Abstract
To increase our understanding of skin, it is important to define the molecular constituents of the cell types and epidermal layers that signify normal skin. We have combined a genome-wide transcriptomics analysis, using deep sequencing of mRNA from skin biopsies, with immunohistochemistry-based protein profiling to characterize the landscape of gene and protein expression in normal human skin. The transcriptomics and protein expression data of skin were compared to 26 (RNA) and 44 (protein) other normal tissue types. All 20,050 putative protein-coding genes were classified into categories based on patterns of expression. We found that 417 genes showed elevated expression in skin, with 106 genes expressed at least five-fold higher than that in other tissues. The 106 genes categorized as skin enriched encoded for well-known proteins involved in epidermal differentiation and proteins with unknown functions and expression patterns in skin, including the C1orf68 protein, which showed the highest relative enrichment in skin. In conclusion, we have applied a genome-wide analysis to identify the human skin-specific proteome and map the precise localization of the corresponding proteins in different compartments of the skin, to facilitate further functional studies to explore the molecular repertoire of normal skin and to identify biomarkers related to various skin diseases.
Keywords: Proteomics, Transcriptomics, Gene expression, RNA-Seq, Antibody, Human Protein Atlas, Immunohistochemistry
Introduction
Human skin represents a physical barrier that separates the interior from the exterior and protects the body against water loss and external physical, chemical and biological insults (Cork et al. 2009; Proksch et al. 2008). The skin barrier is formed by the constantly renewing epidermis, which predominantly consists of keratinocytes. Epidermal homeostasis depends on keratinocyte stem cells that retain the capacity to divide asymmetrically and detach from the basement membrane for subsequent terminal differentiation (Ghadially 2012). During epidermal differentiation, keratinocytes undergo several discrete transcriptional stages as they form the morphologically defined spinous and granular layers before the outermost stratum corneum, the final differentiation stage represented by a layer of flattened corneocytes with compact keratin fibers and an absence of sub-cellular organelles.
The highly specialized functions of the skin and the dynamics of epidermal differentiation are based on patterns of gene expression in the various cell types that constitute the epidermal layers. Numerous studies on normal and diseased skin in both humans and animal models have provided detailed knowledge about the molecular repertoire of the different skin cell types and enhanced our understanding of the complex cellular interactions under normal and pathological conditions (Ghadially 2012; Li et al. 2014; Scharadin and Eckert 2014; Vanbokhoven et al. 2011). However, the expression pattern in skin is still unknown for a large fraction of the human proteome and a readily available compilation of skin-specific gene and protein expression is lacking.
Following the human genome project (Lander et al. 2001; Venter et al. 2001), a multitude of studies, mainly based on microarray technology, have been conducted to map gene expression in various human cell and tissue types, including skin. A majority of these data sets are published in online repositories, such as the Gene Expression Omnibus (Barrett et al. 2013) and ArrayExpress (Rustici et al. 2013), and have been utilized to determine the global map of human gene expression (Lukk et al. 2010). Several transcriptomics studies of normal skin have been performed, including an analysis of developing epidermis (Bazzi et al. 2007) and of individual epidermal layers (Toulza et al. 2007). Global gene expression studies have also compared diseased skin with normal controls, including several microarray-based studies on psoriasis (Gudjonsson et al. 2010; Suarez-Farinas et al. 2010). As an alternative to array-based analyses, next-generation sequencing technologies (RNA-Seq) have empowered a more sensitive and precise analytical approach for the determination of RNA expression levels. Direct comparisons of microarray technology to RNA-seq have concluded that the latter substantially increases the number of identified differentially expressed genes, for instance, in psoriasis as compared to normal skin (Jabbari et al. 2012; Li et al. 2014).
RNA-Seq studies that compared the transcriptomic landscapes between various tissues and organs have identified genes ubiquitously expressed across several different tissue types, as well as genes with some level of tissue-specific expression distribution (Krupp et al. 2012; Ramskold et al. 2009); however, skin was, until now, not incorporated or specifically explored in such large-scale, cross-tissue comparisons using RNA-Seq. Here, we used the RNA-Seq transcriptomics data from an analysis of 27 different tissues and organs in the human body (Fagerberg et al. 2013) to identify and analyze genes with elevated expression in normal skin and combined this analysis with immunohistochemistry-based protein profiling using tissue microarrays, to map the expression of corresponding proteins to the specific cell types and different epidermal layers that constitute human skin.
Materials & Methods
Sample Characteristics
All ‘fresh’-frozen and paraffin-embedded tissue samples used in this study were acquired from the Uppsala Biobank and the Department of Clinical Pathology, Uppsala University Hospital, Uppsala, Sweden. Tissue samples were collected in agreement with approval from the Research Ethics Committee at Uppsala University (Ups 02-577, #2011/473). All human tissues, including three shave biopsies from the normal back skin of healthy volunteers (2 females, 1 male), were embedded in Optimal Cutting Temperature (OCT) compound and stored at -80C. Hematoxylin-eosin (HE) staining of 4-µm sections were examined by a pathologist (FP) to ascertain that samples consisted of skin with normal morphology (Supplementary Fig. 1). The relative percentages of cell types present in the samples (i.e., keratinocytes, melanocytes and cells outside the epidermis) were determined by manually counting nuclei from four representative regions per sample (expressed as the percentage mean ± SD; Supplementary Fig. 1).
Transcript Profiling (RNA-Seq) and Data Analysis
Transcript profiling was performed essentially as previously described (Fagerberg et al. 2013). In brief, total RNA extraction from 95 samples representing 27 different homogenized tissues (Supplementary Table 1) was performed using the RNeasy Mini Kit (Qiagen; Hilden, Germany) and analyzed using the Experion automated electrophoresis system (Bio-Rad Laboratories; Hercules, CA) or the Agilent 2100 Bioanalyzer system (Agilent Technologies; Santa Clara, CA). High-quality RNA samples were sequenced using the Illumina HiSeq2000/2500 (Illumina; San Diego, CA). Raw reads were processed using the Sickle software and mapped to the GRCh37 version of the human genome with Tophat v2.0.3 (Trapnell et al. 2010). To obtain quantification scores for all human genes, fragments per kilobase of exon model per million mapped reads (FPKM) values were calculated using gene models from Ensembl build 69 with Cufflinks v2.0.2 (Trapnell et al. 2010). All data was analyzed in R Statistical Environment (RCoreTeam 2013) and network analysis was performed using Cytoscape 3.0 (Shannon et al. 2003). When a log2-scale of the data was used, pseudo-counts of +1 were added to the data set.
Mean FPKM values of all samples for each tissue was used to estimate gene expression levels in the total set of 20,050 genes. A cut-off value of 1 FPKM (roughly corresponding to 1 mRNA per average cell in the sample) was set as the detection limit (Hebenstreit et al. 2011). Based on relative FPKM levels, genes were classified into six categories depending on expression patterns: (1) skin enriched, (2) group enriched, (3) skin enhanced, (4) expressed in all, (5) mixed and (6) not detected (Table 1). Genes categorized as skin enriched, skin enhanced and group enriched in skin, were together defined as having an elevated expression in skin. A “skin-specific score” was calculated for each gene dividing the skin FPKM by the maximum FPKM value in any of the other 26 tissue types.
