synthesized and pharmacologically evaluated. These two AMPA receptor PAMs, HJC0122 and HJC0124, exhibited high potency and efficacy in preventing neuroapoptosis (Figure 4D) both in vitro and in vivo. This FBDD strategy by using online databases and public algorithms represents an attractive approach for the rational design of potential drug candidates with enhanced potency and efficacy with cost-effectiveness.
Figure 4.
The discovery of HJC0122 and HJC0124 as new -amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor-positive allosteric modulators (PAMs). (A) Overlay analysis of the nine known PAMs with structures available from the Protein Data Bank (PDB). (B) General deconstruction–reconstruction approach for drug design schemes. (C) Predicted binding mode of HJC0122 (pink) to the GluA2 dimer interface. (D) Modulation of the effect of AMPA (1 MM) on caspase-3 activity induced by phencyclidine (PCP) at 3 MM in a concentration-dependent fashion and with higher potency compared with LY451395 (in Phase II clinical trials). Inset, chemical structures of neuroprotective agents HJC0122 and HJC0124. Reproduced, with permission, from [59].
Concluding remarks and future perspectives
Over the past decades, the development of new chemical entities by utilizing traditional HTS and combinatorial chemistry has been demonstrated to be a useful approach, but it is also time consuming, costly and risky. Different from the resource-intensive conventional drug discovery methods, FBDD has emerged as an attractive strategy, and several drug candidates developed by this approach are currently undergoing clinical and preclinical trials [36,37]. For instance, FBDD has yielded vemurafenib, the first drug approved for metastatic melanoma after only 6 years of research and development [38]. Construction of a fragment library is the first step for FBDD. Several fragment-screening technologies using sensitive biophysical techniques have proven beneficial for various challenging drug discovery targets. A fragment library statistically covers more chemical space compared with a hit and/or lead library and, as a consequence, fewer compounds are required to be screened. Given the high sensitivity, FBS tends to deliver high hit rates, providing multiple starting points for further structural optimizations [39]. In addition, because of the exponentially growing amount of information about one certain target, the effective utilization of bioinformatics and chemoinformatics is expected to contribute markedly toward the discovery of new drugs. Recently, a growing number of reported drug discovery campaigns taking advantage of the deconstruction–reconstruction approach suggest that efficient defragmentation of selected ligands as starting frameworks is likely to increase the chances of obtaining a final optimized ligand with high potency and efficacy as well as favorable drug properties in a cost-effective manner.
Application of this promising approach might also provide the recognition of structural features that contribute to potential binding to the target. Generally, pendant substituents are defragmented from the ligand to generate a fragment library, and these fragments are then reconstructed by reintroducing various original structural features together with elaboration to yield unique chemical entities with better drug-likeness. As discussed above, deconstruction of reported ligands appears to be relatively straightforward. However, it is usually more challenging and tricky in the reconstruction procedure, that is, to optimize the fragments by merging, linking, or growing them to develop drug-like molecules with enhanced binding modes. To this end, computer-aided rational drug design and the classic drug discovery guidelines, such as Lipinski’s Rule of Five and Veber’s rules, together with public algorithms, might be helpful to promote the outcomes of reconstruction. Hence, as research tools and drug discovery of novel scaffolds, the appropriate deconstruction of known ligands into several fragments followed by a rational reconstruction approach are likely to facilitate significantly the identification of chemical probes. It is our opinion that the future of this useful approach looks bright with expanded applications into different drug discovery platforms by pharmaceutical industry and academic laboratories in the coming years.
Highlights.
The higher investment on HTS did not result in enhanced productivity of new drugs.
FBDD emerged as an attractive strategy in drug discovery over the past two decades.
Construction of fragment library is the critical step for FBDD.
The pros and cons of various fragment-based screening are discussed.
Deconstruction-reconstruction approach offers promise in current drug discovery.