Table 1.
Classification of All Human Putative Protein-coding Genes into Categories based on Expression Patterns in Skin and Other Human Tissue Types.
| Category | Description | Number of Genes | Fraction (%) |
|---|---|---|---|
| Skin enriched | At least five-fold higher mRNA levels in skin as compared to all other tissues | 106 | 0.5 |
| Group enriched | At least five-fold higher mRNA levels in a group of 2–7 tissues, including skin | 149 | 0.7 |
| Skin enhanced | At least five-fold higher mRNA levels in skin as compared to average levels in all tissues | 162 | 0.8 |
| Expressed in all | Detected in all tissues (FPKM > 1) | 9,222 | 46 |
| Mixed | Detected in 2–27 tissues, but not elevated in skin | 3,558 | 18 |
| Not detected in skin | FPKM less than 1 in skin | 5,131 | 26 |
| Not detected in any tissue | FPKM less than 1 in all tissues | 1,722 | 9 |
| Total | Total number of genes analyzed with RNA-seq | 20,050 | 100 |
| Total elevated in skin | Total number of skin-enriched, group-enriched, and skin-enhanced genes expressed in skin | 417 | 2 |
FPKM, fragments per kilobase of exon model per million mapped reads.
Gene Ontology Analysis
Gene ontology (GO) analysis (Ashburner et al. 2000) was performed using the GOrilla tool (Eden et al. 2009) to determine overrepresented GO categories in the set of genes considered as skin enriched. For cellular component analysis, the GOSlim GOA associations were used to determine whether genes encoded extracellular, intracellular or membrane-bound proteins. The number of genes for each term was counted, allowing a gene to be associated with more than one term. A list of all genes analyzed in this study was used as the background list in GOrilla.
Antibody-based Profiling
Generation of tissue microarrays (TMA), immunohistochemistry and slide scanning was performed in accordance with standards set up within the Human Protein Atlas project, as previously described (Kampf et al. 2012). In brief, TMA sections, containing 1-mm cores of 45 different normal tissue types (Supplementary Table 1), were subjected to automated immunohistochemistry. Immunohistochemically stained and mounted slides were scanned using an Aperio ScanScope XT Slide Scanner (Aperio Technologies; Vista, CA) to generate high-resolution digital images. Web-based annotation of the intensity and fraction of positive cells defined in each tissue type was performed by certified pathologists (Uhlen et al. 2010). Extensive information on the antibodies used in this study (including manufacturer and antibody validation data) is available on the Human Protein Atlas website (www.proteinatlas.org); in this work, antibodies are referred to with the antibody ID that is used in the Human Protein Atlas.
All skin-specific genes with corresponding antibody-based expression profiles in the Human Protein Atlas database were manually and individually analyzed to determine the degree of consistency between RNA-Seq and the protein profile data across all tissues, as well as taking into account the available technical validation of the antibodies. Antibodies with reliable validation and high RNA-consistency were considered for presentation in this work. When two or more antibodies were available per gene, the antibody regarded as the most reliable based on above criteria was selected for presentation.
Data Availability
All FPKM data is available without restrictions (www.proteinatlas.org/about/download). The primary sequencing reads are available through the Array Express Archive (www.ebi.ac.uk/arrayexpress/); accession number E-MTAB-1733. Transcript profiling data for each gene in each cell and tissue type is also available on the Human Protein Atlas website, together with protein profiling data and underlying images of immunohistochemically stained tissues.
Results
Analysis of the Skin Transcriptome
RNA-Seq was performed on 95 fresh-frozen tissue samples from 27 different tissue types, as described earlier (Fagerberg et al. 2013). Skin samples from three healthy individuals were included and the transcriptome of each sample was quantified to determine normalized mRNA levels, calculated as FPKM. Tissue sections from two specimens were stained with HE and used to estimate the relative numbers of different cell types (Supplementary Fig. 1). Keratinocytes constituted approximately 55% to 60% of all cells; melanocytes, approximately 2% to 3%; and other cells located in the dermis (e.g., fibroblasts, smooth muscle and endothelial cells) amounted to approximately 35% to 40%.
Approximately 66% of all protein coding genes were expressed in skin above the cut-off of >1 FPKM, which represented approximately 1 mRNA molecule on average per cell (Hebenstreit et al. 2011). The number of expressed genes was similar among the three individuals: 12,801, 13,044 and 13,329 genes, respectively. To assess the inter-sample variance, pairwise Spearman correlation analyses of FPKM expression levels was performed. All samples showed overall high correlation coefficients of 0.94–0.98 (Fig. 1A–1C). These data demonstrate a high technical reproducibility and overall similarity in gene expression between the individual samples. The mRNA expression level in skin ranged from 9,619 FPKM (mitochondrial-expressed ATP synthase 8, MT-ATP8) yielding a dynamic range of approximately 104 using the >1 FPKM cut-off.
Figure 1.
Global analysis of genes expressed in skin. (A–C) Correlation plots of the three skin samples used for RNA-Seq. High correlations were observed among the three samples, with the best correlation found between the two female samples. (D) Distribution of the number of genes across each of the categories based on transcript abundance. (E) Distribution of the fraction of transcripts across different categories for all genes detected in skin. Color codes in (D) apply to all subfigures (A–E).
Classification of Genes Expressed in Skin
Based on the transcriptomic data, all putative protein-coding genes were classified into six different categories (Table 1; Fig. 1D). Approximately one third of the genes (34%) were not detected in skin (overall, 9% of all genes were not detected in any analyzed tissue type). The largest group of expressed genes (46%) represented genes expressed in all 27 different tissue types. Only 2% of all genes showed an elevated expression pattern in skin. The relative proportion of mRNA molecules representing the different categories showed that over 80% of all transcripts expressed in skin encode for proteins expressed in all tissues (i.e., “housekeeping proteins”), whereas only 13% of transcripts represent genes with elevated expression in skin (Fig. 1E).
The 30 genes with highest expression levels in the skin (Supplementary Table 2) mainly included genes with “housekeeping” functions; e.g., encoding mitochondrial and ribosomal proteins. A subset of the identified, highly expressed genes encoded proteins with enriched expression in the skin or were group enriched as tissues containing squamous epithelia, including keratins (KRT1, KRT10, KRT14 and KRT5) as well as other skin-related proteins; e.g., dermokine (DMKN), keratinocyte differentiation-associated protein (KRTDAP), galectin-7 (LGALS7) and suprabasin (SBSN) (Bazzi et al. 2007; Gendronneau et al. 2008; Madsen et al. 1995; Toulza et al. 2007).
There were 417 genes with elevated expression in skin. These genes were subclassified as skin enriched (n=106), with at least 5-fold higher FPKM level in skin compared to all other tissues; group enriched (n=149; these genes were expressed in only a limited number of tissues, including skin); and skin enhanced (n=162), with an expression in the skin at least 5-fold higher than the average expression in all 27 tissues. The 268 genes in the skin enriched and skin enhanced groups included several expected and previously well-known genes implicated in skin: keratins (n=9), late cornified envelope proteins (n=14), cadherins (n=5), collagens (n=3), IGF-family like members (n=3), kalkrein-related peptidases (n=4), and the WNT-family of proteins (n=5). Interestingly, 12 out of these 268 genes encoded for “uncharacterized proteins”, as defined by Ensembl’s gene annotation (Supplementary Table 3, bold-faced numbers in the first column indicate these genes). A GO-based enrichment analysis of the genes categorized as skin enriched showed that the top gene categories were related to keratinization, epidermal cell development and differentiation, melanin synthesis, and water homeostasis.
Several of the genes with highest levels of enriched expression in the skin encoded for proteins involved in skin barrier function, including squamous differentiation and cornification (for example, proteins from the late cornified envelope protein family (n=12), keratins (n=4), loricin (LOR), corneodesmosin (CDSN), filaggrin family member 2 (FLG2), caspase-14 (CASP14), SERPINB7) and local, immunological defense (such as chemokine ligand CCL27 and c-type lectin CLEC2A). Additional highly expressed genes that were skin enriched were related to melanin production, including melan-A (MLANA), tyrosinase (TYR) and dopachrome tautomerase (DCT). Also in the list of genes enriched in skin were less-well characterized genes, including SERPINA12, which was earlier shown to be linked to the inhibition of the kalkrein-related peptidase, KLK7, in the development of psoriasis (Schultz et al. 2013), and interleukin IL37, which was shown to be associated to atopic dermatitis (Kimura et al. 2013). The most highly expressed gene in the skin enriched group was the poorly characterized C1orf68 gene, which encodes for the skin-specific protein 32 (synonyms: LEP7, xp32) located within the epidermal differentiation complex together with other skin-specific genes, including the late cornified envelope proteins (Toulza et al. 2007; Zhao and Elder 1997). The top 50 genes classified as skin enriched are shown in Table 2.
Table 2.
The Top 50 Skin-enriched Genes.
| # | Gene Name | Description | Cellular Location* | Skin-Specific Score** | Expression in Skin (FPKM)*** | Max FPKM in Other Tissues**** |
|---|---|---|---|---|---|---|
| 1 | C1orf68 | chromosome 1 open reading frame 68 | IC | 3858.3 | 385.8 | 0.1 |
| 2 | LCE2B | late cornified envelope 2B | IC | 3716.6 | 386.5 | 0.1 |
| 3 | FLG2 | filaggrin family member 2 | IC | 2455.4 | 358.5 | 0.1 |
| 4 | LCE2C | late cornified envelope 2C | IC | 1874.1 | 187.4 | 0.1 |
| 5 | CDSN | corneodesmosin | SP | 1430.9 | 253.3 | 0.2 |
| 6 | LCE1A | late cornified envelope 1A | IC | 1275.7 | 164.6 | 0.1 |
| 7 | LCE6A | late cornified envelope 6A | IC | 1116 | 111.6 | 0 |
| 8 | KRT2 | keratin 2 | IC | 1060.1 | 1192.6 | 1.1 |
| 9 | LCE1F | late cornified envelope 1F | IC | 996.4 | 106.6 | 0.1 |
| 10 | LCE1B | late cornified envelope 1B | IC | 980.2 | 98 | 0 |
| 11 | LOR | loricrin | IC | 967 | 177.9 | 0.2 |
| 12 | LCE1C | late cornified envelope 1C | IC | 859.6 | 234.7 | 0.3 |
| 13 | CASP14 | caspase 14, apoptosis-related cysteine peptidase | IC | 758.3 | 260.1 | 0.3 |
| 14 | KRT77 | keratin 77 | IC | 636.6 | 222.2 | 0.3 |
| 15 | SERPINA12 | serpin peptidase inhibitor, clade A, member 12 | SP | 622.7 | 203.6 | 0.3 |
| 16 | LCE2D | late cornified envelope 2D | IC | 619.5 | 90.4 | 0.1 |
| 17 | LCE2A | late cornified envelope 2A | IC | 509.7 | 89.2 | 0.2 |
| 18 | KRT10 | keratin 10 | IC | 504.2 | 4147.2 | 8.2 |
| 19 | LCE1D | late cornified envelope 1D | IC | 474.3 | 55.5 | 0.1 |
| 20 | CCL27 | chemokine (C-C motif) ligand 27 | SP | 369.6 | 298.3 | 0.8 |
| 21 | LCE1E | late cornified envelope 1E | IC | 323 | 36.5 | 0.1 |
| 22 | IL37 | interleukin 37 | IC | 204.9 | 72.5 | 0.4 |
| 23 | LCE5A | late cornified envelope 5A | IC | 168.5 | 46 | 0.3 |
| 24 | KRT1 | keratin 1 | IC | 132.4 | 4698.1 | 35.5 |
| 25 | CLEC2A | C-type lectin domain family 2, member A | TM | 116.2 | 27 | 0.2 |
| 26 | TYR | tyrosinase (oculocutaneous albinism IA) | SP/TM | 115.7 | 28 | 0.2 |
| 27 | DCT | dopachrome tautomerase | SP/TM | 79.8 | 83.7 | 1.1 |
| 28 | MLANA | melan-A | TM | 70.5 | 35.4 | 0.5 |
| 29 | THEM5 | thioesterase superfamily member 5 | IC | 67.1 | 50.7 | 0.8 |
| 30 | PSORS1C2 | psoriasis susceptibility 1 candidate 2 | SP | 64.8 | 44.4 | 0.7 |
| 31 | ASPRV1 | aspartic peptidase, retroviral-like 1 | TM | 58.7 | 186 | 3.2 |
| 32 | WFDC5 | WAP four-disulfide core domain 5 | SP | 53.1 | 251.5 | 4.7 |
| 33 | SERPINB7 | serpin peptidase inhibitor, clade B, member 7 | IC | 50.8 | 112.7 | 2.2 |
| 34 | KRTDAP | keratinocyte differentiation-associated protein | SP | 50.3 | 2176.2 | 43.3 |
| 35 | SPRR2G | small proline-rich protein 2G | IC | 44.9 | 185.4 | 4.1 |
| 36 | FLG | filaggrin | IC | 43.8 | 320.8 | 7.3 |
| 37 | LCE4A | late cornified envelope 4A | IC | 42.9 | 4.3 | 0 |
| 38 | IGFL3 | IGF-like family member 3 | SP | 42.7 | 6.7 | 0.2 |
| 39 | BPIFC | BPI fold containing family C | SP | 37.1 | 48.2 | 1.3 |
| 40 | C6orf15 | chromosome 6 open reading frame 15 | SP | 32.5 | 3.3 | 0.1 |
| 41 | IL1F10 | interleukin 1 family, member 10 (theta) | IC | 30.6 | 3.1 | 0.1 |
| 42 | LIPK | lipase, family member K | SP | 28.8 | 10 | 0.3 |
| 43 | AADACL2 | arylacetamide deacetylase-like 2 | SP | 27.6 | 45.9 | 1.7 |
| 44 | POU2F3 | POU class 2 homeobox 3 | IC | 27.2 | 70.5 | 2.6 |
| 45 | ALOXE3 | arachidonate lipoxygenase 3 | SP | 26.8 | 40.4 | 1.5 |
| 46 | NLRP10 | NLR family, pyrin domain containing 10 | IC | 23.3 | 6.5 | 0.3 |
| 47 | RPTN | repetin | IC | 22.4 | 24 | 1.1 |
| 48 | NKPD1 | NTPase, KAP family P-loop domain containing 1 | TM | 20.6 | 8.2 | 0.4 |
| 49 | KPRP | keratinocyte proline-rich protein | IC | 19.7 | 160.6 | 8.1 |
| 50 | KRT14 | keratin 14 | IC | 19.6 | 4021.3 | 205.7 |
#, Sample number; FPKM, fragments per kilobase of exon model per million mapped reads. *IC=intracellular, SP=predicted signal peptide/secreted protein, TM=predicted transmembrane region/membrane bound protein. **The skin-specific score is the ratio of the mean FPKM value in skin and the maximal mean FPKM value for any other tissue type analyzed in this study. *** Mean expression level (FPKM) in skin, based on three samples. **** Highest expression level (FPKM) in non-skin tissues.