Acknowledgments
This work was supported by the Technology Development Foundation of Fuzhou University (Project Numbers 2011-XY-7, 2013-XQ-8 and 2013-XQ-9), Fujian Natural Science Foundation of China (No. 2012J05155), grants from The State Oceanic Administration of China (No. 201205022), grants P50 CA097007, P30 DA028821, R21 MH093844 from the National Institutes of Health, R.A. Welch Foundation Chemistry and Biology Collaborative Grant from the Gulf Coast Consortia (GCC), John Sealy Memorial Endowment Fund, Institute for Translational Sciences (ITS), and the Center for Addiction Research (CAR) at UTMB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat Rev Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 2.Fattori D, et al. Fragment-based approach to drug lead discovery: overview and advances in various techniques. Drugs R D. 2008;9:217–227. doi: 10.2165/00126839-200809040-00002. [DOI] [PubMed] [Google Scholar]
- 3.Erlanson DA. Introduction to fragment-based drug discovery. Top Curr Chem. 2012;317:1–32. doi: 10.1007/128_2011_180. [DOI] [PubMed] [Google Scholar]
- 4.Hajduk PJ. Puzzling through fragment-based drug design. Nat Chem Biol. 2006;2:658–659. doi: 10.1038/nchembio1206-658. [DOI] [PubMed] [Google Scholar]
- 5.Hajduk PJ. Fragment-based drug design: how big is too big? J Med Chem. 2006;49:6972–6976. doi: 10.1021/jm060511h. [DOI] [PubMed] [Google Scholar]
- 6.Jencks WP. On the attribution and additivity of binding energies. Proc Natl Acad Sci U S A. 1981;78:4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shuker SB, et al. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 8.Erlanson DA, et al. Tethering: fragment-based drug discovery. Annu Rev Biophys Biomol Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 9.Congreve M, et al. A ‘rule of three’ for fragment-based lead discovery? Drug Discov Today. 2003;8:876–877. doi: 10.1016/s1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 10.Jhoti H, et al. The ‘rule of three’ for fragment-based drug discovery: where are we now? Nat Rev Drug Discov. 2013;12:644–645. doi: 10.1038/nrd3926-c1. [DOI] [PubMed] [Google Scholar]
- 11.Neumann T, et al. SPR-based fragment screening: advantages and applications. Curr Top Med Chem. 2007;7:1630–1642. doi: 10.2174/156802607782341073. [DOI] [PubMed] [Google Scholar]
- 12.Leach AR, et al. Fragment screening: an introduction. Mol Biosyst. 2006;2:430–446. doi: 10.1039/b610069b. [DOI] [PubMed] [Google Scholar]
- 13.Schuffenhauer A, et al. Library design for fragment based screening. Curr Top Med Chem. 2005;5:751–762. doi: 10.2174/1568026054637700. [DOI] [PubMed] [Google Scholar]
- 14.Davis BJ, Erlanson DA. Learning from our mistakes: the ‘unknown knowns’ in fragment screening. Bioorg Med Chem Lett. 2013;23:2844–2852. doi: 10.1016/j.bmcl.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 15.Glennon RA. Pharmacophore identification for sigma-1 (sigma1) receptor binding: application of the ‘deconstruction-reconstruction-elaboration’ approach. Mini Rev Med Chem. 2005;5:927–940. doi: 10.2174/138955705774329519. [DOI] [PubMed] [Google Scholar]
- 16.Congreve M, et al. Recent developments in fragment-based drug discovery. J Med Chem. 2008;51:3661–3680. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- 17.Vieth M, et al. Characteristic physical properties and structural fragments of marketed oral drugs. J Med Chem. 2004;47:224–232. doi: 10.1021/jm030267j. [DOI] [PubMed] [Google Scholar]
- 18.Ciulli A, et al. Probing hot spots at protein-ligand binding sites: a fragment-based approach using biophysical methods. J Med Chem. 2006;49:4992–5000. doi: 10.1021/jm060490r. [DOI] [PubMed] [Google Scholar]
- 19.Babaoglu K, Shoichet BK. Deconstructing fragment-based inhibitor discovery. Nat Chem Biol. 2006;2:720–723. doi: 10.1038/nchembio831. [DOI] [PubMed] [Google Scholar]
- 20.Barelier S, et al. Fragment-based deconstruction of Bcl-xL inhibitors. J Med Chem. 2010;53:2577–2588. doi: 10.1021/jm100009z. [DOI] [PubMed] [Google Scholar]
- 21.Krimm I, et al. Binding evaluation of fragment-based scaffolds for probing allosteric enzymes. J Med Chem. 2012;55:1287–1295. doi: 10.1021/jm201439b. [DOI] [PubMed] [Google Scholar]
- 22.Andersen OA, et al. Structure-based dissection of the natural product cyclopentapeptide chitinase inhibitor argifin. Chem Biol. 2008;15:295–301. doi: 10.1016/j.chembiol.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipinski CA, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 24.Veber DF, et al. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, et al. Fragment-based drug design and identification of HJC0123, a novel orally bioavailable STAT3 inhibitor for cancer therapy. Eur J Med Chem. 2013;62:498–507. doi: 10.1016/j.ejmech.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, et al. Discovery of potent anticancer agent HJC0416, an orally bioavailable small molecule inhibitor of signal transducer and activator of transcription 3 (STAT3) Eur J Med Chem. 2014;82:195–203. doi: 10.1016/j.ejmech.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu W, et al. Discovery of novel STAT3 small molecule inhibitors via in silico site-directed fragment-based drug design. J Med Chem. 2013;56:4402–4412. doi: 10.1021/jm400080c. [DOI] [PubMed] [Google Scholar]
- 28.Stahl SM, et al. Serotonergic drugs for depression and beyond. Curr Drug Targets. 2013;14:578–585. doi: 10.2174/1389450111314050007. [DOI] [PubMed] [Google Scholar]
- 29.Capelli AM, Micheli F. Triple monoamine uptake inhibitors. Pharm Pat Anal. 2012;1:469–481. doi: 10.4155/ppa.12.46. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J, et al. Piperidine-based nocaine/modafinil hybrid ligands as highly potent monoamine transporter inhibitors: efficient drug discovery by rational lead hybridization. J Med Chem. 2004;47:5821–5824. doi: 10.1021/jm040117o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caldarone BJ, et al. The novel triple reuptake inhibitor JZAD-IV-22 exhibits an antidepressant pharmacological profile without locomotor stimulant or sensitization properties. J Pharmacol Exp Ther. 2010;335:762–770. doi: 10.1124/jpet.110.174011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao W, et al. Design, synthesis, and pharmacological evaluation of benzamide derivatives as glucokinase activators. Bioorg Med Chem. 2012;20:2982–2991. doi: 10.1016/j.bmc.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Brune S, et al. Structure of the sigma1 receptor and its ligand binding site. J Med Chem. 2013;56:9809–9819. doi: 10.1021/jm400660u. [DOI] [PubMed] [Google Scholar]
- 34.Schlager T, et al. Combination of two pharmacophoric systems: synthesis and pharmacological evaluation of spirocyclic pyranopyrazoles with high sigma(1) receptor affinity. J Med Chem. 2011;54:6704–6713. doi: 10.1021/jm200585k. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, et al. A combined bioinformatics and chemoinformatics approach for developing asymmetric bivalent AMPA receptor positive allosteric modulators as neuroprotective agents. Chem Med Chem. 2013;8:226–230. doi: 10.1002/cmdc.201200554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray CW, et al. Experiences in fragment-based drug discovery. Trends Pharmacol Sci. 2012;33:224–232. doi: 10.1016/j.tips.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Baker M. Fragment-based lead discovery grows up. Nat Rev Drug Discov. 2013;12:5–7. doi: 10.1038/nrd3926. [DOI] [PubMed] [Google Scholar]
- 38.Bollag G, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012;11:873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 39.Keseru GM, Makara GM. Hit discovery and hit-to-lead approaches. Drug Discov Today. 2006;11:741–748. doi: 10.1016/j.drudis.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 40.Lepre CA, et al. Theory and applications of NMR-based screening in pharmaceutical research. Chem Rev. 2004;104:3641–3676. doi: 10.1021/cr030409h. [DOI] [PubMed] [Google Scholar]