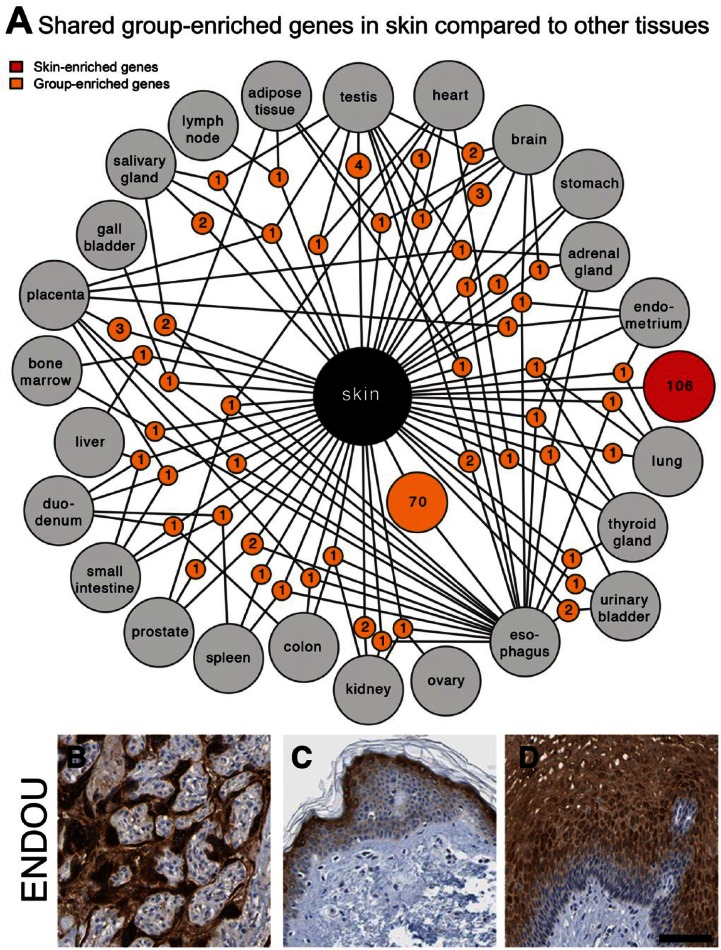
Group Enriched Genes are Mainly Shared with Esophagus
The shared expression of group enriched genes (n=149) is distributed among the other tissue types, with a clear overrepresentation of genes shared with esophagus (Fig. 2A, Supplementary Table 4). Three of the 30 genes with highest expression levels in skin (p53 apoptosis effector related to PMP-22 (PERP), keratin 5 (KRT5) and suprabasin (SBSN)) were also expressed at high levels in esophagus. Not surprisingly, pairwise correlation analyses among all 27 tissues revealed that the esophagus had the highest correlation to skin (0.90; data not shown). In contrast, the testis with most diverse gene expression and highest number of unique tissue-enriched genes (testis-enriched) (Djureinovic et al. 2014), had the lowest correlation to skin (0.69; data not shown). Several genes, expressed in both skin and esophagus, are well characterized or have been described as proteins important for the normal differentiation and function of squamous epithelia. Examples include the keratins KRT5, KRT8, KRT15, KRT17 and KRT31, and genes related to cell adhesion and squamous differentiation, such as desmoplakin (DSP), envoplakin (EVPL), stratifin (SFN), involucrin (IVL) and filaggrin (FLG). A subset of the group enriched genes are less-well characterized, such as the ENDOU gene, which encodes for the poly (U)-specific endoribonuclease protein (placenta protein 11), which has not been previously reported to be expressed in squamous epithelium. This protein has been described as a placenta-specific protein and also shown to be expressed in certain tumors (Inaba et al. 1980). As expected, the protein showed a distinct expression pattern in the placenta (3 FPKM) but was also expressed in differentiated layers of squamous epithelium in both skin (21 FPKM) and esophagus (66 FPKM) (Fig. 2B–2D).
Figure 2.
Group enriched genes in skin. (A) Network plot of skin enriched (red circle) and group enriched (yellow circles) genes showing the relationship between the analyzed tissues. Yellow circle nodes represent group enriched genes in skin and are connected to the respective enriched tissues (grey circles). Only genes group enriched in combinations of up to four tissues are shown. Most tissues share 1-4 group enriched-genes with skin, but esophagus stands out by sharing 70 group enriched genes (Table S4). Many of these correspond to genes known to be important for squamous epithelial biology but a number of genes have less defined function in squamous epithelia as exemplified by the “placenta-specific” Endou protein expressed in placenta (B), skin (C) and esophagus (D). Scale, 100 µm.
A GO-based enrichment analysis was performed on genes shared between skin and esophagus. The top-shared categories were related to epidermal and keratinocyte differentiation, desmosome organization, peptide cross-linking and regulation of lymphocytes. The analysis also revealed that the major difference was that esophagus did not have genes associated with water homeostasis and melanin synthesis.