- 41.Murray CW, Blundell TL. Structural biology in fragment-based drug design. Curr Opin Struct Biol. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 42.Cala O, et al. NMR-based analysis of protein-ligand interactions. Anal Bioanal Chem. 2014;406:943–956. doi: 10.1007/s00216-013-6931-0. [DOI] [PubMed] [Google Scholar]
- 43.Hartshorn MJ, et al. Fragment-based lead discovery using X-ray crystallography. J Med Chem. 2005;48:403–413. doi: 10.1021/jm0495778. [DOI] [PubMed] [Google Scholar]
- 44.Caliandro R, et al. Protein crystallography and fragment-based drug design. Future Med Chem. 2013;5:1121–1140. doi: 10.4155/fmc.13.84. [DOI] [PubMed] [Google Scholar]
- 45.Bartoli S, et al. The fragment-approach: An update. Drug Discovery Today: Technologies. 2006;3:425–431. [Google Scholar]
- 46.Navratilova I, Hopkins AL. Fragment Screening by Surface Plasmon Resonance. ACS Med Chem Lett. 2010;1:44–48. doi: 10.1021/ml900002k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navratilova I, Hopkins AL. Emerging role of surface plasmon resonance in fragment-based drug discovery. Future Med Chem. 2011;3:1809–1820. doi: 10.4155/fmc.11.128. [DOI] [PubMed] [Google Scholar]
- 48.Erlanson DA, et al. Fragment-based drug discovery. J Med Chem. 2004;47:3463–3482. doi: 10.1021/jm040031v. [DOI] [PubMed] [Google Scholar]
- 49.Hofstadler SA, Sannes-Lowery KA. Applications of ESI-MS in drug discovery: interrogation of noncovalent complexes. Nat Rev Drug Discov. 2006;5:585–595. doi: 10.1038/nrd2083. [DOI] [PubMed] [Google Scholar]
- 50.Deng G, Sanyal G. Applications of mass spectrometry in early stages of target based drug discovery. J Pharm Biomed Anal. 2006;40:528–538. doi: 10.1016/j.jpba.2005.08.038. [DOI] [PubMed] [Google Scholar]
- 51.Pantoliano MW, et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 52.Lo MC, et al. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 53.Kranz JK, Schalk-Hihi C. Protein thermal shifts to identify low molecular weight fragments. Methods Enzymol. 2011;493:277–298. doi: 10.1016/B978-0-12-381274-2.00011-X. [DOI] [PubMed] [Google Scholar]
- 54.Leavitt S, Freire E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr Opin Struct Biol. 2001;11:560–566. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 55.Gozalbes R, et al. Contributions of computational chemistry and biophysical techniques to fragment-based drug discovery. Curr Med Chem. 2010;17:1769–1794. doi: 10.2174/092986710791111224. [DOI] [PubMed] [Google Scholar]
- 56.Ladbury JE, et al. Adding calorimetric data to decision making in lead discovery: a hot tip. Nat Rev Drug Discov. 2010;9:23–27. doi: 10.1038/nrd3054. [DOI] [PubMed] [Google Scholar]
- 57.Valkov E, et al. Targeting protein-protein interactions and fragment-based drug discovery. Top Curr Chem. 2012;317:145–179. doi: 10.1007/128_2011_265. [DOI] [PubMed] [Google Scholar]
- 58.Barker J, et al. Fragment screening by biochemical assay. Expert Opin Drug Discov. 2006;1:225–236. doi: 10.1517/17460441.1.3.225. [DOI] [PubMed] [Google Scholar]
- 59.Boyd SM, et al. Fragment library design considerations. Wiley Interdiscip Rev Comput Mol Sci. 2012;2:868–885. [Google Scholar]
- 60.Sancineto L, et al. From small to powerful: the fragments universe and its ‘chem-appeal’. Curr Med Chem. 2013;20:1355–1381. doi: 10.2174/09298673113209990111. [DOI] [PubMed] [Google Scholar]
- 61.Rabal O, et al. Computational medicinal chemistry in fragment-based drug discovery: what, how and when. Future Med Chem. 2011;3:95–134. doi: 10.4155/fmc.10.277. [DOI] [PubMed] [Google Scholar]
- 62.Kumar A, et al. Fragment based drug design: from experimental to computational approaches. Curr Med Chem. 2012;19:5128–5147. doi: 10.2174/092986712803530467. [DOI] [PubMed] [Google Scholar]