Proteins Expressed in the Basal Layers of the Epidermis
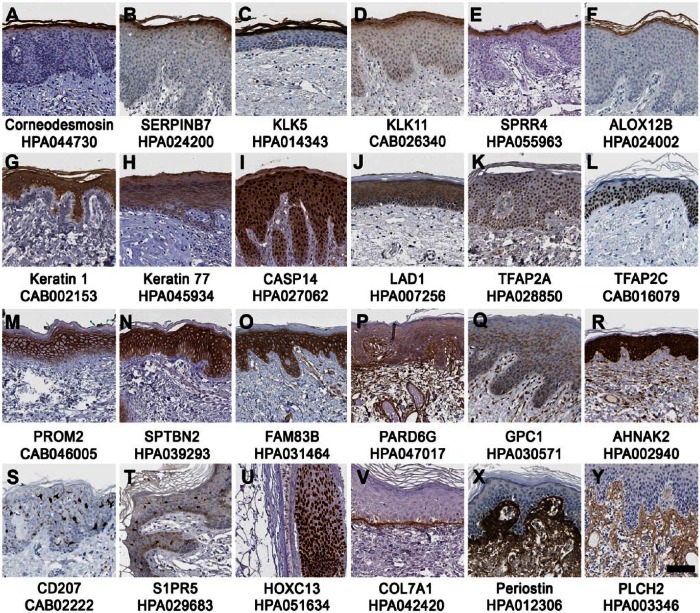
The basal layer of the skin is the site of proliferation and skin renewal, and several well-known skin enriched proteins showed restricted expression in the basal layer, such as keratin-15 (KRT15), collagen 17A1 (COL17A1) and tumor protein 73 (TP73). A novel finding was the poorly studied protein encoded by the FAM83G gene, which showed strong cytoplasmic expression in keratinocytes of the basal cell layer (Fig. 3D) and in sebaceous glands (data not shown). FAM83G has previously been genetically linked to the spontaneous “wooly mutation”, which causes a rough or matted fur phenotype in mice despite its histologically normal-appearing skin and hair (Radden et al. 2013). Enriched expression in the basal layers was also observed for tumor protein p63 (TP63) (Fig. 3E), a master regulator of stratified epithelial development and known to regulate, for example, desmocollin-3 (DSC3) (group enriched in skin and esophagus) and fibroblast growth factor receptor 3 (FGFR3) (Fig. 3F), which both showed enhanced expression in the basal layers of the epidermis (Ferone et al. 2013; Sayan et al. 2010). Another key regulator of epidermal proliferation, differentiation and lipid biosynthesis in the basal layer was the B-cell lymphoma/leukemia 11B transcription factor (BCL11B, Fig. 3G) (Wang et al. 2013).
Figure 3.
Protein expression patterns in different epidermal layers of genes categorized as skin enriched. Examples of immunohistochemistry-based protein expression patterns reflecting genes with elevated expression in skin, including proteins expressed in the basal layers of the epidermis (A–G), proteins expressed predominantly in the upper layers of the epidermis (H–N) and proteins expressed in the stratum granulosum (O–U). The gene/protein name and antibody ID used in the Human Protein Atlas database (www.proteinatlas.org) is shown below each image. Scale, 100 µm.
Proteins Expressed in the Stratum Spinosum
Post-mitotic keratinocytes differentiate and are pushed from the basal layers through the stratum spinosum towards the outer surface of the skin and gradually transform into layers of flattened, enucleated cells that are connected by tight junctions and secrete keratins and lipids to the extracellular matrix. Accordingly, a gradual enrichment in protein expression towards the outer layer of stratum spinosum was evident for genes involved in the differentiation and keratinization processes, including dermokine (DMKN), keratinocyte differentiation-associated protein (KRTDAP), gasdermin A (GSDMA), scillein (SCEL; also detected in hair follicles; data not shown), Galectin-7 (LGALS7) (Fig. 3H), the transcription factor HOP homeobox (HOPX) (Fig. 3I), the cytochrome P450 protein CYP4F22 (Fig. 3J), and keratin-10 (KRT10) (Fig. 3K) (Akiyama 2011; Bazzi et al. 2007; Champliaud et al. 2000; Gendronneau et al. 2008; Toulza et al. 2007; Yang et al. 2010). Similar expression patterns in upper epidermal layers were furthermore detected for several other proteins including grainyhead-like 1 (GRHL1) (Fig. 3L), a transcription factor previously not described in skin but shown to be downregulated after bile exposure in the esophagus (Reveiller et al. 2012); the prostaglandin G/H synthase 1 (PTGS1) (Fig. 3M), which is important for wound healing (Abdou et al. 2013); and the KLK9 (Fig. 3N). KRT10, KLK9 and PTGS1 were expressed in all epidermal layers except the basal layer.
Proteins Expressed in Stratum Granulosum
The stratum granulosum—important for the generation of the skin’s hydrophobic barrier and where organelles and nuclei gradually diminish as keratinocytes and undergo terminal differentiation into non-viable corneocytes—is a thin layer of keratinocytes between the underlying stratum spinosum and the overlying stratum corneum. Consistent with previous reports, restricted expression in stratum granulosum was found for filaggrin (FLG) and FLG2 (data not shown); Ly6/PLAUR domain-containing protein 5 (LYPD5) (Gardsvoll et al. 2013); keratinocyte proline-rich protein (KPRP; Fig. 3O) (Kong et al. 2003); premature ovarian failure 1B protein (POF1B; Fig. 3P), which is also expressed in melanocytes (Rizzolio et al. 2007); deoxyribonuclease I-like 2 (DNASE1L2) (Fig. 3Q); and three prime repair exonuclease 2 (TREX2) (Fig. 3R). DNASE1L2 and TREX2 have previously been shown to be involved in the process of ablating DNA in keratinocytes (Eckhart et al. 2012) and the urokinase receptor homolog encoded by the LYPD5 gene was recently described as a novel differentiation marker of stratum granulosum (Gardsvoll et al. 2013); the functions and expression patterns of KPRP and POF1B in skin are largely unknown. Three proteins previously uncharacterized in skin also showed an expression pattern restricted to stratum granulosum: orthodentacle homeobox 1 protein (OTX1) (Fig. 3S), with a putative role in brain development (Chen et al. 2013); epidermal growth factor receptor kinase substrate 8-like protein 1 (EPS8L1) (Fig. 3T), potentially implicated in actin-reorganization (Offenhauser et al. 2004); and caspase recruitment domain family member 18 (CARD18) (Fig. 3U), which has an unknown function.
Proteins Expressed in Stratum Corneum
The cornified layer, comprising 15–20 layers of tightly coupled, flattened and dead corneocytes, functions as a mechanical barrier to protect against UV radiation, dehydration and pathogens. Genes with enriched expression in the cornified layer of the skin included several well-known proteins, including corneodesmosin (CDSN) (Fig. 4A); SERPINB7 (Fig. 4B); KLK5 (Fig. 4C); KLK11 (Fig. 4D) (Caubet et al. 2004; Komatsu et al. 2003); small proline-rich protein 4 (SPRR4) (Fig. 4E), which is involved in UV induced keratinization (Cabral et al. 2001); and the arachidonate 12-lipoxygenase ALOX12B (Fig. 4F), which has been linked to ichthiyoses (Akiyama 2011). No novel gene with skin-enriched expression confined to the stratum corneum was identified.
Figure 4.
Protein expression patterns in skin of skin enriched genes. Immunohistochemistry-based protein profiling shows examples of proteins with distinct expression in stratum corneum (A–F) and proteins expressed throughout all epidermal layers, including previously well-known proteins (G–L) and less-well studied proteins with respect to skin biology (M–R). Shown are examples of skin enriched genes where the corresponding proteins are expressed in specific compartments of skin: (S–T) Langerhan cells, (U) hair follicles and (V–Y) dermis, including basement membrane, extracellular matrix and dermal fibroblasts. The gene name and corresponding antibody ID used in the Human Protein Atlas database (www.proteinatlas.org) is shown below each image. Scale, 100 µm.
Proteins Expressed throughout the Epidermis
Although several genes with elevated expression in skin showed a corresponding protein expression in restricted epidermal compartments, others were expressed throughout the full thickness of the epidermis. Examples of more widely expressed proteins included the keratins KRT1 and KRT77, caspase-14 (CASP14), ladinin-1 (LAD1), and the transcription factors AP-2 alpha and gamma (TFAP2A and TFAP2C; Fig. 4G–4L) (Bazzi et al. 2007; Motoki et al. 1997; Wang et al. 2008; Yamamoto et al. 2012). An additional six genes expressed throughout the epidermis and not earlier characterized in skin encoded for proteins with poorly known functions: prominin-2 (PROM2), (Fargeas et al. 2003), spectrin beta (SPTBN2) (Stankewich et al. 1998), FAM83B, par-6 family cell polarity regulator gamma (PARD6G), glypican-1 (GPC1), and a nucleoprotein encoded by the AHNAK gene (Fig. 4M–4R). The PARD6G, GPC1 and AHNAK proteins were also expressed in dermal fibroblasts and endothelial cells.
Proteins Expressed in Melanocytes, Langerhans Cells and Fibroblasts
The classical melanocyte markers, including Melan-A (MLANA), tyrosinase (TYR) and dopachrome tautomerase (DCT), as well as the Langerhans cell markers, CD1A (not shown) and CD207 (Fig. 4S), are examples of genes with skin enriched expression where corresponding proteins show expression in well-defined cell types that are relatively abundant in skin. CD1A and CD207 were classified as group enriched due to high expression in both skin and the esophagus (Meyer et al. 2010). The G-protein coupled sphingosine 1-phosphate receptor 5 (S1PR5; Fig. 4T), with evidence of existence only at the transcript level, showed a localization pattern suggestive of expression in Langerhans cells. A smaller subset of skin enriched genes were expressed in non-epidermal compartments, such as the transcription factor homeobox C13 (HOXC13), which is expressed in hair follicle cells (Fig. 4U) (Jave-Suarez et al. 2002); collagen 7A1 (COL7A1), which is expressed in the basement membrane (Fig. 4V) (Nystrom et al. 2013); and periostin (POSTN) and phospholipase C-eta-2 protein (PLCH2), which are mainly expressed in dermal fibroblasts (Fig. 4X–4Y) (Ontsuka et al. 2012).
Discussion
In this study, we provide a comprehensive portrait of gene expression in normal human skin with examples of expression patterns across epidermal layers and cell types for a subset of proteins with enriched expression in the skin. The analyses are based on the synergistic combination of quantitative transcriptomics data (RNA-Seq) using homogenates of skin biopsies and spatial protein data at the single-cell level through in situ protein profiling using immunohistochemistry-based tissue microarrays. Our approach has allowed us to explore in a genome-wide manner the cross-tissue expression levels of these markers, taking advantage of the analytical power of RNA-Seq combined with spatial organization of the epidermis using immunohistochemistry to confer layer-specific properties and in situ visualization of human skin.
We found 417 genes with elevated expression in the skin and, of these, 106 genes were defined as enriched (Table 1). As expected, most skin enriched proteins have functions related to terminal squamous differentiation and cornification; e.g., late cornified envelope proteins and filaggrin family members. In addition to previously well-characterized genes expressed in skin, we identified a number of genes with unknown functions and expression patterns in skin. Interestingly, the gene with the highest degree of skin specificity was C1orf68, which encodes for skin-specific protein 32, for which the precise function is unknown; although, skin barrier function and enriched expression in the granular cell layer has previously been suggested [10, 30].
The detection of novel skin-related genes with enriched expression patterns of corresponding proteins in different epidermal layers and cell types can provide contextual clues to its function. It is well recognized that specific gene mutations and altered proteins are linked to specific forms of skin diseases, such as ichthyosis, reflecting abnormal squamous differentiation and acantholytic disorders, including pemphigus with epidermal blistering at different levels. Several genes with elevated expression in skin and specific expression patterns in different epidermal layers have earlier been linked to specific forms of skin disease; e.g., (i) KRT15 (alopecia) (Al-Refu 2012) and COL17A1 (bullous pemphigoid and epidermolysis bullosa) (Powell et al. 2005) expressed in the basal layer; (ii) FLG (ichthyosis vulgaris (Thyssen et al. 2013), which is expressed in the granular layer; (iii) KLK5 (rosacea) (Meyer-Hoffert and Schroder 2011), which is expressed in stratum corneum; and (iv) HOXC13 (ectodermal dysplasia) (Lin et al. 2012), which is expressed in hair follicles. Although specific expression patterns do not per se provide functional data, clues to function and possible involvement in disease are given. The presented lists of genes with elevated expression in skin and data regarding the specific expression patterns of corresponding proteins appear to offer an advantageous starting point for further in-depth studies to link genes with elevated expression in skin to various forms of disease.
In conclusion, we provide a comprehensive analysis of the skin transcriptome with examples of specific expression patterns of the corresponding proteins. The combination of quantitative transcriptomics and immunohistochemistry-based protein profiling could be extended to include comparisons of normal skin from various regions as well as comparing normal skin with various forms of skin disease, to gain further insight into both normal skin biology and the plethora of appearances that underlie the multitude of defined forms of skin disease. We anticipate that the presented data and proposed strategy will be a useful tool for both basic and clinical dermatological research.
Acknowledgments
The staff at the Department of Clinical Pathology, Uppsala University Hospital are acknowledged for providing samples, particularly Dr. Anders Grängsjö for skin biopsies and Simin Tahmasebpoor for excellent technical assistance.
Footnotes
Supplementary material for this article is available on the Journal of Histochemistry & Cytochemistry Web site at http://jhc.sagepub.com/supplemental.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Knut and Alice Wallenberg Foundation.
References
- Abdou AG, Maraee AH, Abd-Elsattar Saif HF. (2014). Immunohistochemical Evaluation of COX-1 and COX-2 Expression in Keloid and Hypertrophic Scar. Am J Dermatopathol 36:311-317. [DOI] [PubMed] [Google Scholar]
- Akiyama M. (2011). Updated molecular genetics and pathogenesis of ichthiyoses. Nagoya J Med Sci 73:79-90. [PMC free article] [PubMed] [Google Scholar]
- Al-Refu K. (2012). Stem cells and alopecia: a review of pathogenesis. Br J Dermatol 167:479-484. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. (2013). NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 41:D991-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi H, Fantauzzo KA, Richardson GD, Jahoda CA, Christiano AM. (2007). Transcriptional profiling of developing mouse epidermis reveals novel patterns of coordinated gene expression. Dev Dyn 236:961-970. [DOI] [PubMed] [Google Scholar]
- Cabral A, Sayin A, de Winter S, Fischer DF, Pavel S, Backendorf C. (2001). SPRR4, a novel cornified envelope precursor: UV-dependent epidermal expression and selective incorporation into fragile envelopes. J Cell Sci 114:3837-3843. [DOI] [PubMed] [Google Scholar]
- Caubet C, Jonca N, Brattsand M, Guerrin M, Bernard D, Schmidt R, Egelrud T, Simon M, Serre G. (2004). Degradation of corneodesmosome proteins by two serine proteases of the kallikrein family, SCTE/KLK5/hK5 and SCCE/KLK7/hK7. J Invest Dermatol 122:1235-1244. [DOI] [PubMed] [Google Scholar]
- Champliaud MF, Baden HP, Koch M, Jin W, Burgeson RE, Viel A. (2000). Gene characterization of sciellin (SCEL) and protein localization in vertebrate epithelia displaying barrier properties. Genomics 70:264-268. [DOI] [PubMed] [Google Scholar]
- Chen J, Lin M, Foxe JJ, Pedrosa E, Hrabovsky A, Carroll R, Zheng D, Lachman HM. (2013). Transcriptome comparison of human neurons generated using induced pluripotent stem cells derived from dental pulp and skin fibroblasts. PLoS One 8:e75682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cork MJ, Danby SG, Vasilopoulos Y, Hadgraft J, Lane ME, Moustafa M, Guy RH, Macgowan AL, Tazi-Ahnini R, Ward SJ. (2009). Epidermal barrier dysfunction in atopic dermatitis. J Invest Dermatol 129:1892-1908. [DOI] [PubMed] [Google Scholar]
- Djureinovic D, Fagerberg L, Hallstrom B, Danielsson A, Lindskog C, Uhlen M, Ponten F. (2014). The human testis-specific proteome defined by transcriptomics and antibody-based profiling. Mol Hum Reprod 20:476-488. [DOI] [PubMed] [Google Scholar]
- Eckhart L, Fischer H, Tschachler E. (2012). Mechanisms and emerging functions of DNA degradation in the epidermis. Front Biosci (Landmark Ed) 17:2461-2475. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13:397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargeas CA, Florek M, Huttner WB, Corbeil D. (2003). Characterization of prominin-2, a new member of the prominin family of pentaspan membrane glycoproteins. J Biol Chem 278:8586-8596. [DOI] [PubMed] [Google Scholar]
- Ferone G, Mollo MR, Thomason HA, Antonini D, Zhou H, Ambrosio R, De Rosa L, Salvatore D, Getsios S, van Bokhoven H, et al. (2013). p63 control of desmosome gene expression and adhesion is compromised in AEC syndrome. Hum Mol Genet 22:531-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardsvoll H, Kriegbaum MC, Hertz EP, Alpizar-Alpizar W, Ploug M. (2013). The urokinase receptor homolog Haldisin is a novel differentiation marker of stratum granulosum in squamous epithelia. J Histochem Cytochem 61:802-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendronneau G, Sidhu SS, Delacour D, Dang T, Calonne C, Houzelstein D, Magnaldo T, Poirier F. (2008). Galectin-7 in the control of epidermal homeostasis after injury. Mol Biol Cell 19:5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadially R. (2012). 25 years of epidermal stem cell research. J Invest Dermatol 132:797-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson JE, Ding J, Johnston A, Tejasvi T, Guzman AM, Nair RP, Voorhees JJ, Abecasis GR, Elder JT. (2010). Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J Invest Dermatol 130:1829-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebenstreit D, Fang M, Gu M, Charoensawan V, van Oudenaarden A, Teichmann SA. (2011). RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol Syst Biol 7:497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba N, Renk T, Wurster K, Rapp W, Bohn H. (1980). Ectopic synthesis of pregnancy specific beta 1-glycoprotein (SP1) and placental specific tissue proteins (PP5, PP10, PP11, PP12) in nontrophoblastic malignant tumours. Possible markers in oncology. Klin Wochenschr 58:789-791. [DOI] [PubMed] [Google Scholar]
- Jabbari A, Suarez-Farinas M, Dewell S, Krueger JG. (2012). Transcriptional profiling of psoriasis using RNA-seq reveals previously unidentified differentially expressed genes. J Invest Dermatol 132:246-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jave-Suarez LF, Winter H, Langbein L, Rogers MA, Schweizer J. (2002). HOXC13 is involved in the regulation of human hair keratin gene expression. J Biol Chem 277:3718-3726. [DOI] [PubMed] [Google Scholar]
- Kampf C, Olsson I, Ryberg U, Sjostedt E, Ponten F. (2012). Production of tissue microarrays, immunohistochemistry staining and digitalization within the human protein atlas. J Vis Exp 31; pii:3620. doi: 10.3791/3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Sugaya M, Suga H, Morimura S, Miyamoto A, Kai H, Kagami S, Yanaba K, Fujita H, Asano Y, et al. (2013). Variations in Serum TARC and I-TAC Levels Reflect Minor Changes in Disease Activity and Pruritus in Atopic Dermatitis. Acta Derm Venereol 94:331-332. [DOI] [PubMed] [Google Scholar]
- Komatsu N, Takata M, Otsuki N, Toyama T, Ohka R, Takehara K, Saijoh K. (2003). Expression and localization of tissue kallikrein mRNAs in human epidermis and appendages. J Invest Dermatol 121:542-549. [DOI] [PubMed] [Google Scholar]
- Kong W, Longaker MT, Lorenz HP. (2003). Molecular cloning and expression of keratinocyte proline-rich protein, a novel squamous epithelial marker isolated during skin development. J Biol Chem 278:22781-22786. [DOI] [PubMed] [Google Scholar]
- Krupp M, Marquardt JU, Sahin U, Galle PR, Castle J, Teufel A. (2012). RNA-Seq Atlas–a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics 28:1184-1185. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. (2001). Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, Ding J, Stuart PE, Xing X, Kochkodan JJ, et al. (2014). Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol 134:1828-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Chen Q, Shi L, Lee M, Giehl KA, Tang Z, Wang H, Zhang J, Yin J, Wu L, et al. (2012). Loss-of-function mutations in HOXC13 cause pure hair and nail ectodermal dysplasia. Am J Hum Genet 91:906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukk M, Kapushesky M, Nikkila J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A. (2010). A global map of human gene expression. Nat Biotechnol 28:322-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen P, Rasmussen HH, Flint T, Gromov P, Kruse TA, Honore B, Vorum H, Celis JE. (1995). Cloning, expression, and chromosome mapping of human galectin-7. J Biol Chem 270:5823-5829. [DOI] [PubMed] [Google Scholar]
- Meyer W, Hornickel I, Schoennagel B. (2010). A note on langerhans cells in the oesophagus epithelium of domesticated mammals. Anat Histol Embryol 39:160-166. [DOI] [PubMed] [Google Scholar]
- Meyer-Hoffert U, Schroder JM. (2011). Epidermal proteases in the pathogenesis of rosacea. J Investig Dermatol Symp Proc 15:16-23. [DOI] [PubMed] [Google Scholar]
- Motoki K, Megahed M, LaForgia S, Uitto J. (1997). Cloning and chromosomal mapping of mouse ladinin, a novel basement membrane zone component. Genomics 39:323-330. [DOI] [PubMed] [Google Scholar]
- Nystrom A, Velati D, Mittapalli VR, Fritsch A, Kern JS, Bruckner-Tuderman L. (2013). Collagen VII plays a dual role in wound healing. J Clin Invest 123:3498-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offenhauser N, Borgonovo A, Disanza A, Romano P, Ponzanelli I, Iannolo G, Di Fiore PP, Scita G. (2004). The eps8 family of proteins links growth factor stimulation to actin reorganization generating functional redundancy in the Ras/Rac pathway. Mol Biol Cell 15:91-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontsuka K, Kotobuki Y, Shiraishi H, Serada S, Ohta S, Tanemura A, Yang L, Fujimoto M, Arima K, Suzuki S, et al. (2012). Periostin, a matricellular protein, accelerates cutaneous wound repair by activating dermal fibroblasts. Exp Dermatol 21:331-336. [DOI] [PubMed] [Google Scholar]
- Powell AM, Sakuma-Oyama Y, Oyama N, Black MM. (2005). Collagen XVII/BP180: a collagenous transmembrane protein and component of the dermoepidermal anchoring complex. Clin Exp Dermatol 30:682-687. [DOI] [PubMed] [Google Scholar]
- Proksch E, Brandner JM, Jensen JM. (2008). The skin: an indispensable barrier. Exp Dermatol 17:1063-1072. [DOI] [PubMed] [Google Scholar]
- Radden LA, 2nd, Child KM, Adkins EB, Spacek DV, Feliciano AM, King TR. (2013). The wooly mutation (wly) on mouse chromosome 11 is associated with a genetic defect in Fam83g. BMC Res Notes 6:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Wang ET, Burge CB, Sandberg R. (2009). An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RCoreTeam (2013). R: A language and environment for statistical computing. In Vienna, Austria [Google Scholar]
- Reveiller M, Ghatak S, Toia L, Kalatskaya I, Stein L, D’Souza M, Zhou Z, Bandla S, Gooding WE, Godfrey TE, et al. (2012). Bile exposure inhibits expression of squamous differentiation genes in human esophageal epithelial cells. Ann Surg 255:1113-1120. [DOI] [PubMed] [Google Scholar]
- Rizzolio F, Bione S, Villa A, Berti E, Cassetti A, Bulfone A, Tribioli C, Toniolo D. (2007). Spatial and temporal expression of POF1B, a gene expressed in epithelia. Gene Expr Patterns 7:529-534. [DOI] [PubMed] [Google Scholar]
- Rustici G, Kolesnikov N, Brandizi M, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Ison J, Keays M, et al. (2013). ArrayExpress update–trends in database growth and links to data analysis tools. Nucleic Acids Res 41:D987-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayan AE, D’Angelo B, Sayan BS, Tucci P, Cimini A, Ceru MP, Knight RA, Melino G. (2010). p73 and p63 regulate the expression of fibroblast growth factor receptor 3. Biochem Biophys Res Commun 394:824-828. [DOI] [PubMed] [Google Scholar]
- Scharadin TM, Eckert RL. (2014). TIG3: an important regulator of keratinocyte proliferation and survival. J Invest Dermatol 134:1811-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S, Saalbach A, Heiker JT, Meier R, Zellmann T, Simon JC, Beck-Sickinger AG. (2013). Proteolytic activation of prochemerin by kallikrein 7 breaks an ionic linkage and results in C-terminal rearrangement. Biochem J 452:271-280. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankewich MC, Tse WT, Peters LL, Ch’ng Y, John KM, Stabach PR, Devarajan P, Morrow JS, Lux SE. (1998). A widely expressed betaIII spectrin associated with Golgi and cytoplasmic vesicles. Proc Natl Acad Sci U S A 95:14158-14163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Farinas M, Lowes MA, Zaba LC, Krueger JG. (2010). Evaluation of the psoriasis transcriptome across different studies by gene set enrichment analysis (GSEA). PLoS One 5:e10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyssen JP, Godoy-Gijon E, Elias PM. (2013). Ichthyosis vulgaris: the filaggrin mutation disease. Br J Dermatol 168:1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulza E, Mattiuzzo NR, Galliano MF, Jonca N, Dossat C, Jacob D, de Daruvar A, Wincker P, Serre G, Guerrin M. (2007). Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol 8:R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Bjorling L, Ponten F. (2010). Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28:1248-1250. [DOI] [PubMed] [Google Scholar]
- Vanbokhoven H, Melino G, Candi E, Declercq W. (2011). p63, a story of mice and men. J Invest Dermatol 131:1196-1207. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. (2001). The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- Wang X, Pasolli HA, Williams T, Fuchs E. (2008). AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. J Cell Biol 183:37-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kirkwood JS, Taylor AW, Stevens JF, Leid M, Ganguli-Indra G, Indra AK. (2013). Transcription factor Ctip2 controls epidermal lipid metabolism and regulates expression of genes involved in sphingolipid biosynthesis during skin development. J Invest Dermatol 133:668-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Miyai M, Matsumoto Y, Tsuboi R, Hibino T. (2012). Kallikrein-related peptidase-7 regulates caspase-14 maturation during keratinocyte terminal differentiation by generating an intermediate form. J Biol Chem 287:32825-32834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JM, Sim SM, Kim HY, Park GT. (2010). Expression of the homeobox gene, HOPX, is modulated by cell differentiation in human keratinocytes and is involved in the expression of differentiation markers. Eur J Cell Biol 89:537-546. [DOI] [PubMed] [Google Scholar]
- Zhao XP, Elder JT. (1997). Positional cloning of novel skin-specific genes from the human epidermal differentiation complex. Genomics 45:250-258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All FPKM data is available without restrictions (www.proteinatlas.org/about/download). The primary sequencing reads are available through the Array Express Archive (www.ebi.ac.uk/arrayexpress/); accession number E-MTAB-1733. Transcript profiling data for each gene in each cell and tissue type is also available on the Human Protein Atlas website, together with protein profiling data and underlying images of immunohistochemically stained tissues.